Human CD34+ hematopoietic progenitor cells obtained from bone marrow (BM), umbilical cord blood (UCB), and mobilized peripheral blood (MPB) were purified and investigated for the expression of the chemokine receptor CXCR4 and its ligand, stromal cell–derived factor-1 (SDF-1). CXCR4 was found present on the cell surface of all CD34+ cells, although it was expressed at lower density on MPB with respect to BM CD34+ cells. Freshly isolated and in vitro–cultured CD34+ cells also coexpressed SDF-1 mRNA, as determined by reverse transcriptase-polymerase chain reaction (RT-PCR). Of interest, CD34+/CD38+ committed progenitor cells, unlike primitive CD34+/CD38− cells, expressed SDF-1 mRNA. Supernatants from in vitro–cultured CD34+ cells contained substantial (3 to 8 ng/mL) amounts of SDF-1 by enzyme-linked immunosorbent assay and induced migration of CD34+ cells. Because CD34+ cells express low levels of CD4, the primary receptor of the human immunodeficiency virus (HIV), and CXCR4 is a coreceptor for T-cell tropic (X4) HIV strains, we investigated the susceptibility of CD34+cells to infection by this subset of viruses. Lack of productive infection was almost invariably observed as determined by a conventional RT activity in culture supernatants and by real-time PCR for HIV DNA in CD34+ cells exposed to both laboratory adapted (LAI) and primary (BON) X4 T-cell tropic HIV-1 strain. Soluble gp120 Env (sgp120) from X4 HIV-1 efficiently blocked binding of the anti-CD4 Leu3a monoclonal antibody (MoAb) to either human CD4+ T cells or CD34+ cells. In contrast, sgp120 interfered with an anti-CXCR4 MoAb binding to human T lymphocytes, but not to CD34+ cells. However, CXCR4 on CD34+ cells was downregulated by SDF-1. These results suggest that CXCR4 and its ligand SDF-1 expressed in CD34+ progenitors may play an important role in regulating the local and systemic trafficking of these cells. Moreover, these findings suggest multiple and potentially synergistic mechanisms at the basis of the resistance of CD34+ cells to X4 HIV infection, including their ability to produce SDF-1, and the lack of CXCR4 internalization following gp120 binding to CD4.
HEMATOPOIETIC PROGENITOR CELLS normally reside in the bone marrow (BM) in close contact with cells of the stromal microenvironment that provide a rich milieu of cytokines, extracellular matrix proteins, and adhesion molecules. Progenitor cells are likely compartmentalized in different areas of the BM, based on their degree of commitment and lineage differentiation.1They home to and emigrate from the BM under experimental procedures such as transplantation or peripheral blood (PB) mobilization.2 However, little is known about the mechanisms and molecules that regulate the homing, retention, and emigration of progenitor cells in hematopoietic organs. By analogy with mature leukocytes, these processes likely involve chemoattractant molecules and their receptors, which are known to regulate the trafficking of leukocytes under both physiological and pathological conditions.3
It has been recently reported that human CD34+hematopoietic progenitor cells are capable of responding to a gradient of chemoattractant(s) produced by stromal cells.4 A chemotactic factor was isolated and identified as stromal cell–derived factor-1 (SDF-1), a CXC chemokine previously cloned from mouse BM stromal cells.5 SDF-1 has been also shown to be a low-potency, high-efficacy chemoattractant for human T lymphocytes and monocytes.6 In addition, this chemokine is likely constitutively expressed in a wide variety of tissues.5 The receptor for SDF-1 was subsequently identified on human lymphocytes as CXCR4 (previously named fusin, HUMSTR, or LESTR),7,8 a seven-transmembrane-domain protein member of the α chemokine receptor family. Before its identification as the receptor for SDF-1, CXCR4 was shown to act, along with CD4, as a receptor for human immunodeficiency virus (HIV)-1 T-cell tropic strains,9 recently classified as X4 viruses.10 Indeed, SDF-1 inhibits cell fusion and infection by HIV strains with a syncytium-inducing (SI) phenotype typically emerging during the late, symptomatic stages of the disease.7 8
Recent studies have demonstrated the presence on CD34+cells of both CD4 antigens (Ags),11,12 and of mRNA encoding for CXCR4,13 raising the possibility that these cells may be susceptible to HIV-1 infection by X4 viral strains. This issue of CD34+ progenitor cell infection is of particular relevance for understanding the frequent cytopenias, dysmyelopoiesis, and impaired colony growth that affect HIV-1 patients in their advanced stages14 as well as in the view of an anti-HIV gene therapy approach of progenitor cells.15 Conflicting studies have been reported regarding the susceptibility of human CD34+cells to HIV-1 infection both in vivo and in vitro. However, most studies indicate that progenitor cells are not a major viral reservoir in HIV-1–infected individuals at different stages of disease.16-19 Susceptibility of human CD34+cells to in vitro HIV-1 infection has been reported, although with variable efficiency.21-25 However, no systematic attempts have been made thus far of comparing the susceptibility to HIV infection of progenitor cells obtained from different sources, such as BM, umbilical cord blood (UCB), or mobilized PB (MPB). This would be of particular relevance in the view of the different implications for adult or in utero infection of progenitor cells as well as for the proposed use of MPB CD34+ cells as targets for gene therapy.26 In addition, a substantial heterogeneity exists among progenitor cells obtained from different sources in terms of their phenotypes,27 content in primitive and committed populations,27-29 proliferative capacity,29-31and cell-cycle status,32 all factors that may also affect the ability of these cells to support HIV replication.
In the present study, we report the simultaneous expression of a functional CXCR4 receptor and its ligand, SDF-1, on human BM, UCB, and MPB CD34+ progenitor cells. Despite the expression of both primary (CD4) and accessory (CXCR4) viral receptors, we almost invariably observed lack of productive infection with X4 HIV-1 strains. Our results suggest multiple and potentially synergistic mechanisms at the basis of the resistance of CD34+ cells to X4 HIV-1 infection.
MATERIALS AND METHODS
Human CD34+ cell sources.
UCB was collected after normal deliveries according to institutional guidelines for discarded material. BM was collected, after informed consent, from normal healthy donors at the time of harvest for allogeneic transplantation. Granulocyte colony-stimulating factor (G-CSF) MPB was collected, after informed consent, by leukapheresis from normal healthy donors after subcutaneous injections of G-CSF (12 μg/kg) starting from day +4. Cyclophosphamide (CY)/G-CSF MPB was collected from patients undergoing autologous BM transplantation for multiple myeloma, breast cancer, or non-Hodgkin’s lymphoma treated with CY (7 g/m2, day 0) plus G-CSF (5 μg/kg) starting from day +3. PB stem cells were collected on consecutive days at the time of hematological recovery after the chemotherapy-induced leukopenia.
Mononuclear cell (MNC) and CD34+ cell purification.
MNCs were obtained after Ficoll (Lymphoprep; Nycomed Pharma, Oslo, Norway) gradient separation from BM samples, UCB samples, G-CSF or CY/G-CSF–mobilized apheresis products. CD34+cells were purified by a two-round procedure using magnetic-cell sorting (MINIMACS; Milteny Biotech, Bergisch Gladbach, Germany) following the manufacturer’s guidelines. This protocol consistently yielded CD34+ cells with a purity greater than 95%.
Flow cytometry and cell sorting.
For immunophenotyping, all cell stainings were performed in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) (Sigma, St Louis, MO), 0.1% sodium azide (PBS-FACS) in 100-μL volume for 30 minutes at +4°C. Staining of BM, UCB, and MBP MNC samples was performed as follows: 3 × 105 low-density cells were incubated with either purified anti-CXCR4 12.G5 MoAb (5 μg/mL) (a kind gift of Dr J. Hoxie, University of Pennsylvania Medical Center, Philadelphia) or mouse IgG2a control antibody (Ab). After washing with PBS-FACS, cells were labeled with goat anti-mouse phycoerythrin (PE)-conjugated polyclonal Ab (Southern Biotechnology, Birmingham, AL), washed, and the free binding-site blocked by an excess of mouse IgG (1 mg/mL; Sigma) for 10 minutes at room temperature. Finally, cells were incubated with anti-CD34 fluorescein isothiocyanate (FITC)-conjugated monoclonal Ab (MoAb) (Becton Dickinson, Mountain View, CA) and anti-CD45 TC MoAb (Caltag, Burlingame, CA). The analysis of CXCR4 expression was performed on low side scatter (SSC), CD34+/CD45low gated cells, as described.33 Fluorescence intensity of CXCR4 was quantitated in terms of number of molecules of equivalent soluble fluorochrome using Quantum-26 PE beads (Flow Cytometry Standards, San Juan, PR). The fluorescence intensity (MESF, molecules of equivalent soluble fluorochrome) of cells stained with the control MoAb were subtracted from the CXCR4 values. Staining of freshly purified or in vitro cultured CD34+ cells was performed as follows: 0.5 × 105 cells were labeled with biotinylated anti-CXCR4 (Pharmingen, San Diego, CA) or biotinylated mouse IgG2a (Caltag). After washing, cells were incubated with anti-CD34 FITC MoAb, streptavidin TC (Caltag), and either anti-CD4 PE MoAb (Becton Dickinson) or mouse IgG1 PE, washed again, and then analyzed using a FACScan apparatus (Becton Dickinson). All data were acquired in listmode with the Cell Quest software (Becton Dickinson). To separate primitive (CD34+/CD38−) from committed (CD34+/CD38+) subpopulations of progenitor cells, immunoaffinity-purified CD34+ cells were labeled with anti-CD34 FITC MoAb (Becton Dickinson) and anti-CD38 PE MoAbs (Coulter, Hialeah, FL). Cells were washed, resuspended in cold PBS 0.3% BSA at the concentration of 1 × 106/mL, and sorted on a FACStarPLUS (Becton Dickinson). Nonspecific background fluorescence was determined using cells stained with isotype-matched control MoAbs. Sorted cells were collected in 10% fetal calf serum (FCS) Iscove’s Modified Dulbecco’s Medium (IMDM), whereas a fraction of the cells was restained and analyzed to verify the purity of the sorted population. For cell sorting performed to isolate the CD34+/CXCR4+/CD4+subpopulation, cells were labeled with anti-CD34 FITC MoAb, anti-CD4 PE MoAb, and biotinylated anti-CXCR4 MoAb, the latter revealed by streptavidin TC. Sorted CD34+/CXCR4+/CD4+ cells were cultured 24 hours after cell sorting to release remaining bound MoAbs and thereafter washed two times before infection.
Modulation of CD4/CXCR4 receptors.
Purified CD34+ cells or CD4+ T cells from UCB (2.5 × 105 /test) were washed once with RPMI 0.2% BSA 10 mmol/L HEPES and resuspended in 0.15 mL of the same medium containing either: (1) HIV-1LAI/IIIB soluble (s) gp120 (5 μg/mL) (a kind gift of P. Lusso, DIBIT, Scientific Institute H.S. Raffaele, Milan, Italy); this concentration of sgp120 Env was previously found to be optimal for CD34+ cells binding assays (A.A., unpublished observations, January 1998); (2) SDF-1 (1 mg/mL); (3) no ligands. Cells were incubated in continuous agitation conditions for 2 hours at 4°C or 37°C and then washed once with RPMI 0.2% BSA. Subsequently, the cells were incubated for 30 minutes in PBS-FACS with anti-CD4 PE MoAb and anti-CXCR4 TC MoAb. Cells were then analyzed by flow cytometry for the relative expression and determination of the mean fluorescence intensity (MFI) of the CD4 and CXCR4 Ags.
Ex vivo culture of CD34+ cells.
CD34+ cells (1 × 105/well) were seeded in a 24-well plastic plate (Falcon; Becton Dickinson Labware, Lincoln Park, NJ) in IMDM (GIBCO-BRL, Life Technology s.r.l., Milan, Italy) containing 20% FCS or X-VIVO 20 serum-free medium (Biowhittaker, Walkersville, MD) containing 2 mmol/L L-glutamin, 50 U/mL penicillin, 50 μg/mL streptomycin, in the presence of interleukin-3 (IL-3) (10 ng/mL), IL-6 (10 ng/mL), and stem cell factor (SCF) (10 ng/mL) (all from R&D Systems, Minneapolis, MN), at a concentration previously determined to be optimal for CD34+ cell expansion.
Colony-forming cell (CFC) assay.
Cells were seeded in duplicate cultures in 1 mL methylcellulose assay-based media containing recombinant human (rHu) erythropoietin (2.5 U/mL) (Janssen-Cilag, Schaffausen, Switzerland), rHu SCF (10 ng/mL), rHuG-CSF (10 ng/mL) (Amgen, Thousand Oaks, CA), rHu granulocyte macrophage (GM)-CSF (10 ng/mL) (Schering-Plough, Milan, Italy), rHu IL-3 (10 ng/mL). Colonies were scored for colony-forming units (CFU)-GM, CFU-MIX, and burst-forming units-erythroid (BFU-E) after 2 weeks of incubation at 37°C, 5% CO2.
Reverse transcription-polymerase chain reaction (RT-PCR) for chemokines and chemokine receptor expression.
Aliquots (0.2 to 0.4 × 106 cells) of uninfected cells were pelleted either immediately after purification or after 3, 6, and 11 days of culture by centrifuging at 12,000 rpm for 5 minutes at 4°C. Total RNA was extracted by the RNA-zol method (Duotech, Milan, Italy) and resuspended in different volumes of water depending on cell numbers. The same amount of RNA was reverse transcribed in the presence of 1X RT buffer (GIBCO-BRL), 800 μmol/L of each dNTPs (Pharmacia Biotech, Milan, Italy), 20 μg/mL random hexamers (Promega, Madison, WI), 4 mmol/L dithiothreitol (GIBCO-BRL), 16 U RNA guard (Promega), and 400 U of murine-Moloney leukemia virus (M-MLV) RT (GIBCO-BRL). The reaction mixture (50 μL) was incubated at 68°C for 5 minutes, 37°C for 60 minutes, heated at 94°C for 5 minutes, and cooled on ice. Aliquots corresponding to 1/40 of the cDNA obtained were amplified in the presence of 0.4 μmol/L primer pairs (PRIMM s.r.l., Milan, Italy), 200 μM dNTPs (Pharmacia), 1X PCR buffer, and 1.25 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Corp, Norwalk, CN) in a 50-μL reaction mixture. The mRNA for the cellular housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was used as control. The following primers were used for amplifying mRNAs for: GAPDH sense,34 5′CCA TGG AGA AGG CTG GGG 3′, antisense 5′CAA AGT TGT CAT GGA TGA CC 3′ (generating a 195-bp fragment); SDF-1 sense,35 5′ ACG AAT TCG CGC CAT GAA CGC CAA GGT CGT 3′, antisense, 5′CAG GAT CCT GCA AAC CTC AGG CCC GAT C 3′ (kind gift of Dr S. Polo, DIBIT) (generating a 451-bp length fragment); CXCR4 sense, 5′ GCC AAC GTC AGT GAG GCA GAT G 3′; antisense 5′ GAG GAT GAC TGT GGT CTT GAG G 3′ (generating a 209-bp fragment); PCR products were analyzed by electrophoresis in 2% agarose gel and visualized by ethidium bromide staining.
SDF-1.
Murine SDF-1 was purified from MS-5 conditioned medium (kind gift of Dr K. Itoh, Kyoto University, Kyoto, Japan)36by affinity chromatography and high-performance liquid chromatography (HPLC) according to a modification37 of a previous protocol.6 Synthetic human SDF-1 was kindly donated by Iain Clark-Lewis (Biomedical Research Centre, University of British Columbia, Vancouver, Canada). The biochemical, antigenic, and functional properties of human synthetic SDF-1 and native murine SDF-1 were comparable based on capillary electrophoreses, enzyme-linked immunosorbent (ELISA), and chemotaxis assays (C. Arcelloni et al, submitted).
SDF-1 ELISA.
Supernatants of cells grown in X-VIVO 20 serum-free medium (50 μL), or serial dilutions of SDF-1 in serum-free medium (from 500 ng/mL to 1 ng/mL) were incubated overnight in polycarbonate 96-well microtiter plates (Nunc, Roskilde, Denmark). All samples were set up in duplicate. The remaining binding sites were blocked by a 60-minute incubation with PBS containing 2% BSA. Wells were then incubated for 90 minutes with a goat polyclonal Ab (1 μg/mL) against human SDF-1 (R&D Systems). After four washes, they were incubated for 90 minutes with a rabbit anti-goat IgG polyclonal Ab conjugated to alkaline phosphatase (AP), affinity isolated, adsorbed with human proteins (SIGMA, Milan, Italy) and washed again. The bound immunocomplexes were detected with 4-Nitrophenyl phosphate (1 mg/mL) (Boehringer Mannheim, Mannheim, Germany) in developing buffer pH 9.8 (Boehringer Mannheim) and the optical density (OD) measured at 405 nm in a microplate reader. The standard curve set up with SDF-1 allowed the detection of SDF-1 Ag in serum-free supernatants ranging between 2 and 500 ng/mL.
Chemotaxis.
This assay was performed in duplicate using 5-μm pore polycarbonate Transwell culture insert (Costar, Cambridge, MA). Immediately before each assay, filters were rinsed in RPMI containing 0.3% human serum albumin (SA). One hundred thousand CD34+ cells (in 100 μL) were loaded into each transwell filter. Filters were then carefully transferred to another well containing human synthetic or mouse native SDF-1 (at concentrations ranging from 1.5 to 0.01 μg/mL). After 3 hours, the upper chamber was carefully removed and the cells in the bottom chamber resuspended and transferred to tubes for determining the proportion of migrated CD34+ cells. Cytofluorimetric analysis was performed on a FACScan for a constant, predetermined period of time and counts obtained for each sample were compared with flow cytometric counts obtained from control wells containing 10% of the input population. Data were expressed as a percentage of the input population or as the ratio of cells migrated to SDF-1 to cells migrated to assay medium (chemotactic index).
HIV-1 infection and coreceptor usage assay.
HIV-1 LAI/IIIB is known to infect CD4+ cells expressing CXCR4 (fusin).38 The viral stock was expanded on phytohemagglutinin (PHA)-stimulated PB mononuclear cells (PBMC) and titrated by an Mg2+-dependent RT activity assay39 and by ID50 quantitation according to the Reed and Muench formula.40 The primary isolate BON was isolated from the plasma of an HIV-1+individual onto a mixture of PHA-stimulated PBMC of two seronegative donors.39 The primary viral stock was aliquoted and preserved at −80°C. The usage of HIV-1 coreceptors was determined on U87 astrocytoma cell line permanently transfected with human CD4 either alone or together with one of the following chemokine receptor–expressing plasmids: CCR2b, CCR3, CCR5, or CXCR4, as reported.41 By this assay, BON HIV-1 selectively infected U87 cells coexpressing CD4 and CXCR4 (generous gift of Dr Dan Littman, The Skirball Institute of Biomolecular Medicine, New York University Medical Center, New York, NY), but not other coreceptors. CD34+ cells (2.5 × 105/mL) were incubated for 1 hour at 37°C with either HIV-1LAI/IIIB or BON at the multiplicities of infection (m.o.i.) of 1, 0.1, and 0.01, respectively. Cells were washed twice in PBS and then seeded in a 48-well plate (Falcon; Becton Dickinson Labware) in duplicate cultures, and maintained in IMDM medium supplemented with 20% FCS, IL-3 (10 ng/mL), IL-6 (10 ng/mL), and SCF (10 ng/mL) (complete medium) throughout the period of infection (approximately 4 weeks). Culture supernatants were harvested every 3 days, stored at −80°C, and replaced with fresh complete medium.
HIV DNA copy number quantification by real-time PCR.
HIV-1LAI/IIIB was treated with 50 U/mL of RQ1 RNase-free Dnase (Boehringer) for 30 minutes at room temperature (rt°) before infection. Cells (0.4 to 2 × 105) were harvested 1, 24, 48, 72, and 168 hours after infection, centrifuged at 12,000 rpm for 5 minutes, and the pellets were stored at −80°C until DNA extraction. DNA quantitation was performed simultaneously from all the stored samples. Briefly, cell pellets were lysed by incubation at 50°C for 4 hours with 200 μL of a buffer (0.25% Triton X, 0.25% sodium dodecyl sulfate [SDS] in 1X Tris EDTA, TE) containing 600 μg/mL of proteinase K; DNA extraction was performed by the phenol-chloroform protocol followed by ethanol precipitation; DNA was resuspended in 10 μL of water. DNA proviral synthesis quantitation was performed by the “real-time” quantitative PCR method.42-44 The quantitation is based on the cleavage of fluorescent dye labeled probe by the 5′-3′ endonuclease activity of Taq DNA polymerase during PCR and measurement of fluorescence intensity by the ABI Prism 7700 Sequence Detector System (PE Applied Biosystems, Foster City, CA). A standard curve was generated by serial dilutions of the chronically HIV-infected ACH-2 T-cell line (containing one proviral copy per cell)45 in PBMC of a seronegative donor. Primer pair and probe spanning gag were used: for 5′-ACA TCA AGC AGC CAT GCA AAT-3′; rev 5′-ATC TGG CCT GGT GCA ATA GG-3′; probe 5′(FAM) CAT CAA TGA GGA AGC TGC AGA ATG GGA TAG A (TAMRA)-3′. Amplification reactions (25 μL) contained 1X buffer A, 2.5 mmol/L MgCl2, 200 μmol/L dATP, dCTP, and dGTP, 400 μmol/L dUTP, 0.625 U of AmpliTaq Gold, 0.25 U of AmpErase UNG (PE Applied Biosystems), 0.25 μmol/L of each primer, and 75 nmol/L of probe. The thermal cycling conditions were 50°C for 2 minutes, 95°C for 12 minutes, and 40 cycles of 95°C for 15 seconds and 65°C for 1 minute.
Statistics.
Results of experimental points from multiple experiments (n) were expressed, as the mean ± SD, where applicable, or the range if n = 2. Significance levels were determined by two-sided Student’st-test analysis.
RESULTS
CXCR4 is expressed on human CD34+ progenitor cells from different sources and acts as a receptor for SDF-1.
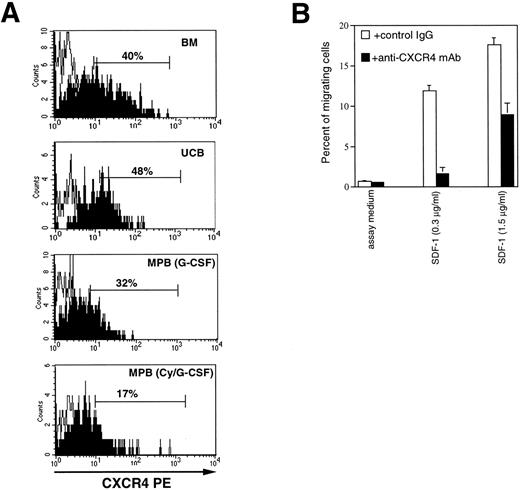
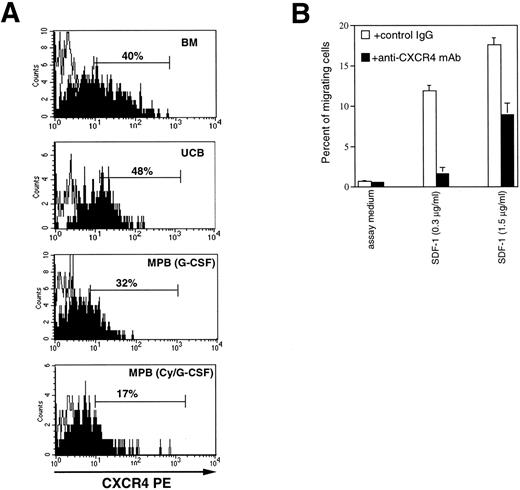
The anti-CXCR4 12.G5 MoAb46 was used to analyze the expression of the receptor on CD34+/CD45dullcells from various hematopoietic sources (Fig 1A). On average, 39.2% ± 7% of adult BM (n = 11), and 43.8% ± 5.8% of UCB (n = 7) expressed CXCR4 on the cell surface. In comparison to BM, the proportion of CXCR4+ expressing cells was reduced on CD34+cells from Cy/G-CSF MPB (9.88 ± 6.8; n = 6; P < .005) and, to a lesser extent, on CD34+ cells from G-CSF MPB (30.3% ± 16%; n = 6). Furthermore, the MFI of CXCR4 expression was considerably decreased both in G-CSF (1,068 ± 780, MESF) and CY/G-CSF (784 ± 389) MPB CD34+ cells with respect to BM CD34+ cells (2,280 ± 1,325, P < .05). To confirm the specificity of the receptor-ligand interaction in human CD34+ cells, we tested whether the anti-CXCR4 12.G5 MoAb, previously shown to partially inhibit migration of lymphocytes and CXCR4-transfected cells,47 could block SDF-1–mediated chemotaxis of BM CD34+ cells. As shown in Fig 1B, when the 12.G5 MoAb (10 μg/mL) was added to CD34+ cells before migration, the chemotactic response to optimal and suboptimal concentrations of SDF-1 were reduced to approximately 50% and 10%, respectively; no inhibition was observed in the presence of a control Ab. A similar level of inhibition was obtained with CD34+cells obtained from UCB or MPB. Thus, CXCR4 mediates SDF-1–induced chemotaxis of CD34+ cells of different origin.
CXCR4 is expressed on CD34+cells and mediates SDF-1–induced responses. (A) Flow cytometric analysis of CXCR4 expression on CD34+ cells from bone marrow (BM), umbilical cord blood (UCB), G-CSF mobilized peripheral blood (MPB) cells, or cyclophosphamide plus G-CSF (CY/G-CSF) MPB. Low-density cells were stained with CD34 FITC, CD45TC, and either a control mouse antibody IgG2a (empty histograms) or anti-CXCR4 MoAb (filled histograms), revealed by goat anti-mouse PE antibody, as described in Materials and Methods. The analysis shows the staining of a gated population of low SSC, CD34+, CD45dull cells. (B) Inhibition of SDF-1–dependent chemotaxis by anti-CXCR4 antibody. Migration of BM CD34+ cells in response to SDF-1 (1.5 and 0.3 μg/mL, respectively) and in the presence of 10 μg/mL of either 12.G5 anti-CXCR4 MoAb (▪), or an isotype-matched control mouse IgG (□).
CXCR4 is expressed on CD34+cells and mediates SDF-1–induced responses. (A) Flow cytometric analysis of CXCR4 expression on CD34+ cells from bone marrow (BM), umbilical cord blood (UCB), G-CSF mobilized peripheral blood (MPB) cells, or cyclophosphamide plus G-CSF (CY/G-CSF) MPB. Low-density cells were stained with CD34 FITC, CD45TC, and either a control mouse antibody IgG2a (empty histograms) or anti-CXCR4 MoAb (filled histograms), revealed by goat anti-mouse PE antibody, as described in Materials and Methods. The analysis shows the staining of a gated population of low SSC, CD34+, CD45dull cells. (B) Inhibition of SDF-1–dependent chemotaxis by anti-CXCR4 antibody. Migration of BM CD34+ cells in response to SDF-1 (1.5 and 0.3 μg/mL, respectively) and in the presence of 10 μg/mL of either 12.G5 anti-CXCR4 MoAb (▪), or an isotype-matched control mouse IgG (□).
CD34+ cells express and secrete SDF-1.
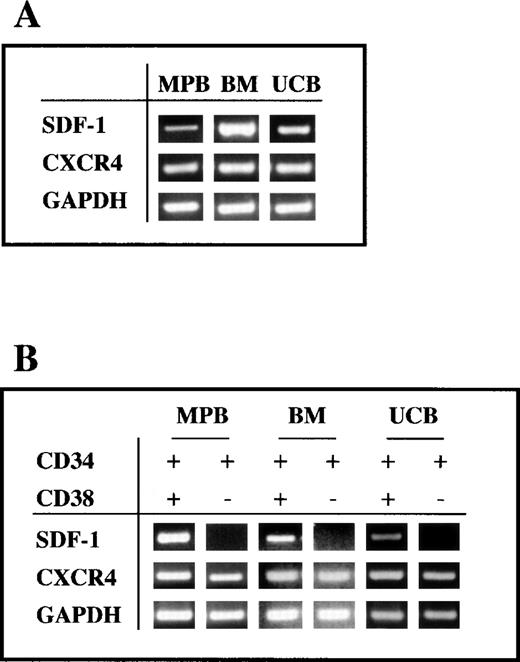
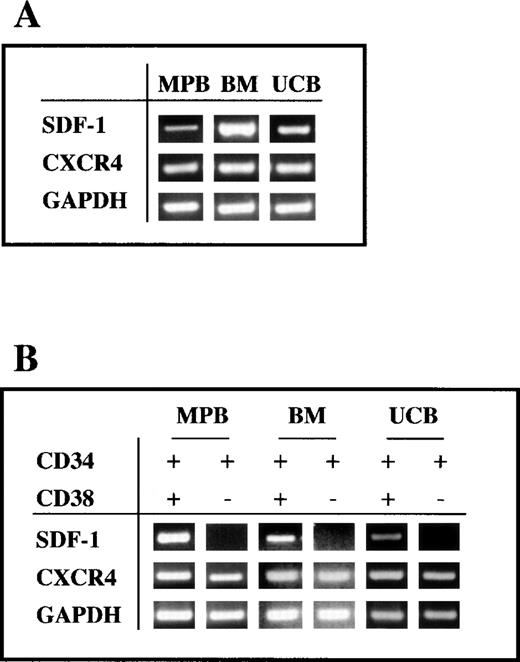
Increasing evidence suggests that CD34+ progenitor cells secrete, in an autocrine fashion, hematopoietic growth factors, such as erythropoietin,48 IL-3, GM-CSF,49 and transforming growth factor-β (TGF-β).50 Therefore, we investigated whether CD34+ cells expressed the CXCR4 ligand SDF-1 by means of RT-PCR. Total RNA was extracted from freshly purified CD34+ cells obtained from BM, UCB, and MPB, reverse transcribed, and amplified as described in Materials and Methods. SDF-1 mRNA transcripts of the expected length (451 bp) were present in all BM CD34+ cells tested (n = 3), in all UCB samples (n = 4), and in 2 of 3 MPB tested (Fig 2A, and data not shown). No products were amplified in the absence of RT, thus excluding contamination by genomic DNA (data not shown).
RT-PCR analysis of the expression of SDF-1 and CXCR4 by CD34+ cells. (A) Expression of CXCR4 and SDF-1 in freshly isolated CD34+ cells from UCB, BM, and G-CSF MPB. The RT-PCR was performed as indicated in Materials and Methods. GAPDH is shown as a control. (B) RT-PCR analysis of CXCR4 and SDF-1 expression in sorted CD34+/CD38− and CD34+/CD38+ cells from UCB, BM, and MPB.
RT-PCR analysis of the expression of SDF-1 and CXCR4 by CD34+ cells. (A) Expression of CXCR4 and SDF-1 in freshly isolated CD34+ cells from UCB, BM, and G-CSF MPB. The RT-PCR was performed as indicated in Materials and Methods. GAPDH is shown as a control. (B) RT-PCR analysis of CXCR4 and SDF-1 expression in sorted CD34+/CD38− and CD34+/CD38+ cells from UCB, BM, and MPB.
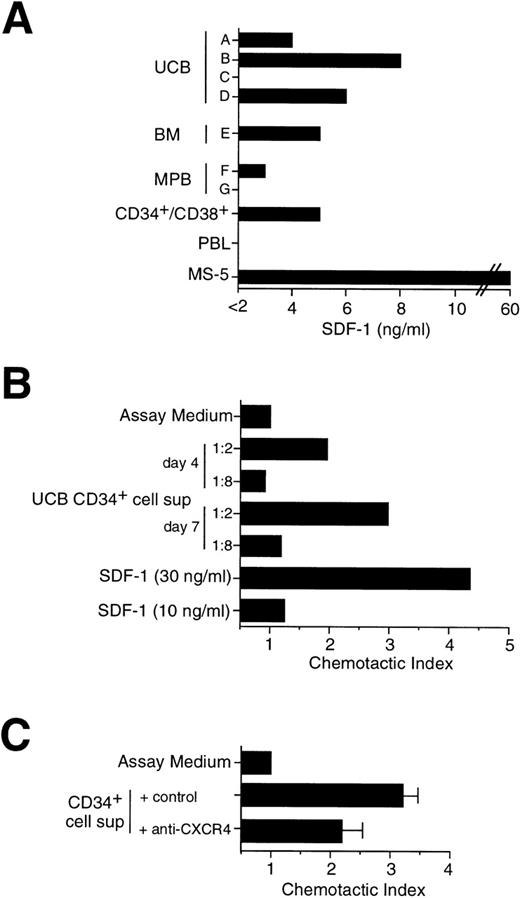
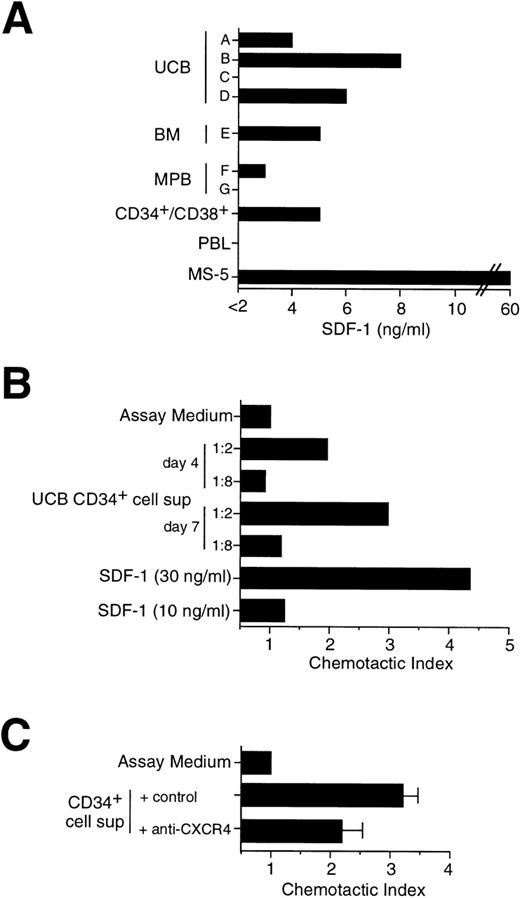
The CD34+/CD38− immunophenotype defines a rare, quiescent subpopulation of primitive progenitor cells that can be functionally distinguished from committed CD34+/CD38+ progenitor cells by sustained clonogenicity in long-term culture,51 and the ability to repopulate immunodeficient mice.52 Thus, CD34+cells from UCB, BM, and MPB were sorted by flow cytometry into CD34+/CD38− and CD34+/CD38+ cell subpopulations and analyzed by RT-PCR for the expression of SDF-1 and CXCR4. As shown in Fig 2B, SDF-1 was found expressed by CD34+/CD38+ progenitor cells, but not by CD34+/CD38− cells. In contrast, both CD34+/CD38− and CD34+/CD38+ subpopulations expressed CXCR4 by RT-PCR (Fig 2B) and FACS analysis (data not shown). The kinetics of both SDF-1 and CXCR4 expression were tested by RT-PCR in UCB CD34+ cells grown in culture for up to 17 days in medium containing IL-3, SCF, and IL-6. No substantial changes in chemokine or chemokine receptor mRNA were observed over time (data not shown). To confirm SDF-1 expression at the protein level, we set up a direct ELISA using an anti–SDF-1 polyclonal Ab that allowed us to detect the presence of SDF-1 Ag in serum-free medium with a lower detection limit of 2 ng/mL. Cell supernatants were obtained from seven different CD34+ cell samples purified either from BM, UCB, and MPB and cultured for 5 days in serum-free medium in the presence of IL-3, SCF, and IL-6. The presence of SDF-1 was shown in five samples, including sorted CD34+/CD38+ cells, in a range variable between 3 and 8 ng/mL (Fig 3A). SDF-1 Ag was detectable also in CD34+ cell supernatants collected after 3 days of culture (data not shown). SDF-1 Ag was below the detection level of this ELISA in two CD34+ cell supernatants and in the supernatant of a polyclonal T-cell line. Moreover, 50 to 100 ng/mL of SDF-1 Ag was detected in the supernatant of MS-5 stromal cell line, which is known to constitutively produce SDF-1 (Fig 3A).
Secretion of SDF-1 by cultured CD34+ cells. (A) Determination of SDF-1 Ag in CD34+ cell supernatant by ELISA. CD34+ cells (2 × 105/mL) were cultured for 5 days in serum-free medium in the presence of cytokines. Cell supernatants were obtained from: different donors (A through G) and sources (UCB, BM, MPB) of CD34+ cells; sorted MPB CD34+/CD38+ cells; a human polyclonal T-cell line (PBL); the mouse stromal cell line MS-5. Supernatants were assayed for the presence of SDF-1 by an Ag capture ELISA, as indicated in Materials and Methods. (B) Migration of CD34+ cells in response to serum-free supernatant from UCB CD34+ cells cultured in the presence of IL-3, SCF, and IL-6 for 4 days and 7 days, respectively. Chemotaxis to serum-free medium containing cytokines (assay medium) and to low doses of SDF-1 (10 ng/mL and 30 ng/mL) are shown as controls. (C) Inhibition of chemotaxis induced by supernatant from CD34+ cells. Chemotaxis of CD34+ cells in response to 5 days supernatant from UCB CD34+ cells in the presence of an anti-CXCR4 MoAb or of a control MoAb.
Secretion of SDF-1 by cultured CD34+ cells. (A) Determination of SDF-1 Ag in CD34+ cell supernatant by ELISA. CD34+ cells (2 × 105/mL) were cultured for 5 days in serum-free medium in the presence of cytokines. Cell supernatants were obtained from: different donors (A through G) and sources (UCB, BM, MPB) of CD34+ cells; sorted MPB CD34+/CD38+ cells; a human polyclonal T-cell line (PBL); the mouse stromal cell line MS-5. Supernatants were assayed for the presence of SDF-1 by an Ag capture ELISA, as indicated in Materials and Methods. (B) Migration of CD34+ cells in response to serum-free supernatant from UCB CD34+ cells cultured in the presence of IL-3, SCF, and IL-6 for 4 days and 7 days, respectively. Chemotaxis to serum-free medium containing cytokines (assay medium) and to low doses of SDF-1 (10 ng/mL and 30 ng/mL) are shown as controls. (C) Inhibition of chemotaxis induced by supernatant from CD34+ cells. Chemotaxis of CD34+ cells in response to 5 days supernatant from UCB CD34+ cells in the presence of an anti-CXCR4 MoAb or of a control MoAb.
To further evaluate the secretion of SDF-1, cell supernatants from UCB CD34+ cells cultured for 4 to 7 days were tested for their ability to induce migration of freshly isolated CD34+ cells in a chemotactic chamber (Fig 3B). Consistent with the production of SDF-1 by CD34+ cells, their supernatant was able to induce migration of CD34+ cells (Fig 3B), and the chemotactic activity was inhibited by 45% when an anti-CXCR4 MoAb was added before migration (Fig 3C).
Coexpression of the HIV-1 X4 strain receptors CXCR4 and CD4 on CD34+ cells.
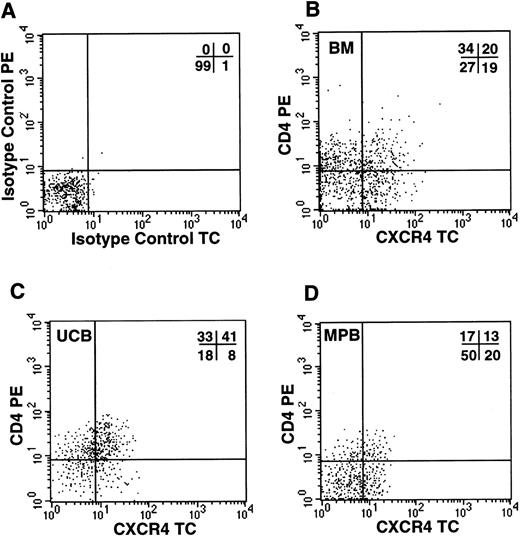
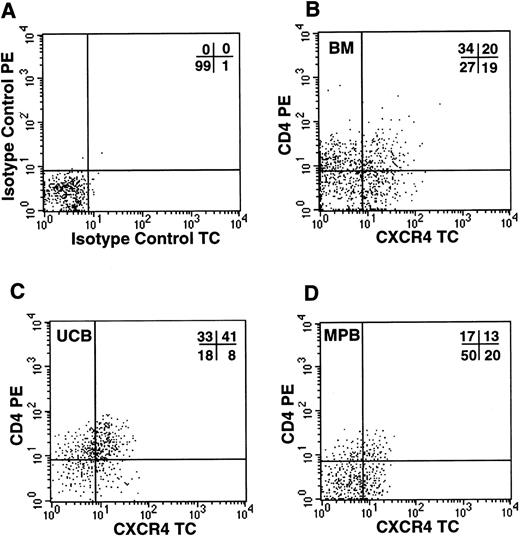
Because the simultaneous expression of functional CXCR4 and CD4 molecules on the cell surface should render CD34+ cells susceptible to infections by X4 HIV strains, we investigated by FACS analysis whether CD34+ cells coexpressed the two viral receptors. In all samples tested, including UCB, BM, and G-CSF MPB, we detected a significant proportion of CD34+ cells coexpressing both molecules (Fig 4). UCB cells showed the highest proportion of CD34+/CD4+/CXCR4+ cells (31%, n = 4), in comparison to BM (20%, n = 3) and MPB (12.4%, n = 5). To further confirm that this subpopulation contains hematopoietic progenitor cells, CD34+ cells from UCB were sorted for CXCR4 and CD4 coexpression and seeded in a standard methylcellulose colony assay. We observed that both CXCR4+/CD4+and CXCR4−/CD4− cells yielded comparable numbers of CFU-GM (110 and 104 colonies per 400 cells plated, respectively), BFU-E (102 and 85), and CFU-MIX (12.5 and 13), respectively (data not shown).
Coexpression of HIV-1 X4 coreceptors on BM, UCB, and MPB CD34+ cells. Flow cytometric analysis of purified CD34+ cells labeled with isotype control PE/TC (A) or anti-CD4 PE and anti-CXCR4 TC (B, bone marrow; C, umbilical cord blood; D, G-CSF–mobilized peripheral blood).
Coexpression of HIV-1 X4 coreceptors on BM, UCB, and MPB CD34+ cells. Flow cytometric analysis of purified CD34+ cells labeled with isotype control PE/TC (A) or anti-CD4 PE and anti-CXCR4 TC (B, bone marrow; C, umbilical cord blood; D, G-CSF–mobilized peripheral blood).
CD34+ cells are resistant to X4 HIV-1LAI/IIIB infection.
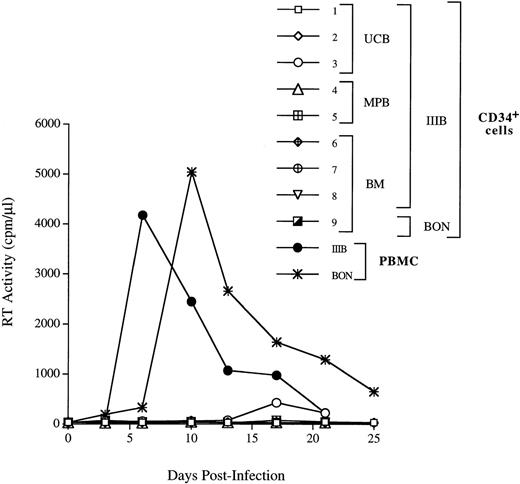
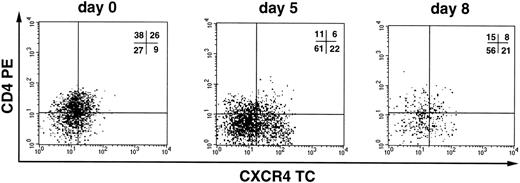
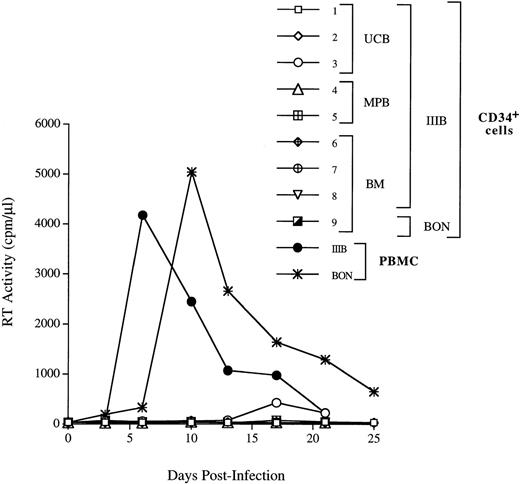
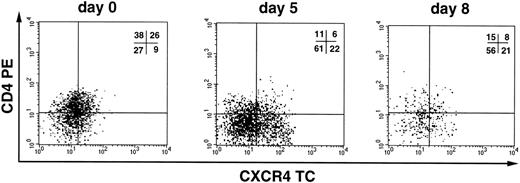
We next investigated whether CD34+ cells derived either from normal adult BM (n = 3), UCB (n = 4), or G-CSF MPB (n = 2) were susceptible to infection by X4 HIV-1. CD34+ cells were isolated from PBMC cells by a two-round immunoaffinity procedure that yielded cells with a purity greater than 95%. In all samples used for HIV infection studies, the proportion of CD4+/CXCR4+ cells was greater than 15%. Cells were infected for 1 hour at 37°C with HIV-1LAI/IIIB, washed, and then cultured for up to 3 weeks in complete medium containing IL-3, IL-6, and SCF (routinely used for ex vivo expansion and gene transfer into progenitor cells). HIV-1 replication was assessed by an Mg2+-dependent RT-assay on cell supernatant every 3 to 4 days. We did not observe productive HIV infection of CD34+ cells (Fig 5), except in one experiment with UCB cells in which low levels of replication were observed after 2 weeks of culture (Fig 5, exp. 1). In one additional experiment, MPB CD34+ cells were sorted by flow cytometry for coexpression of both CD4 and CXCR4 receptors (purity = 99.5%), cultured for 24 hours before infection, and, again, no HIV-1 production was observed (Fig 5, exp. 5). Experiments were performed also using the primary X4 HIV isolate BON that similarly failed to replicate in BM CD34+ cells (Fig 5, exp. 9). No substantial changes in the levels of CXCR4 and SDF-1 mRNA were observed in CD34+ cells exposed or unexposed to HIV, up to 11 days after incubation with the virus (data not shown). FACS analysis confirmed the persistence of CXCR4 on these cultured CD34+ cells. In addition, there were no significant differences in the proportion of CD4+cells in uninfected and HIV-1 infected CD34+ cells after 3 days of culture (with 16.9% v 19.5%, and 23.5% v24.6% in uninfected v HIV-infected BM and UCB CD34+ cells, respectively; data not shown). However, the proportion of CD34+ cells bearing CD4 alone or both HIV-1 receptors decreased over time in CD34+ cells during culture (Fig 6).
CD34+ progenitor cells are not able to productively sustain X4 HIV-1 infection. The UCB-, BM-, and MPB-derived CD34+ cells were incubated for 1 hour with the HIV-1LAI/IIIB strain of HIV-1 virus at the m.o.i. of 1. BM cells were infected with BON primary isolate diluted 1/30 (exp. 9). In exp. 5, MPB CD34+ cells were sorted for the presence of both CD4 and CXCR4 and cultured for 24 hours before infection, as indicated in Materials and Methods. After infection, cells were washed twice and plated in a 48-well plate. Culture supernatants were harvested every 3 days and the particles production was monitored by RT activity. A kinetic of HIV-1 IIIB replication in PBMC is shown as control of the HIV-1LAI/IIIB and BON infectivity.
CD34+ progenitor cells are not able to productively sustain X4 HIV-1 infection. The UCB-, BM-, and MPB-derived CD34+ cells were incubated for 1 hour with the HIV-1LAI/IIIB strain of HIV-1 virus at the m.o.i. of 1. BM cells were infected with BON primary isolate diluted 1/30 (exp. 9). In exp. 5, MPB CD34+ cells were sorted for the presence of both CD4 and CXCR4 and cultured for 24 hours before infection, as indicated in Materials and Methods. After infection, cells were washed twice and plated in a 48-well plate. Culture supernatants were harvested every 3 days and the particles production was monitored by RT activity. A kinetic of HIV-1 IIIB replication in PBMC is shown as control of the HIV-1LAI/IIIB and BON infectivity.
CXCR4 and CD4 expression during in vitro culture of CD34+ cells. Flow cytometric analysis of CXCR4 and CD4 expression on UCB CD34+ cells cultured for 0, 5, and 8 days, respectively. Cells were stained with anti-CD34 FITC, anti-CD4 PE, and anti-CXCR4 TC and analyzed for HIV-1 coreceptor expression on the subpopulation of cells that maintain the CD34 antigen during the culture. Quadrant regions were set using isotype matched-control antibodies. Similar results were obtained for uninfected and HIV-1–infected cells.
CXCR4 and CD4 expression during in vitro culture of CD34+ cells. Flow cytometric analysis of CXCR4 and CD4 expression on UCB CD34+ cells cultured for 0, 5, and 8 days, respectively. Cells were stained with anti-CD34 FITC, anti-CD4 PE, and anti-CXCR4 TC and analyzed for HIV-1 coreceptor expression on the subpopulation of cells that maintain the CD34 antigen during the culture. Quadrant regions were set using isotype matched-control antibodies. Similar results were obtained for uninfected and HIV-1–infected cells.
To elucidate whether impaired HIV-1 infection in CD34+cells was caused by inefficient entry of HIV into these cells, BM CD34+ cells were infected with DNase-treated HIV-1LAI/IIIB and harvested 1, 18, 48, 72, and 168 hours after infection; cell lysates were subjected to HIV-1 DNA PCR to detect proviral DNA synthesis. The levels of HIV-1 gag DNA in CD34+ cells, although detectable, were consistently very low (range, 1 to 25 copies per 105 cells) at the first three time points and became undetectable (<1 copy) after 3 days of culture (data not shown). By this method, productive HIV infection, as determined by detection of RT activity in culture supernatants,34 is associated with levels of proviral DNA equal to or above approximately 1,000 copies of DNA/105cells.34 Similar results were obtained with sorted MPB CD34+ cells analyzed between 1 and 72 hours as well as with CB CD34+ cells sorted for the simultaneous surface expression of both CD4 and CXCR4 receptors before infection. These results suggest that HIV-1 infection is curtailed in CD34+cells at early stages of the viral life cycle, consistent with a poor ability of the virus to enter CD34+ cells.
HIV-1 gp120 does not cause the formation of trimeric complexes with CD4 and CXCR4 in CD34+ cells.
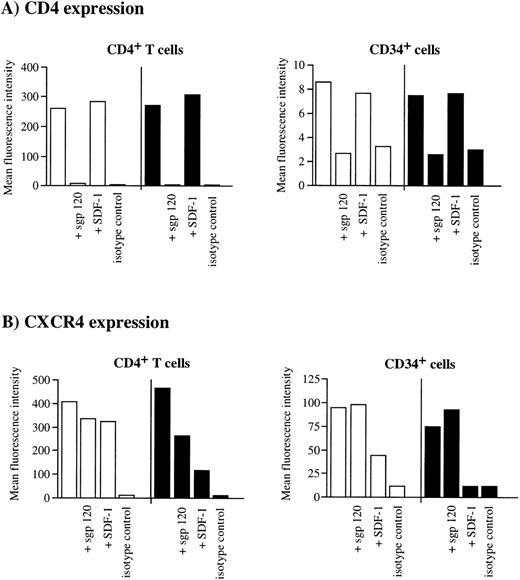
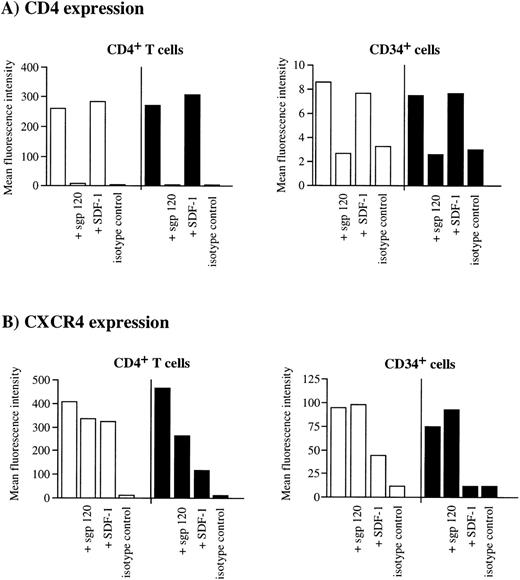
It has been recently shown that the binding of sgp120 to CD4+ T cells leads to an interaction among CXCR4, CD4, and gp120 that masks anti-CXCR4 epitopes and eventually induces the cointernalization of these trimolecular complexes.53 To investigate whether gp120Env was able to interact with CXCR4 on the surface of CD34+/CD4+ cells, we tested the ability of X4 HIV-1 sgp120 to interfere with MoAb binding to CD4 and CXCR4. As expected, sgp120 bound efficiently to both human CD4+ T cells and CD34+ cells, resulting in the potent inhibition of the anti-CD4 Leu3A MoAb (recognizing an epitope of CD4 required for HIV infection) staining (Fig 7A). Binding of anti-CXCR4 12G5 MoAb to CD4+ T cells was reduced by 20% at +4°C, and by 48% at 37°C, respectively, in the presence of prebound sgp120 (Fig7B). In contrast, sgp120 did not interfere with the anti-CXCR4 MoAb binding to CD34+ cells, either at +4°C or at 37°C (Fig 7B). On the other hand, when cells were pretreated with the natural ligand SDF-1, CXCR4 was downmodulated from the cell surface of both CD4+ T cells (22% reduction of mean fluorescence intensity at +4°C, 80% reduction at +37°C, respectively), and CD34+ cells (70% reduction at +4°C, 100% at +37°C, respectively) (Fig 7B). These results confirm that CXCR4 expressed on CD34+ cells can be downmodulated by its ligand, as recently reported for other cell types including human lymphocytes,54 55 but not by HIV-1 gp120/CD4 complexes. Taken together, these observations suggest that the binding of gp120 to CD4 expressed on human CD34+ cells, although efficient, does not lead to the association with CXCR4 and consequent internalization of the virus, providing a potential mechanism, in addition to SDF-1 expression, for explaining the inefficient susceptibility of these cells to X4 HIV-1 infection.
Failure of sgp120/CD4 to induce CXCR4 downmodulation in CD34+ progenitor cells. Purified CD34+cells or CD4+ cells from UCB were incubated without gp120, with sgp120 (5 μg/mL), or SDF-1 (1 μg/mL) for 2 hours at either +4°C (□) or 37°C (▪) in serum-free medium. Cells were then washed once, labeled with anti-CD4 PE in combination with anti-CXCR4 TC, and analyzed by flow cytometry. An aliquot of cells was incubated with PE- and TC-conjugated isotype control-matched antibodies. Columns represent the mean fluorescence intensity for CD4 expression (A) and CXCR4 expression (B) on CD4+ T cells (left) and CD34+ cells (right), respectively.
Failure of sgp120/CD4 to induce CXCR4 downmodulation in CD34+ progenitor cells. Purified CD34+cells or CD4+ cells from UCB were incubated without gp120, with sgp120 (5 μg/mL), or SDF-1 (1 μg/mL) for 2 hours at either +4°C (□) or 37°C (▪) in serum-free medium. Cells were then washed once, labeled with anti-CD4 PE in combination with anti-CXCR4 TC, and analyzed by flow cytometry. An aliquot of cells was incubated with PE- and TC-conjugated isotype control-matched antibodies. Columns represent the mean fluorescence intensity for CD4 expression (A) and CXCR4 expression (B) on CD4+ T cells (left) and CD34+ cells (right), respectively.
DISCUSSION
In the present study, we have observed that CD34+ precursor cells obtained from BM, UBC, or MPB express the CXCR4 chemokine receptor. In addition, CD34+/CD38+ cells also expressed and secreted SDF-1, the natural ligand of CXCR4. Because this chemokine receptor serves, together with CD4, as coreceptor for T-cell tropic HIV strains, we investigated the susceptibility of CD34+ cells to this subset of viruses. No evidence of productive X4 HIV infection was obtained, either with the laboratory-adapted LAI/IIIB strain or with a primary X4 isolate (BON), likely as a consequence of a very poor entry of the virus despite cell-surface expression of both CD4 and CXCR4. This apparent discrepancy is likely explained by multiple mechanisms. In addition to the potential role of endogenous SDF-1 as coreceptor blocker, we observed that sgp120 failed to downregulate CXCR4 from the surface of CD34+ cells, in contrast to its effect on productively infected adult CD4+ T cells, and despite the fact that the chemokine receptor was susceptible to internalization after binding to SDF-1.
We found that circulating CD34+ cells from individuals treated with CY/G-CSF showed a considerable reduction both in the proportion of cells expressing CXCR4 and in the cell-surface density of the receptor in comparison to BM or UCB CD34+ cells. These data are consistent with the diminished chemotactic responsiveness to SDF-1 previously reported for CY/G-CSF MPB CD34+cells.4 When G-CSF was administered alone, the reduction in the percentage of CXCR4-expressing cells was less pronounced, but the receptor cell-surface density was still significantly lower in comparison to BM CD34+ cells. These observations may help to explain the exit of BM progenitor cells to the PB compartment under experimental mobilization. It is possible that chemotherapy and/or G-CSF induce a downregulation of the CXCR4 receptor on BM CD34+ cells, which, in turn, may facilitate their exit from the stromal microenvironment compartment into the blood sinusoids. Alternatively, cells that already express low levels of CXCR4 may preferentially leave the BM after mobilization. All these events may be preceded, accompanied, or followed by changes either in the levels of expression or in the function of adhesion receptors that have been implicated in the mobilization process, such as α4 integrin, αLβ2, and c-kit.27,56 57 Our results suggest a role for CXCR4 in regulating the exit of progenitors from the BM; however, they do not exclude the possibility that MPB CD34+ cells may have a reduced capacity to migrate in response to SDF-1 due to an impaired cell migration or alteration in the signal transduction machinery.
The reduced expression of CXCR4 may render these cells less prone to home back to the BM extravascular space when subsequently transplanted. In this view, it is noteworthy that CY/G-CSF–mobilized mouse stem cells show a reduced capacity to home to the BM with respect to their normal BM counterpart.58 Yet, mouse mobilized cells do not differ with respect to BM cells for in vitro clonogenic assays and CFU-spleen activity.58 In addition, no evidence exists thus far that this putative homing defect may affect the outcome of autologous or allogeneic transplantation of MPB stem cells in humans, because rapid neutrophil and platelet engraftment are routinely obtained with these cell sources. Therefore, while the reduced expression of CXCR4 may contribute to the mobilization of CD34+ cells to the periphery, it remains to be established whether it affects the homing potential of MPB stem cells during human cell transplantation.
Surprisingly enough, CD34+ cells, both freshly isolated and in vitro–cultured, express SDF-1 mRNA by RT-PCR analysis. SDF-1 was differentially expressed in progenitor cells, being produced by committed (CD34+/CD38+), but not more primitive, progenitor cells (CD34+/CD38−), whereas the CXCR4 receptor was equally expressed on both cell subsets. In addition, when CD34+ cells were cultured in vitro in the presence of cytokines, SDF-1 production was demonstrated by ELISA, as well as by the ability to induce the chemotaxis of other CD34+ cells via the CXCR4 receptor. Because culture of CD34+ cells is invariably associated with their differentiation, we cannot rule out the possibility that SDF-1 detected in CD34+ cell supernatant was produced also by their progeny of differentiated cells.
To our knowledge, this is the first report of a chemokine endogenously produced by human CD34+ cells. The expression of SDF-1 by CD34+ cells was, however, not totally unexpected because this chemokine was previously found constitutively expressed in several tissues. In addition, production of chemokines in an autocrine fashion, both under steady state and activated conditions, has been described for several blood cell types including monocytes (monocyte chemotactic protein-1 and macrophage inflammatory protein-1α, MIP-1α59,60), eosinophils (regulated upon activation normal T-cell expressed and secreted, RANTES, and eotaxin61,62), B cells (MIP-1α63), and megakaryocytes (platelet factor-464). Moreover, the endogenous production of the CC chemokines RANTES, MIP-1α, and MIP-1β by CD4+ T cells from both long-term nonprogressor HIV-1–infected subjects and from some highly exposed but uninfected individuals was shown to be responsible for the reduced infectability of their PBMC to HIV strains using the CCR5 chemokine receptor.65 66
We cannot formally exclude the possibility that contaminating BM mesenchymal stromal cell precursors,67 or endothelial cells expressing CD34,68 may contribute to SDF-1 production in CD34+ cell cultures. However, this event seems very unlikely, in that the frequency of these cells in the BM is rare,67,69 and it is even more rare in CB70 and PB samples.71 In addition, stromal cell precursors and endothelial cells do not express CD38,69 whereas we observed that the expression of SDF-1 was restricted to the CD34+/CD38+ subset. Finally, in light of the occasional event that some adherent cells remained in the cultures of CD34+ cells, SDF-1 was found to be expressed in the nonadherent cell population (data not shown).
The secretion of SDF-1 may allow progenitor cells to sense each other, cluster together, and influence their respective migratory behavior within the BM microenvironment. In support of this hypothesis, a recent study of time-lapse microscopy showed that CD34+cells exhibit in vitro a coordinated migratory pattern and the capacity to aggregate together,72 two events that could be attributed to the endogenous production by CD34+ cells of a chemotactic factor. Finally, the expression of SDF-1 by CD34+ progenitors may represent a mechanism by which these cells downregulate or desensitize their receptor, in response to different stimuli.54
Our findings indicate that CD34+ cells derived from BM, UCB, and MPB are not efficiently infected by X4 HIV-1 strains. HIV infection did not spread efficiently, as indicated both in terms of RT activity and proviral DNA accumulation. The block in HIV-1 infection seems to occur early in the viral cell cycle. This finding is in agreement with earlier observations of BM-derived CD34+cells21,23,73,74 and it is in contrast with others.20 It should be noted, however, that in this latter study incubation of CD34+ cells with HIV-1 was maintained up to 24 hours before removal of the viral excess, allowing for a prolonged contact of the virus with CD34+ cells undergoing differentiation along the mononuclear phagocytic lineage.20Endogenous SDF-1 production by CD34+ cells may compete with X4 HIV-1 for binding to the coreceptor. In this regard, it has been reported that the simple occupancy of HIV coreceptors by chemokines is sufficient for inhibition of HIV infection, even in the absence of G-protein signaling.75 Indeed, lymphocytic cell lines transfected with native SDF-1 show a considerable reduction in their susceptibility to HIV-1 infection, likely due to partial saturation of the binding sites on CXCR4 caused by SDF-1 secretion.76Although the concentrations of SDF-1 produced by CD34+ cell cultures are substantially lower than those required to block in vitro infection, its local accumulation at the cell surface may be sufficient to interfere with HIV-1 and to prevent its entry and/or its spreading. In addition, endogenous SDF-1 may downregulate CXCR4 expression by inducing its endocytosis.54 However, the fact that CXCR4 is rapidly recirculating from and to the plasma membrane,55and that we have not observed a significant reduction of CXCR4 expression on cultured CD34+ cells, makes this mechanism quite unlikely. An alternative explanation is that CXCR4 on CD34+ cells may be in a conformation that does not allow the binding of the gp120/CD4 complexes or that receptor internalization upon gp120 binding may be reduced or impaired in CD34+cells. In this regard, we have reported here that sgp120 failed to downregulate CXCR4 from CD34+ cells, although it was fully effective on activated PB-derived CD4+ T lymphocytes and it interfered with the ability of an anti-CD4 MoAb to stain both lymphocytes and CD34+ cells. Finally, the decreased proportion of cells expressing both CD4 and CXCR4 during CD34+ cell culture may have contributed to further reduce the viral spreading.
Of interest, a mutation at the 3′ untranslated region of SDF-1 has been recently described to delay the onset of acquired immunodeficiency syndrome (AIDS) symptoms of HIV-infected individuals.77 Although the mechanism of the protective role of SDF-1 has yet to be uncovered, it is likely that it may involve regulation of endogenous levels of expression and/or of tissue-specificity of this chemokine, therefore emphasizing the importance for SDF-1 and CXCR4 in HIV infection.
In conclusion, this study provides novel elements relevant for both the physiological trafficking and homing of CD34+ cells and for their potential use in a gene therapy approach for HIV disease. The ability of CD34+/CD38+ cells to produce SDF-1, the ligand of a viral coreceptor, may represent an important determinant in the natural resistance displayed by CD34+cells to be infected by T-cell tropic HIV-1 strains.
ACKNOWLEDGMENT
We thank Dr E. Zappone for providing clinical samples, Dr M. Salomoni for experimental help, and G. Torriani for cell sorting.
A.A. and L.T. equally contributed to this study.
L.T. is the recipient of a fellowship of the Istituto Superiore di Sanità, Rome, Italy. This research was funded by grants from Telethon, EU-BIO4-CT95-0284, and by grants from the National Program for AIDS Research, Istituto Superiore di Sanità, Rome, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Alessandro Aiuti, MD, PhD, Telethon Institute for Gene Therapy (TIGET), Scientific Institute H.S. Raffaele, Via Olgettina 58, 20132, Milan, Italy; e-mail: aiuti@tigem.it.