Abstract
Proper regulation of the human CD34 gene requires a combinatorial action of multiple proximal and long-range, ciselements. This report shows that, like the murine CD34 5′ untranslated region (UTR), the corresponding region of the human CD34 gene is necessary for optimal promoter activity. We localized the most critical element of this region to base pairs +48/+75. Through oligonucleotide competition and antibody supershift experiments in electrophoretic mobility shift assays, we found that this sequence contains a binding site (CCAAT box) for the transcription factor NFY (nuclear factor Y), a factor mediating cell type-specific and cell-cycle regulated expression of genes. Mutating this site led to a 5-fold decrease in CD34 promoter activity in transient transfection experiments. Interestingly, NFY binds adjacently to the earlier identified c-myb binding site. Here we show that both binding sites are important for CD34 promoter function: mutating either site alone decreased CD34 promoter-driven reporter gene activity 4-fold. We also show that the integrity of the c-myb binding site is necessary for stabilization of NFY binding to its site. Such cooperation between c-myb, which is expressed in early hematopoietic cells, and NFY, which is expressed in many cell types, might contribute to specific activation of CD34 in stem cells. The CCAAT box motif was also noted in the 5′ UTR of the murine CD34 gene, however, NFY did not bind to this region. Thus, our results indicate that the functional similarities between the human and murine CD34 5′ UTRs are achieved through different molecular mechanism(s).
ALTHOUGH MUCH PROGRESS has been made in the field of tissue-specific gene regulation, the molecular mechanisms underlying gene expression during the earliest stages of hematopoietic development remain largely unknown. CD34 is an antigen expressed on the surface of hematopoietic stem cells and early progenitors, and its expression declines rapidly as cells differentiate.1 Such restricted expression of CD34 prompted several groups to study regulatory mechanisms acting on the CD34 gene. We and others isolated and characterized the promoter regions of human and murine CD34,2-5 identified a cell type-specific enhancer element at the 3′ end of the human gene,2and an enhancer at the 5′ end of the murine gene.5,6Although the murine CD34 5′ enhancer was described to play an important role in the context of chromatin,6 the in vivo function of the human 3′ enhancer is still undefined.7 Most recently we provided the evidence that multiple regulatory cis elements, including long-range elements located as far as 110 kb upstream of the human CD34 gene, are necessary to provide proper control of expression.7
Identification of the human and murine CD34 promoters revealed the presence of potential binding sites for a number of transcription factors.2,3 Subsequently, c-myb, ets-2,8,9 MZF-1,10,10a Sp1, and Sp34 were demonstrated to bind to their cognate sites and modulate the promoter activities. Although c-myb and ets-2 were shown to independently induce endogenous human CD34 expression,8 9 the functional importance of the remaining protein-binding motifs is not clear.
In the present report, we describe yet another level of human CD34 regulation that occurs through the 5′ UTR of the gene and is mediated by a novel regulator of CD34, nuclear factor Y (NFY). NFY is a heterotrimeric complex highly conserved among eukaryotes11-13 that has been shown to regulate genes in a cell type specific and cell-cycle dependent fashion.13-17The NFY site identified in this report is adjacent to a previously described c-myb binding site.8 9 Potential cooperation between c-myb, which is expressed in early myeloid cells, and NFY could contribute to specific activation of CD34 in stem cells.
MATERIALS AND METHODS
Cell lines and primary cells.
M1 (ATCC No. TIB-192) murine myeloid leukemia cells, KG1a (ATCC No. CCL 246.1) human myelogenous leukemia, T-lymphoblastic lines RPMI-840218 and Jurkat (ATCC No. TIB 152), B-cell lines BJA-B,19 and Raji (ATCC No. CCL 86) were grown in RPMI 1604 medium (GIBCO, Grand Island, NY) containing 10% fetal bovine serum (FBS; Hyclone, Logan, UT) and 2 mmol/L L-glutamine. Human cervical carcinoma line HeLa (ATCC No. CCL2) and the murine myeloid progenitor line 416B20 were cultured in Dulbecco’s modified Eagle’s medium with 10% or 15% FBS, respectively.
Human in vitro differentiated neutrophilic cells were a gift of Dr Jeff Marx (Baxter Healthcare Corporation, Round Lake, IL). CD34+ cells of healthy donors were mobilized with granulocyte colony stimulating factor (G-CSF), isolated by using a magnetic cell separation system (Isolex, Baxter Healthcare Corporation), and cultured for 22 days in the presence of G-CSF. The cells were determined by flow cytometry to be 75% CD15+.
Plasmids.
The promoter-less luciferase vector pXP221 was used for all CD34 promoter/luciferase constructs. Plasmid −391 bp/CD34-Luc was described.2 It contains 391 bp of the 5′ flanking sequence of human CD34 and 175 bp of the 5′ untranslated region, and it is referred to as the −319/+175CD34-luc.
To generate the −391/+300CD34-luc construct, human CD34 genomic BamHI-ApaI fragment (extending from base pairs −1,089 to +300) was subcloned into pXP2. The 4.6-kb BglI fragment containing CD34 5′UTR (base pairs +16/+300) and most of the pXP2 vector was isolated and ligated to the 2.3-kb BglI fragment of −391/+175CD34-luc containing the CD34 5′ flanking sequences and the rest of the pXP2 vector. The −391/+75CD34-luc construct was made by ligating the 466 bp BamHI-SstI promoter fragment from −391/+175-luc (with BamHI site being in the multiple cloning region, upstream from the promoter, and the SstI site being at the base pair +75 of CD34) to BamHI-SstI digested pXP2. To construct the −391/+12CD34-luc plasmid, a 400 bp BamHI-TaqI fragment (TaqI site being at the base pair +12 of CD34) from −391/+175CD34-luc was first subcloned into BamHI-AccI cut pBluescript II KS-, excised as BamHI-XhoI fragment, and ligated into BamHI-XhoI digested pXP2.
Generation of the −68/+175CD34-Luc construct was described earlier.2 To make the −32/+175CD34-luc deletion mutant, human CD34 promoter fragment was amplified under standard polymerase chain reaction (PCR) conditions22 with a sense primer (5′-AACGAGGCATCTGGAGCCCGAACAAACCTCCA-3′; base pairs −32/−1), an anti-sense primer (5′-GCTACTAACTTGAGCTC-3′; base pairs +87/+72), and −68/+175CD34-luc as a template. The resulting PCR product was digested with SstI and subcloned into SstI and HindIII-digested HindIII-SmaI/CD34-Luc construct,2 where the HindIII end was made blunt with Klenow polymerase (New England Biolabs, Beverly, MA). The −10/+175CD34-luc construct was made similarly, with the exception that the sense primer for PCR corresponded to base pairs −10/+20 of CD34 (5′-CAAACCTCCACCTTTTTTGGCCTCGACGGC-3′). A sense primer (5′-GGGAATTCCTTTTTTGGCCTCGACGG-3′; base pairs +1/+19 of CD34) and the anti-sense primer +87/+72 (see above) were used to amplify the first 72 bp of the 5′ untranslated region from the −68/+175CD34-luc construct. The PCR product was digested with SstI and subcloned into HindIII (blunt) and SstI-cut HindIII-SmaI/CD34-Luc,2 thus creating the +1/+175CD34-luc. For +24/+175CD34-luc and +48/+175CD34-luc constructs, sense primers +24/+40 (5′-ccaagcttAACCCAGCCTCCCTCCT-3′; CD34-derived sequence is capitalized and the engineered HindIII site is underlined) or +48/+64 (5′-ccaagctTCCGCCTTTGGGACCAA-3′; CD34-derived sequence is in capital letters and HindIII site is underlined), respectively, were employed together with an anti-sense primer (5′-CTCTAGAGGATAGAATGGCG-3′), which annealed to base pairs +53/+34 of luciferase gene in pXP2, where the A nucleotide in ATG is denoted +1. The -68/+175CD34-luc plasmid2 served as a template. Each PCR product was digested with HindIII and BglII, gel purified, and subcloned into HindIII-BglII digested pXP2. The HindIII-SmaI/CD34-Luc construct2 was digested with HindIII and SstI, the ends were made blunt with T4 DNA polymerase, and plasmid was relegated to make the +81/+175CD34-luc construct. The internal deletion of base pairs +40/+75 was created as follows. BamHI-SmaI/CD34-Luc2 served as a template for PCR with a sense primer spanning base pairs 1 to 17 of the pXP2 vector (5′-ggGGATCCAGCTCAGATCC-3′; BamHI site is underlined) and an anti-sense primer corresponding to base pairs +46/+25 of CD34 (5′-GGCGTTAGGAGGGAGGCTGGGT-3′) to amplify the −391/+46 region of CD34. On digestion with BamHI, the PCR product was ligated to BamHI-SstI digested BamHI-SmaI/CD34-Luc.2 Sequence analysis showed that the deletion spanned base pairs +40/+75 rather than predicted +46/+75. All PCR generated mutations were verified by sequencing.
Site-directed mutagenesis.
Point mutations were introduced into NFY and c-myb binding sites essentially as described.23,24 Two complementary “internal” primers, sense (C) and antisense (B), containing point mutation(s) in NFY site alone, c-myb site alone, or both sites together8,9 were synthesized (see Fig 5A for primer sequence information). The human CD34 promoter construct −391/+300CD34-luc plasmid was used as a template. The “external” primer A containing partial sequence of the multiple cloning site and comprising the BamHI restriction site (5′-ggGGATCCAGCTCAGATCC-3′; BamHI site is underlined) from the pXP2 vector21 was used with the primer B to generate fragment 1 (40 PCR cycles: 94°C, 1′; 55°C, 1′; 72°C, 1′; 50-μL reaction volume). Fragment 2 was amplified by using the same PCR conditions and the primer C and the “external” primer D (5′-CTCTAGAGGATAGAATGGCG-3′), which spans base pairs +53/+34 (+1 being assigned to A in ATG codon) in an anti-sense direction of the luciferase gene in pXP2. Fragment 2 contains a SmaI site, which occurs naturally in the CD34 promoter (at position +175). Fragments 1 and 2 have a 36-bp overlap and were used as primers for each other in 3 cycles of PCR (94°C, 1′; 55°C, 1′; 72°C, 1.5′). Subsequently, they were amplified with the “external” primers A and D (40 PCR cycles: 94°C, 1′; 55°C, 1′; 72°C, 1.5′; 50 μL reaction volume).
All PCR-generated mutants were digested with BamHI and SmaI, gel-purified, subcloned into BamHI-SmaI cut pXP2, and verified by DNA sequencing.
Transfections and luciferase assay.
Transient transfections were performed as described previously.25 RPMI-8402 and 416B cells were electroporated at 270 V and 960 μF whereas KG1a cells were electroporated at 350 V and 960 μF. Cells were harvested 5 hours after transfection and luciferase activity was measured. Transfection efficiencies were normalized for the production of cotransfected human growth hormone expressed from the cytomegalovirus (CMV) promoter, as described earlier,2 and measured by radioimmunoassay (Nichol’s Institute, San Juan Capistrano, CA). DNA for transfections was purified on cesium chloride gradients. Luciferase assays were performed as previously described.25 The lysates were quantitated by the Bradford method26 by using the Protein Assay reagent (Bio-Rad Laboratories, Hercules, CA). The results shown in Fig 5D were normalized for the Renilla luciferase activity expressed from the cotransfected pCMV-RL plasmid and assayed by using Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Individual transfections were repeated at least 3 times by using at least 2 independent DNA preparations.
Nuclear extracts.
Nuclear extracts were prepared as described24,27 with minor modifications. After the cells were washed in 1 × phosphate-buffered saline, the volume of the cell pellet was determined, the cells were resuspended in equal volume of ice-cold buffer A (10 mmol/L HEPES pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol (DTT), 0.5 mmol/L phenylmethylsulfonyl fluoride [PMSF]), and allowed to swell on ice for 15 minutes. The cell membranes were disrupted by applying 5 to10 rapid strokes in and out of a 1 mL syringe attached to a 25-gauge needle. The nuclei were pelleted in a microfuge at 12,000g for 20 seconds, resuspended in two thirds of the original cell pellet volume in buffer C (20 mmol/L HEPES pH 7.9, 25% glycerol, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, 0.5 mmol/L PMSF), and incubated at 4°C for 30 minutes with occasional flicking. The nuclei were pelleted for 5 minutes at 4°C at 12,000g in a microfuge. The supernatant was aliquoted and stored at −80°C. Concentration of the nuclear extracts was determined by Bradford assay26 by using the Protein Assay reagent (Bio-Rad Laboratories).
Electrophoretic mobility shift assay (EMSA).
Double-stranded oligonucleotides (+48/+80, Y+M+, or Y+M-; sequences are shown in Figs 2A and 5A) were end-labeled with 32P-ATP and Polynucleotide Kinase to a specific activity of 108 to 109 cpm/μg. Nuclear extracts (about 10 to 20 μg protein) were incubated for 20 minutes on ice with 5 × 104 cpm of labeled DNA. The reaction mixture also contained 2 μg of poly dI-dC, 10 mmol/L Tris-HCl (pH 7.5), 50 mmol/L NaCl, 1 mmol/L DTT, 1 mmol/L EDTA, and 5% glycerol in a total volume of 25 μL. Where indicated, 1 to 3 μL of antibody was added to binding reactions. Antibodies against NFY were: rabbit anti-rat CBF-A (NFYB), a gift of Dr Benoit de Crombrugghe28 (M.D. Anderson Cancer Center, Houston, TX), mouse monoclonal anti-murine NFYA and rabbit polyclonal anti-murine NFYB sera kindly provided by Dr Roberto Mantovani29 (Universita degli Studi di Milano, Milan, Italy), and rabbit anti-human NFYA (Rockland Immunochemicals, Gilbertville, PA). Anti-Sp1 antibody was a generous gift of Dr Steve Jackson (Wellcome Labs, Cambridge, UK). Unlabeled competitor oligonucleotides (identified in figure legends) were added to the binding reactions at a 300- to 600-fold excess immediately before the addition of the radioactive probe. Unbound probe and DNA-protein complexes were separated by electrophoresis through 4% polyacrylamide gels in 0.5 × TBE (45 mmol/L Tris-borate, 1 mmol/L EDTA, pH 8) at 4°C, at 150 V. A 19:1 acrylamide:bis-acrylamide mix was used, except as specified in Fig 2C and D, in which a 29:1 mix was used.
RESULTS
Base pairs +48 to +75 are important for human CD34 promoter activity.
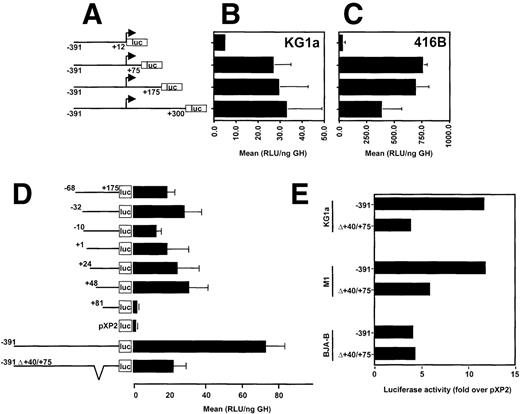
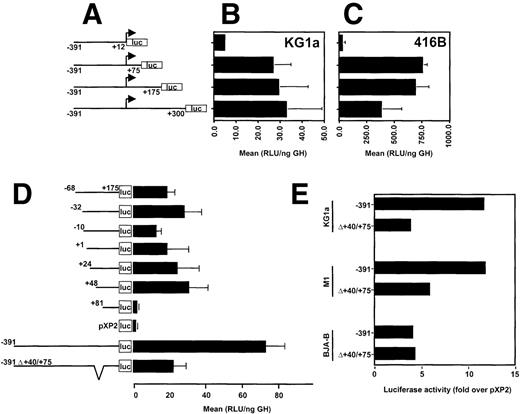
Earlier promoter studies showed that as little as 68 bp upstream from the transcription start site along with 175 bp of the 5′ UTR of CD34 were sufficient to direct expression of the luciferase reporter gene in transiently transfected CD34+ RPMI-8402 cells. Moreover, the activity of this region was about 10-fold higher in RPMI-8402 cells than in HeLa cells.2 Recent studies of the murine CD34 promoter show the importance of the 5′ UTR for promoter activity.4 To determine the role of the 5′ UTR of the human CD34 gene, we generated a series of promoter deletion mutants, which contained 391 bp of the 5′ flanking sequences and had their 3′ endpoints at base pairs +12, +75, +175, or +300, as diagramed in Fig 1A. In transiently transfected human CD34+ KG1a cells, maximal luciferase activity was achieved with the −391/+300CD34-luc construct (Fig1B). Nearly complete deletion of the 5′ UTR from the CD34 promoter, with preservation of the natural transcription initiation site (-391/+12CD34-luc construct), resulted in about 6-fold reduction of luciferase activity. Luciferase activity was nearly restored when the 5′-terminal 75 bp of the 5′ UTR (−391/+75CD34-luc construct) were added to the promoter. The same 75 bp of the 5′ UTR conferred maximal activity in the murine CD34+ cell line 416B (Fig 1C), although an additional 100 bp of the 5′ UTR (−391/+175CD34-luc construct) had no significant influence. The sequences +175 to +300 had a somewhat inhibitory effect. The promoter construct lacking the 5′ UTR (−391/+12CD34-luc construct) showed only background values in 416B cells (Fig 1C).
The 5′ untranslated region of human CD34 is required for maximum promoter activity. (A) A diagram of CD34/luciferase constructs used in transient transfections shown in (B) and (C). All constructs have the same 5′ flanking sequences of CD34 (391 bp), but the 3′ end points are at the base pairs +12, +75, +175, and +300. (B) CD34+ human KG1a and (C) murine 416B cells were transiently transfected with 20 μg of each plasmid depicted in (A) and luciferase activity determined. (D) CD34 promoter/luciferase constructs containing various 5′ end truncations and identical 3′ ends at the base pair +175 (diagrammed on the left) were analyzed in transient transfection assays in human CD34+ RPMI-8402 cells. For comparison, a promoterless pXP2 luciferase vector is included. The internal deletion of base pairs +40/+75 in the context of the −391/+175 promoter is shown on the bottom. (E) The cell type specific effect of the internal deletion of the +40/+75 region in the context of the −391/+175 promoter was tested in transiently transfected human CD34+, KG1a and RPMI-8402, murine CD34+ M1, and human CD34− BJA-B cells. The error bars indicate the standard deviations. The data are presented as mean relative light units (RLU) normalized to the expression of a cotransfected pCMV-human growth hormone (ng GH).
The 5′ untranslated region of human CD34 is required for maximum promoter activity. (A) A diagram of CD34/luciferase constructs used in transient transfections shown in (B) and (C). All constructs have the same 5′ flanking sequences of CD34 (391 bp), but the 3′ end points are at the base pairs +12, +75, +175, and +300. (B) CD34+ human KG1a and (C) murine 416B cells were transiently transfected with 20 μg of each plasmid depicted in (A) and luciferase activity determined. (D) CD34 promoter/luciferase constructs containing various 5′ end truncations and identical 3′ ends at the base pair +175 (diagrammed on the left) were analyzed in transient transfection assays in human CD34+ RPMI-8402 cells. For comparison, a promoterless pXP2 luciferase vector is included. The internal deletion of base pairs +40/+75 in the context of the −391/+175 promoter is shown on the bottom. (E) The cell type specific effect of the internal deletion of the +40/+75 region in the context of the −391/+175 promoter was tested in transiently transfected human CD34+, KG1a and RPMI-8402, murine CD34+ M1, and human CD34− BJA-B cells. The error bars indicate the standard deviations. The data are presented as mean relative light units (RLU) normalized to the expression of a cotransfected pCMV-human growth hormone (ng GH).
We also constructed a set of deletion mutants that extended 3′ to base pair +175 and had their 5′ endpoints ranging from base pairs −68 to +81 (Fig 1D). No profound change in luciferase activity was seen in CD34+ RPMI-8402 cells even after deletion of the major transcription start site down to base pair +48 (+48/+175CD34-luc). Further deletion to base pair +81 (+81/+175CD34-luc) caused a 13-fold decrease in luciferase activity, completely extinguishing promoter function in this cell line. No significant differences were noted between the +48 and the +81 deletion constructs in CD34-HeLa cells (4.6 v 2.6 RLU/ng hGH in HeLa [n=2] as opposed to 31.1 +/−10 v 2.4 +/−0.8 RLU/ng hGH [n=5] in RPMI-8402).
Previous studies showed that the majority of CD34 promoter activity resided between base pairs −391 and +175 (−391CD34-luc2). To determine the functional importance of the +48/+80 region in the context of this minimal promoter, a deletion of base pairs +40 to +75 was incorporated into the −391CD34-luc construct. Transient transfection experiments showed that the internal +40/+75 deletion caused a 3-fold decrease in luciferase activity in CD34+ T lymphoblastic RPMI-8402 cells (73.5 +/− 10.4 RLU/ng hGH v 22.5 +/− 6.5 RLU/ng hGH [n=4]; Fig 1D). A similar decrease of luciferase activity was seen in another human CD34+ cell line, myelogenous leukemia KG1a (Fig 1E), whereas in murine CD34+ myeloid leukemia M1 cells, a 50% decrease was noted. In contrast, no effect was observed in human CD34-BJA-B B cells upon +40/+75 deletion from the −391/+175 promoter. Taken together, these results suggest that the element centered between base pairs +48 and +75 is important for CD34 promoter activity and contributes to cell type specificity.
Nuclear proteins bind to the +48/+80 region.
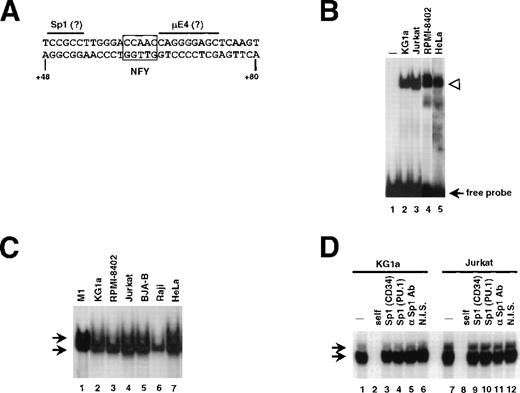
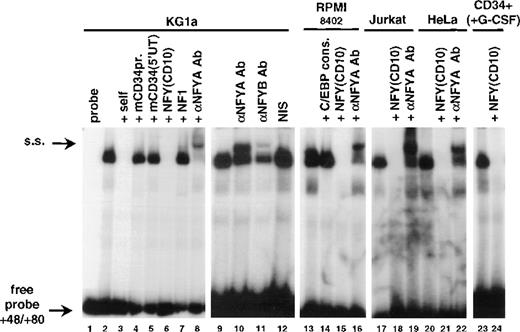
To identify protein(s) interacting with the +48/+80 sequences, an EMSA was performed by using a probe spanning base pairs +48/+80 (Fig 2A). A complex that binds to this region in a sequence specific manner was detected in extracts from murine CD34+ M1 cells (Fig 2C; lane 1), as well as human CD34+ cell lines KG1a and RPMI-8402 (Fig 2B, lanes 2 and 4, respectively; and Fig 2C, lanes 2 and 3, also see Fig 2D). A complex with the same mobility was also seen in nuclear extracts from CD34-cell lines Jurkat, HeLa (Fig 2B, lanes 3 and 5, respectively; Fig 2C, lanes 4 and 7, respectively; also see Fig 2D), BJAB, Raji, and U937 (Fig 2C; lanes 5 and 6; data not shown). Moreover, the binding was detected in purified primary human CD34+, which were differentiated in vitro with G-CSF (Fig 4; lane 23). We asked whether changing the conditions of EMSA would reveal any tissue-specific differences in complex formation. Alterations in temperature of the binding reaction, electrophoresis (+4°C versus room temperature), or composition of the running buffer (1× Tris-glycine v 0.5 × TBE) had no apparent effect. Extending the time of electrophoresis in conjunction with increase of acrylamide:bis-acrylamide ratio (19:1 to 29:1) resulted in separating the retarded band to 2 complexes (Fig 2C; also see Fig 2D). Nevertheless, no lineage-restricted pattern was observed.
A nuclear factor binds to the critical element in the human CD34 5′ UTR. (A) Sequence of the oligonucleotide +48/+80 used as a probe in EMSA. Putative Sp1 and μE4 sites are overlined. The potential Y box motif is boxed. (B) Nuclear extracts (10 μg) from CD34+ (KG1a and RPMI-8402; lanes 2 and 4, respectively) and CD34− (Jurkat and HeLa; lanes 3 and 5, respectively) were incubated with radiolabeled +48/+80 probe. The protein/DNA complexes (marked with an arrowhead) were resolved on native acrylamide gel. No protein was added to the reaction in lane 1. (C) EMSA with the +48/+80 probe and 10 μg of nuclear extract prepared from cell lines indicated above lanes. The gel contained 4% acrylamide:bis-acrylamide at the ratio 29:1 to allow the separation of 2 closely migrating complexes (marked with arrows). The electrophoresis proceeded for 4 hours at 4°C, thus the unbound probe migrated out of the gel. (D) Sp1 is not involved in the binding to the +48/+80 region. Probe shown in (A) was incubated with 10 μg of nuclear extracts of KG1a (lanes 1 to 6) or Jurkat (lanes 7 to 12) with electrophoresis conditions as in (C). Lanes 2 and 8 contained unlabeled self-competitor, lanes 3 and 9 contained the Sp1-binding oligonucleotide from the human CD34 promoter (base pairs −68/−44) as competitor, and lanes 4 and 10 contained the Sp1-binding oligonucleotide from the murine PU.1 promoter (base pairs −114/−90).36 An antiserum raised against Sp1 or nonimmune serum were added to the binding reactions shown in lanes 5 and 11, or 6 and 12, respectively.
A nuclear factor binds to the critical element in the human CD34 5′ UTR. (A) Sequence of the oligonucleotide +48/+80 used as a probe in EMSA. Putative Sp1 and μE4 sites are overlined. The potential Y box motif is boxed. (B) Nuclear extracts (10 μg) from CD34+ (KG1a and RPMI-8402; lanes 2 and 4, respectively) and CD34− (Jurkat and HeLa; lanes 3 and 5, respectively) were incubated with radiolabeled +48/+80 probe. The protein/DNA complexes (marked with an arrowhead) were resolved on native acrylamide gel. No protein was added to the reaction in lane 1. (C) EMSA with the +48/+80 probe and 10 μg of nuclear extract prepared from cell lines indicated above lanes. The gel contained 4% acrylamide:bis-acrylamide at the ratio 29:1 to allow the separation of 2 closely migrating complexes (marked with arrows). The electrophoresis proceeded for 4 hours at 4°C, thus the unbound probe migrated out of the gel. (D) Sp1 is not involved in the binding to the +48/+80 region. Probe shown in (A) was incubated with 10 μg of nuclear extracts of KG1a (lanes 1 to 6) or Jurkat (lanes 7 to 12) with electrophoresis conditions as in (C). Lanes 2 and 8 contained unlabeled self-competitor, lanes 3 and 9 contained the Sp1-binding oligonucleotide from the human CD34 promoter (base pairs −68/−44) as competitor, and lanes 4 and 10 contained the Sp1-binding oligonucleotide from the murine PU.1 promoter (base pairs −114/−90).36 An antiserum raised against Sp1 or nonimmune serum were added to the binding reactions shown in lanes 5 and 11, or 6 and 12, respectively.
Sequence analysis of the +48/+80 region showed areas of similarity to the binding motifs of several transcription factors known to act as regulators in hematopoietic cells, for example MZF-1, Ikaros-2, or c-myb. However, because of the ubiquitous nature of the binding activity to the +48/+80 region, we focused our attention on the factors expressed ubiquitously, such as Sp1, E2A gene products (E12 and E47), and NFY, which are reported to be involved in cell type-specific expression of hematopoietic genes.13,30-34 A sequence resembling an Sp1 binding site was noted at the 5′ end of the oligonucleotide (Fig 2A; overlined). Developing hematopoietic cells are among the tissues expressing the highest levels of Sp1.35Moreover, Sp1 was found to a play critical role in tissue-specific gene regulation30,31 and, in particular, its binding to a site in the murine CD34 5′ untranslated region is required for maximum promoter activity.4 Therefore, we investigated the possibility that Sp1 binds to the +49/+54 region. As shown in Fig 2D, oligonucleotides containing Sp1 binding sites between base pairs −68 to −44 of the human CD34 promoter (A.B. Satterthwaite and D.G. Tenen, unpublished data, August 1992) (lanes 3 and 9) or base pairs −114 to −90 of the murine PU.1 promoter36 (lanes 4 and 10) were used in competition experiments. Both Sp1 oligonucleotides had no effect on the pattern of binding to +48/+80 sequence. When a radiolabeled Sp1-containing oligonucleotide (from the murine PU.1 promoter36) was used as a probe, the unlabeled +48/+80 oligonucleotide did not compete for binding. Moreover, addition of anti-Sp1 antiserum (lanes 5 and 11) did not affect any of the 2 retarded bands, suggesting that Sp1 is not the factor binding to the +48/+80 region.
A region of similarity to one of the E2A gene-encoded basic-helix-loop-helix (b-HLH) protein (E12/E47) binding motifs, namely μE4,37-39 was found between base pairs +65 and +73 (Fig2A; overlined). The μE4 site is located adjacent to the octamer motif in the murine IgH chain intronic enhancer.37,38 When a 51 bp IgH enhancer-derived DNA fragment containing μE4 and octamer sites40 was added to the binding reaction, no change in appearance of the complexes was seen (data not shown). Therefore, we conclude that b-HLH E2A-encoded proteins most likely do not interact with this sequence.
Within this critical domain of the 5′ UTR, there is a centrally located potential Y box motif (Fig 2A; base pairs +60 to +64, boxed). It contained a sequence CCAAC, which matched 4 of 5 bases found in the consensus CCAAT box known to bind a trimeric nuclear factor NFY.13 In gel shift experiments (Fig 4), the oligonucleotide derived from the human CD10 promoter and containing a perfect CCAAT box motif41 competed away the +48/+80 binding activity in nuclear extracts of KG1a, RPMI-8402, Jurkat, and HeLa cells with the efficiency comparable with the self-competition (Fig 4; compare lanes 6, 15, 18, 21, and 24 with lane 3). Furthermore, addition of the specific antibodies raised against NFYA or NFYB subunits of the NFY complex resulted in formation of supershifted complexes (Fig 4; lanes 8, 10, 11, 16, 19, and 22) and nonimmune serum did not affect the complex formation (lane 12). NFY is one of several unrelated families of proteins binding to CCAAT sequences. Oligonucleotides containing binding sites specifically recognized by members of these other families, NF1 or C/EBP(α/β/δ), did not compete for the +48/+80 binding activity (Fig 4; lanes 7 and 14). Conversely, complexes bound to the C/EBP consensus probe were not competed by the cold +48/+80 oligonucleotide (H.S. Radomska and D.G. Tenen, data not shown).
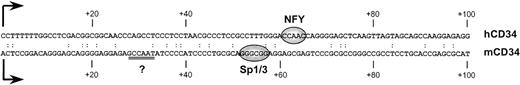
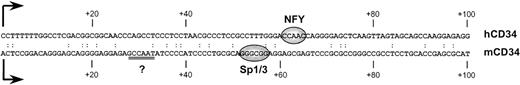
Human and murine CD34 genes exhibit analogous patterns of expression.1,42 Although no long stretches of significant similarity were noted between the sequences of the proximal promoter and 5′ untranslated regions of both genes2 3 (also see Fig 3), some sequence similarities to the CCAAT box motif were noted in the promoter (base pairs −42/−38) and 5′ untranslated region (base pairs +28/+334; double underlined in Fig 3) of the murine CD34 gene. However, oligonucleotides corresponding to those regions did not compete the binding activity to the +48/+80 sequence (Fig 4; lane 4). Similarly, an unrelated oligonucleotide (base pairs +48/+119 from the murine CD34 5′ UTR4) had no effect on the formation of the binding complex (Fig 4; lane 5).
Comparison of sequences from the 5′ UTRs of the human (top strand; hCD34) and murine (lower strand; mCD34) CD34 genes. Transcription start sites are marked with arrows. The regions critical for human (NFY) and murine (Sp1/3) gene activity are indicated as ovals. A potential CCAAT box in the murine sequence is double underlined.
Comparison of sequences from the 5′ UTRs of the human (top strand; hCD34) and murine (lower strand; mCD34) CD34 genes. Transcription start sites are marked with arrows. The regions critical for human (NFY) and murine (Sp1/3) gene activity are indicated as ovals. A potential CCAAT box in the murine sequence is double underlined.
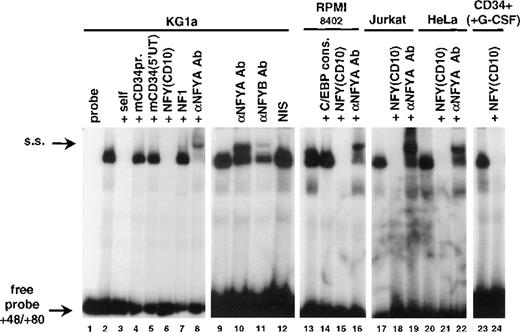
Identification of the +48/+80 binding activity as NFY. Binding reactions were performed in the absence (lane 1) or presence of 20 μg of nuclear extract from KG1a (lanes 2 to 12), RPMI-8402 (lanes 13 to 16), Jurkat (lanes 17 to 19), HeLa (lanes 20 to 22), and CD34+ peripheral blood cells cultured with G-CSF for 22 days (75% CD15+; lanes 23 and 24). The unlabeled competitors were as follows: self-competitor in lane 3; oligonucleotide −78/−28 from the murine CD34 promoter in lane 4; 5′ untranslated region from the murine CD34 (base pairs +48/+119) in lane 5; NFY binding site from the human CD10 promoter41 in lanes 6, 15, 18, 21, and 24; NF1 binding site41 in lane 7; C/EBP consensus oligonucleotide61 in lane 14. The reactions in lanes 8, 16, 19, and 22 included anti-NFYA antibodies provided by Benoit de Crombrugghe.28 Lanes 10 and 11 contained, respectively, anti-NFYA and anti-NFYB antibodies supplied by Roberto Mantovani.29 Supershifted complexes (s.s.) as well as free probe are indicated by arrows.
Identification of the +48/+80 binding activity as NFY. Binding reactions were performed in the absence (lane 1) or presence of 20 μg of nuclear extract from KG1a (lanes 2 to 12), RPMI-8402 (lanes 13 to 16), Jurkat (lanes 17 to 19), HeLa (lanes 20 to 22), and CD34+ peripheral blood cells cultured with G-CSF for 22 days (75% CD15+; lanes 23 and 24). The unlabeled competitors were as follows: self-competitor in lane 3; oligonucleotide −78/−28 from the murine CD34 promoter in lane 4; 5′ untranslated region from the murine CD34 (base pairs +48/+119) in lane 5; NFY binding site from the human CD10 promoter41 in lanes 6, 15, 18, 21, and 24; NF1 binding site41 in lane 7; C/EBP consensus oligonucleotide61 in lane 14. The reactions in lanes 8, 16, 19, and 22 included anti-NFYA antibodies provided by Benoit de Crombrugghe.28 Lanes 10 and 11 contained, respectively, anti-NFYA and anti-NFYB antibodies supplied by Roberto Mantovani.29 Supershifted complexes (s.s.) as well as free probe are indicated by arrows.
Functional interaction of NFY and c-myb binding sites.
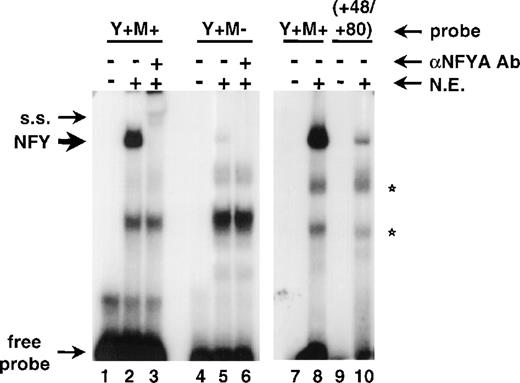
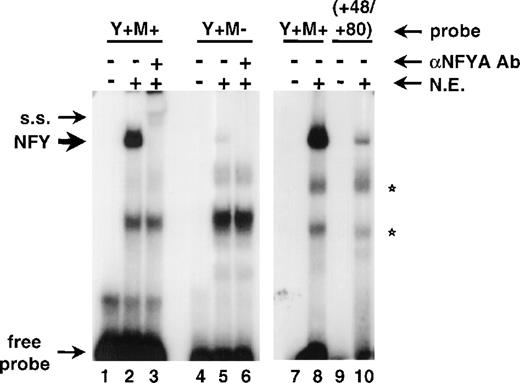
It has been reported earlier that nucleotides +54/+89 of the human CD34 gene contain two sites that bind a bacterially-expressed recombinant c-myb protein and act as a myb responsive element.8 9Interestingly, one of those sites maps precisely to the NFY binding site (base pairs +60/+64) identified herein. We generated an oligonucleotide probe (shown in Fig 5A; Y+M+) containing base pairs +54 to +89 and tested it in gel shift assays. Figure 5B shows that addition of either of the two anti-NFYA antibodies to the binding reactions supershifted a prominent complex (lanes 3 and 4) and the anti-NFYB antibody diminished binding significantly (lane 5). The complex was not formed when the binding reactions contained unlabeled NFY-binding oligonucleotides from the CD10 gene, region +48/+80 from the human CD34 gene, or the Y+M+ oligonucleotide itself (lanes 6, 9, and 10, respectively). Oligonucleotides containing point mutations in the NFY site (Y-M+ and Y-M-) did not abolish NFY binding (Fig 5B; lanes 11 and 12). It should be noted that under gel shift conditions optimal for NFY binding we have not observed binding of c-myb to the Y-M+ probe in nuclear extracts (Fig 5B; lane 14). Also, competition of binding to the Y+M+ probe with oligonucleotides containing only the NFY site was complete (lanes 6 and 9). The competition with an oligonucleotide containing only a potential c-myb binding site did not have a significant effect on the formation or migration of the NFY/DNA complex (lane 11).
The NFY site positively regulates the human CD34 promoter and functionally interacts with c-myb binding site. (A) A comparison of oligonucleotide probes used in Fig 2 (+48/+80) and the +54/+89 sequence (Y+M+) shown previously to bind c-myb.8,9 NFY and c-myb sites are boxed. Arrows under the Y+M+ sequence indicate point mutations introduced into either the Y box or the c-myb site individually (to generate Y−M+ or Y+M− oligonucleotides) or to both sites simultaneously (to generate the Y−M− oligonucleotide). (B) EMSA with Y+M+ probe and no protein (lanes 1 and 7) or 20 μg of RPMI-8402 nuclear extract (N.E.; lanes 2 to 6 and 8 to 12). Reactions in lanes 3 and 4 contained antibodies raised to NFYA [NFYA(1) was from Rockland Immunochemicals and NFYA(2) was described29], and anti-NFYB antibody29 was added in lane 5. Competitors were added to the reactions as follows: CD10-derived NFY binding oligonucleotide41 to lane 6; +48/+80 region of human CD34 to lane 9; self-competitor (Y+M+) to lane 10; +54/+89 oligonucleotide with Y box mutated (Y−M+) to lane 11; +54/+89 oligonucleotide with both the Y box and c-myb site mutated (Y−M−) to lane 12. Reactions in lanes 13 and 14 contain Y−M+ probe incubated in the absence or presence of nuclear extract, respectively. The NFY binding complex is indicated by an arrow. Supershifted complexes (s.s.) and free probe are marked with brackets. Stars indicate nonspecific complexes. (C) A schematic drawing of constructs used in transient transfection assays shown in (D). All constructs contained the human CD34 promoter (base pairs −391/+175) with wild-type Y box and c-myb site (Y+M+), Y box mutated (Y−M+), c-myb site mutated (Y+M−), or both sites mutated (Y−M−). (D) CD34 promoter/luciferase constructs in (C) were transfected to KG1a cells together with an internal control plasmid, CMV-driven Renilla luciferase (pCVM-RL). The results are presented as mean firefly luciferase relative light units (FL RLU) and normalized to Renilla luciferase relative light units (RL RLU). Standard deviations are represented by error bars.
The NFY site positively regulates the human CD34 promoter and functionally interacts with c-myb binding site. (A) A comparison of oligonucleotide probes used in Fig 2 (+48/+80) and the +54/+89 sequence (Y+M+) shown previously to bind c-myb.8,9 NFY and c-myb sites are boxed. Arrows under the Y+M+ sequence indicate point mutations introduced into either the Y box or the c-myb site individually (to generate Y−M+ or Y+M− oligonucleotides) or to both sites simultaneously (to generate the Y−M− oligonucleotide). (B) EMSA with Y+M+ probe and no protein (lanes 1 and 7) or 20 μg of RPMI-8402 nuclear extract (N.E.; lanes 2 to 6 and 8 to 12). Reactions in lanes 3 and 4 contained antibodies raised to NFYA [NFYA(1) was from Rockland Immunochemicals and NFYA(2) was described29], and anti-NFYB antibody29 was added in lane 5. Competitors were added to the reactions as follows: CD10-derived NFY binding oligonucleotide41 to lane 6; +48/+80 region of human CD34 to lane 9; self-competitor (Y+M+) to lane 10; +54/+89 oligonucleotide with Y box mutated (Y−M+) to lane 11; +54/+89 oligonucleotide with both the Y box and c-myb site mutated (Y−M−) to lane 12. Reactions in lanes 13 and 14 contain Y−M+ probe incubated in the absence or presence of nuclear extract, respectively. The NFY binding complex is indicated by an arrow. Supershifted complexes (s.s.) and free probe are marked with brackets. Stars indicate nonspecific complexes. (C) A schematic drawing of constructs used in transient transfection assays shown in (D). All constructs contained the human CD34 promoter (base pairs −391/+175) with wild-type Y box and c-myb site (Y+M+), Y box mutated (Y−M+), c-myb site mutated (Y+M−), or both sites mutated (Y−M−). (D) CD34 promoter/luciferase constructs in (C) were transfected to KG1a cells together with an internal control plasmid, CMV-driven Renilla luciferase (pCVM-RL). The results are presented as mean firefly luciferase relative light units (FL RLU) and normalized to Renilla luciferase relative light units (RL RLU). Standard deviations are represented by error bars.
To test the functional effect of NFY binding to the +60/+64 region, we introduced point mutations abolishing the binding of NFY to its site (see Fig 5A and Fig 5B; lane 14) and incorporated them into a luciferase plasmid containing base pairs −391/+175 from the human CD34 promoter (constructs diagramed in Fig 5C). As shown in Fig 5D, mutating the NFY site (Y-M+ construct) resulted in a 5-fold decrease in luciferase activity in transiently transfected KG1a cells and a 4-fold decrease in RPMI-8402 cells, as compared with the wild-type construct (Y+M+ construct).
NFY has been described to interact with a number of transcription factors and exert a cell type-specific regulatory effect.13To test whether NFY interacts with the c-myb site, we introduced point mutations (shown in Fig 5A) into c-myb site alone or in combination with the NFY site mutations and incorporated them into the −391/+175CD34-luc plasmid (diagramed in Fig 5C). Figure 5D shows that mutating the c-myb site (Y+M-construct) resulted in a 4-fold decrease in luciferase activity of transiently transfected KG1a cells. The same decrease was observed when both sites, c-myb and NFY, were mutated simultaneously (in Y-M-construct).
We conclude that NFY specifically binds to sequences +61/+65 of the human CD34 promoter and positively regulates it. In contrast, NFY does not bind to the putative CCAAT boxes in the proximal sequences of the murine CD34 gene. Moreover, although our gel shift experiments failed to show the binding of c-myb to its site, our data support the idea that functional cooperativity between NFY and c-myb sites is likely.
Integrity of the c-myb binding site facilitates binding of NFY to the adjacent site.
To determine the mechanism(s) of NFY and c-myb site cooperation, we performed co-immunoprecipitation studies. We used several experimental procedures, but in no case could we show the physical interaction of NFY and c-myb in the absence of DNA (data not shown). Because our data strongly suggested the functional cooperation of both factors, we asked if one factor affects the DNA binding of the other one. Accordingly, we performed a gel shift assay by using either the Y+M+ or Y+M− oligonucleotides as probes, both of which were labeled to equal specific activities (5.3 × 109 cpm/μg and 5.2 × 109 cpm/μg, respectively). Figure 6 shows that mutation of the c-myb site resulted in significantly weaker (24-fold, as quantitated by ImageQuant software by Molecular Dynamics, Sunnyvale, CA) binding of NFY to its site in the Y+M-oligonucleotide (lane 5), as compared with the binding of NFY to the wild-type probe (Y+M+; lane 2). Both complexes reacted with anti-NFYA antibody (lanes 3 and 6). In another experiment, the formation of the NFY complex on the Y+M+ probe was compared with the original +48/+80 probe, which lacked a c-myb binding site. The NFY complex formed with the +48/+80 probe was significantly weaker than the corresponding complex with the Y+M+ probe (Fig 5; compare lane 10 with lane 8). Thus, we conclude that the integrity of the c-myb binding site is necessary for stabilization of the binding of NFY to its imperfect site.
Binding of NFY is stabilized by the integrity of the adjacent c-myb binding site. EMSA was performed with the Y+M+ probe (lanes 1 to 3, 7, and 8), Y+M− probe (lanes 4 to 6), and +48/+80 probe (lanes 9 and 10). The reactions contained no protein (lanes 1, 4, 7, and 9) or 20 μg of RPMI-8402 nuclear extract (N.E.). In addition, reactions shown in lanes 3 and 6 contained anti-NFYA antibody (NFYA Ab; from Rockland Immunochemicals). The migration of NFY binding complex (NFY), supershifted complexes (s.s.), or free probe is shown by arrows. Nonspecific complexes are marked by stars.
Binding of NFY is stabilized by the integrity of the adjacent c-myb binding site. EMSA was performed with the Y+M+ probe (lanes 1 to 3, 7, and 8), Y+M− probe (lanes 4 to 6), and +48/+80 probe (lanes 9 and 10). The reactions contained no protein (lanes 1, 4, 7, and 9) or 20 μg of RPMI-8402 nuclear extract (N.E.). In addition, reactions shown in lanes 3 and 6 contained anti-NFYA antibody (NFYA Ab; from Rockland Immunochemicals). The migration of NFY binding complex (NFY), supershifted complexes (s.s.), or free probe is shown by arrows. Nonspecific complexes are marked by stars.
DISCUSSION
Human and murine CD34 genes exhibit similar patterns of expression, which is achieved by the action of multiple elements.4-7Recently, we showed that the proper expression of the human CD34 requires presence of distal elements.7 In the current study, we present data that human CD34 expression, like that of its murine counterpart, is also regulated through the 5′ UTR. However, although both human and murine 5′ UTRs are important for promoter function, they are not conserved in terms of sequence or in terms of binding of common transcription factors. This is in contrast to the promoters for other hematopoietic genes, for example PU.1,36 G-CSF receptor,24 or c-kit,43 which exhibit overall significant (more than 80%) interspecies sequence similarities with the highest degree of conservation within functionally important elements.
The 5′ UTR of murine CD34 gene was described to be necessary for maximum promoter activity. The expression from the murine CD34 promoter was 7- to 10-fold higher in the presence of this DNA element.4 The transcriptional regulators Sp1 and Sp3 bind to a GC-rich element at position +51/+73 of the murine gene.4 Functional activity of the human counterpart is mediated by different mechanism(s): the most critical region of the human CD34 5′ UTR sequence appears to be occupied by the DNA-binding factor NFY. Mutating this site had a dramatic effect on the activity of the linked luciferase reporter gene. A 5-fold decrease in luciferase activity was observed, thus showing that the NFY site positively regulates the human CD34 promoter. Therefore, it appears that similar functional effects can be mediated by different factors acting on homologous regions. Interestingly, in both cases (human versus murine CD34 genes) the critical factors, Sp1/3 and NFY, have ubiquitous patterns of expression. Nevertheless, they are reported to regulate genes in cell type-specific fashion.
NFY (CBF) is a transcription factor composed of three subunits (NFYA, NFYB, and NFYC) that are highly conserved through evolution between yeast and mammals11-13 and all of which are necessary for DNA binding.28 NFY regulates a number of cell type-specific genes by binding to a CCAAT motif in their regulatory regions.13,44 Interaction of NFY with other transcription factors [for example, Sp1,45,46 C/EBP family members,47,48 SREBP-1,49,50 octamer binding factors,51 HMG-I(Y),52 PC414] might explain its cell type-specific mode of action. In addition, NFY activity was demonstrated to be modulated during serum starvation,53 depletion of intracellular calcium,54 differentiation,14-16 and proliferation.17 One characteristic feature of NFY is that its site occupies a relatively fixed position in the promoter, usually between base pairs −80 to −100.13,44 However, some exceptions to this rule exist.45,55 For example, very recently NFY was described to bind to the myeloperoxidase gene upstream enhancer and mediate G-CSF responsiveness.55 Here we show that NFY binds to a site located in the 5′ UTR. This is the first report on binding of NFY to such a region. One of the three subunits of NFY, NFYA, has multiple splicing forms that result in subtle changes in mobility of the NFY/DNA complex in EMSA.56 Therefore, the formation of double complexes in EMSA shown in Fig 2C and D most likely represents alternatively spliced forms of the NFYA subunit. Presumably, NFY enhances CD34 transcription, but posttranscriptional effects cannot be ruled out.
Probably the best studied targets of NFY are MHC class II genes. Binding of NFY to the promoters of these genes increases the DNA affinity of another critical transcription factor, RFX, to its cognate sites located at rather remote positions away from the NFY sites.57,58 Similarly, cooperative binding of Sp1 and NFY was shown on the proximal promoter region of the human MHC class II–associated invariant chain (Ii)45 and the rat fatty acid synthase promoter.46 Stabilization of NFY interaction with CCAAT box DNA by interacting factors, HMG-I(Y) and PC4, was also shown.14,52 In the former case, the interaction with HMG-I(Y) did not affect the mobility characteristics of the NFY/DNA complex in gel shift experiments.52 A similar mechanism of functional collaboration between NFY and c-myb might be involved: binding of NFY to its imperfect site in the 5′ UTR of CD34 was stabilized by the presence of an adjacent c-myb binding site.
How does NFY contribute to lineage-specific expression of CD34? Binding of this factor may be necessary to attract the basal transcription machinery along with cell type-restricted factors to the transcription start site. Alternatively, the major role of NFY could be that it binds to the +61/+65 site constitutively and, as a result of its association with histone acetyltransferases,59 it maintains the chromatin structure in an open configuration. This hypothesis is consistent with the observation of a ubiquitous DNaseI hypersensitive site at the transcription initiation site.7,60 However, efficient transcription of CD34 in stem cells may occur only in the presence of c-myb or other cell type-specific trans-acting factors. Both hypotheses are in agreement with the studies in which overexpression of c-myb activated the endogenous CD34 gene in CD34-cells.8 9
How is the CD34 gene expression abolished as the cells differentiate? NFY is widely expressed but the protein levels of one of its subunits, NFYA, are subject to cell-cycle dependent regulation.15 16Such fluctuations affect the DNA binding activity of NFY and, therefore, might allow for the access of the repressively acting factors to the promoter. Alternatively, the primary cause of downregulation of CD34 transcription might be differential chromatin methylation in the vicinity of the promoter, which might lead to displacement of the positively acting factors (NFY being one of them) from the cis regulatory elements.
ACKNOWLEDGMENT
The authors are thankful to Benoit de Crombrugghe, Sankar N. Maity, Roberto Mantovani, Linda Shapiro, Bruno Calabretta, Danilo Perrotti, Steve Jackson, Jeff Marx, Laurel Eckhardt, Margaret A. Shipp, and Robert Sackstein for providing reagents. We are also indebted to David Gonzalez for synthesis of oligonucleotides and Laura Smith, Martha Petrovick, Pu Zhang, and Atsushi Iwama for helpful discussions and suggestions.
Supported by fellowship award No. DK09721 (to H.S.R.) from the National Institutes of Health (NIH) and grants Nos. DK48660 (to D.G.T.) and 5-R01-53037 (to D.S.K.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Daniel G. Tenen, MD, Harvard Institutes of Medicine, Room 954, 77 Ave Louis Pasteur, Boston, MA 02115; e-mail:dtenen@caregroup.harvard.edu.

![Fig. 5. The NFY site positively regulates the human CD34 promoter and functionally interacts with c-myb binding site. (A) A comparison of oligonucleotide probes used in Fig 2 (+48/+80) and the +54/+89 sequence (Y+M+) shown previously to bind c-myb.89 NFY and c-myb sites are boxed. Arrows under the Y+M+ sequence indicate point mutations introduced into either the Y box or the c-myb site individually (to generate Y−M+ or Y+M− oligonucleotides) or to both sites simultaneously (to generate the Y−M− oligonucleotide). (B) EMSA with Y+M+ probe and no protein (lanes 1 and 7) or 20 μg of RPMI-8402 nuclear extract (N.E.; lanes 2 to 6 and 8 to 12). Reactions in lanes 3 and 4 contained antibodies raised to NFYA [NFYA(1) was from Rockland Immunochemicals and NFYA(2) was described29], and anti-NFYB antibody29 was added in lane 5. Competitors were added to the reactions as follows: CD10-derived NFY binding oligonucleotide41 to lane 6; +48/+80 region of human CD34 to lane 9; self-competitor (Y+M+) to lane 10; +54/+89 oligonucleotide with Y box mutated (Y−M+) to lane 11; +54/+89 oligonucleotide with both the Y box and c-myb site mutated (Y−M−) to lane 12. Reactions in lanes 13 and 14 contain Y−M+ probe incubated in the absence or presence of nuclear extract, respectively. The NFY binding complex is indicated by an arrow. Supershifted complexes (s.s.) and free probe are marked with brackets. Stars indicate nonspecific complexes. (C) A schematic drawing of constructs used in transient transfection assays shown in (D). All constructs contained the human CD34 promoter (base pairs −391/+175) with wild-type Y box and c-myb site (Y+M+), Y box mutated (Y−M+), c-myb site mutated (Y+M−), or both sites mutated (Y−M−). (D) CD34 promoter/luciferase constructs in (C) were transfected to KG1a cells together with an internal control plasmid, CMV-driven Renilla luciferase (pCVM-RL). The results are presented as mean firefly luciferase relative light units (FL RLU) and normalized to Renilla luciferase relative light units (RL RLU). Standard deviations are represented by error bars.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/11/10.1182_blood.v94.11.3772/4/m_blod42319005aw.jpeg?Expires=1766016084&Signature=eXEmARbC9lIqhvrpWiaua6tS8d-NyQIlLupkMiJ24onIkU~O5Ruw1m4eZkkasX9T3Iqq5ZjEpNoZ0okGS-pKv7ChEDNl4-Y5dNLfmIrlSh9P0DZczp09H-vqMkPVmwOJLDajDzRjfs8BgxnJQ4RoUwoll3xFVUGdCwHB4OH0Zu0yiSfIazAUZiv87vHfTvrEzZ5RBbRROFP6JQAvADUD86EwZ086XYZ8GEF2t7a4aqQXXCDshxUlHk4ZcE6L~I0gGnNJ-C0E-cWU1lkA8NgbD9H5wsCwGwUsM766lDfQdXM7JB9cNv5AMau25geJ3Ju8VilG~xXkRHL~jqawDBnLQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. The NFY site positively regulates the human CD34 promoter and functionally interacts with c-myb binding site. (A) A comparison of oligonucleotide probes used in Fig 2 (+48/+80) and the +54/+89 sequence (Y+M+) shown previously to bind c-myb.89 NFY and c-myb sites are boxed. Arrows under the Y+M+ sequence indicate point mutations introduced into either the Y box or the c-myb site individually (to generate Y−M+ or Y+M− oligonucleotides) or to both sites simultaneously (to generate the Y−M− oligonucleotide). (B) EMSA with Y+M+ probe and no protein (lanes 1 and 7) or 20 μg of RPMI-8402 nuclear extract (N.E.; lanes 2 to 6 and 8 to 12). Reactions in lanes 3 and 4 contained antibodies raised to NFYA [NFYA(1) was from Rockland Immunochemicals and NFYA(2) was described29], and anti-NFYB antibody29 was added in lane 5. Competitors were added to the reactions as follows: CD10-derived NFY binding oligonucleotide41 to lane 6; +48/+80 region of human CD34 to lane 9; self-competitor (Y+M+) to lane 10; +54/+89 oligonucleotide with Y box mutated (Y−M+) to lane 11; +54/+89 oligonucleotide with both the Y box and c-myb site mutated (Y−M−) to lane 12. Reactions in lanes 13 and 14 contain Y−M+ probe incubated in the absence or presence of nuclear extract, respectively. The NFY binding complex is indicated by an arrow. Supershifted complexes (s.s.) and free probe are marked with brackets. Stars indicate nonspecific complexes. (C) A schematic drawing of constructs used in transient transfection assays shown in (D). All constructs contained the human CD34 promoter (base pairs −391/+175) with wild-type Y box and c-myb site (Y+M+), Y box mutated (Y−M+), c-myb site mutated (Y+M−), or both sites mutated (Y−M−). (D) CD34 promoter/luciferase constructs in (C) were transfected to KG1a cells together with an internal control plasmid, CMV-driven Renilla luciferase (pCVM-RL). The results are presented as mean firefly luciferase relative light units (FL RLU) and normalized to Renilla luciferase relative light units (RL RLU). Standard deviations are represented by error bars.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/11/10.1182_blood.v94.11.3772/4/m_blod42319005cx.jpeg?Expires=1766016084&Signature=sYH47y9BHHHPM2vr8OBE3nFQT2Y8nkrIW0HL7OzCyYh~48wTfeKNdhMyHtS2Qi-WJbDyz5pBxazxORarG9gm3ijXuUG3NxGu9LlsQOUY~SNDYgN5iSFhfKvdB53ycxEEhgqgMmQpwsCLE-2qbb36B0p-BpvBxSJiazvTWQgCxmix1YuJ1zu0TnEycNcIKR0QWBrgOuhxMpICUziUQG4pueOHvIGkOIjmOHellQas3r7vXyHjPEOIHZ3~n7ZYL01l8a8xvyz1Lz2C0kkthEDmYxRr644-7tonbku2KwEjM3O4ysvOzXkUm~JgLq~2drJp3EiA7FH2LyPA0B~pTTkmqg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Fig. 5. The NFY site positively regulates the human CD34 promoter and functionally interacts with c-myb binding site. (A) A comparison of oligonucleotide probes used in Fig 2 (+48/+80) and the +54/+89 sequence (Y+M+) shown previously to bind c-myb.89 NFY and c-myb sites are boxed. Arrows under the Y+M+ sequence indicate point mutations introduced into either the Y box or the c-myb site individually (to generate Y−M+ or Y+M− oligonucleotides) or to both sites simultaneously (to generate the Y−M− oligonucleotide). (B) EMSA with Y+M+ probe and no protein (lanes 1 and 7) or 20 μg of RPMI-8402 nuclear extract (N.E.; lanes 2 to 6 and 8 to 12). Reactions in lanes 3 and 4 contained antibodies raised to NFYA [NFYA(1) was from Rockland Immunochemicals and NFYA(2) was described29], and anti-NFYB antibody29 was added in lane 5. Competitors were added to the reactions as follows: CD10-derived NFY binding oligonucleotide41 to lane 6; +48/+80 region of human CD34 to lane 9; self-competitor (Y+M+) to lane 10; +54/+89 oligonucleotide with Y box mutated (Y−M+) to lane 11; +54/+89 oligonucleotide with both the Y box and c-myb site mutated (Y−M−) to lane 12. Reactions in lanes 13 and 14 contain Y−M+ probe incubated in the absence or presence of nuclear extract, respectively. The NFY binding complex is indicated by an arrow. Supershifted complexes (s.s.) and free probe are marked with brackets. Stars indicate nonspecific complexes. (C) A schematic drawing of constructs used in transient transfection assays shown in (D). All constructs contained the human CD34 promoter (base pairs −391/+175) with wild-type Y box and c-myb site (Y+M+), Y box mutated (Y−M+), c-myb site mutated (Y+M−), or both sites mutated (Y−M−). (D) CD34 promoter/luciferase constructs in (C) were transfected to KG1a cells together with an internal control plasmid, CMV-driven Renilla luciferase (pCVM-RL). The results are presented as mean firefly luciferase relative light units (FL RLU) and normalized to Renilla luciferase relative light units (RL RLU). Standard deviations are represented by error bars.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/11/10.1182_blood.v94.11.3772/4/m_blod42319005aw.jpeg?Expires=1766254056&Signature=H4OCgtsa6nSu62HRozJBJXOU78CfEyc1a6sORJSlkdDt7yxmohma6slPujozEVqpKU856qnYFxmucVkIzmzIyvNsIBc0SifyQdlzNDGTm5F4Rm~bfnvccupoSVPBOxzfilmPoFzsr3nW5bvyB1VpKufNX8ddNB5p6~5n7bb03DbWghX1~3-th67LT6QlpJfJhXxws08708ZnXALfPgrLG1OKCi7dbDWQm3qwq11SGabEMDmHQQBqfLpzOhjwtAekAXD-tPbhAALtWS6IarVxBnrsJP77HWkNaG62UWGl2D3wCeaoRW~VQ1x2RMbovF3DEOrtSXFo4jWbwJ0DkZe1CQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. The NFY site positively regulates the human CD34 promoter and functionally interacts with c-myb binding site. (A) A comparison of oligonucleotide probes used in Fig 2 (+48/+80) and the +54/+89 sequence (Y+M+) shown previously to bind c-myb.89 NFY and c-myb sites are boxed. Arrows under the Y+M+ sequence indicate point mutations introduced into either the Y box or the c-myb site individually (to generate Y−M+ or Y+M− oligonucleotides) or to both sites simultaneously (to generate the Y−M− oligonucleotide). (B) EMSA with Y+M+ probe and no protein (lanes 1 and 7) or 20 μg of RPMI-8402 nuclear extract (N.E.; lanes 2 to 6 and 8 to 12). Reactions in lanes 3 and 4 contained antibodies raised to NFYA [NFYA(1) was from Rockland Immunochemicals and NFYA(2) was described29], and anti-NFYB antibody29 was added in lane 5. Competitors were added to the reactions as follows: CD10-derived NFY binding oligonucleotide41 to lane 6; +48/+80 region of human CD34 to lane 9; self-competitor (Y+M+) to lane 10; +54/+89 oligonucleotide with Y box mutated (Y−M+) to lane 11; +54/+89 oligonucleotide with both the Y box and c-myb site mutated (Y−M−) to lane 12. Reactions in lanes 13 and 14 contain Y−M+ probe incubated in the absence or presence of nuclear extract, respectively. The NFY binding complex is indicated by an arrow. Supershifted complexes (s.s.) and free probe are marked with brackets. Stars indicate nonspecific complexes. (C) A schematic drawing of constructs used in transient transfection assays shown in (D). All constructs contained the human CD34 promoter (base pairs −391/+175) with wild-type Y box and c-myb site (Y+M+), Y box mutated (Y−M+), c-myb site mutated (Y+M−), or both sites mutated (Y−M−). (D) CD34 promoter/luciferase constructs in (C) were transfected to KG1a cells together with an internal control plasmid, CMV-driven Renilla luciferase (pCVM-RL). The results are presented as mean firefly luciferase relative light units (FL RLU) and normalized to Renilla luciferase relative light units (RL RLU). Standard deviations are represented by error bars.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/11/10.1182_blood.v94.11.3772/4/m_blod42319005cx.jpeg?Expires=1766254056&Signature=3rjLQ5uAii-7vmK9I~KUhWNfHX~UtbZI72-sYyEbMdrEdxwdNLTnqWQx8ckGIFGNGd77wGRWfi4DN4IZiNEPphECxTlCuQTqTvdihzh7FrgLmoOC0ifmnMm6FmPCqSbM6334JRMqPpV68-xpciK9C3dyvu6CkylzsoYIRN6MFxgbNyEfzibe-94ZYETcJXD07-Us8wtrIKXumiiYmswhzKdNQkhaKOSTr7UHQMU1HbwBPuQMyiSSaZQty2ri8V-lRVukoLgNFwCFj8KSlkmefwLomUHA~UVMZdVrhOf352tg7I2~oMKuMMciVdsfHrM9knfdUC~~NS3ZALByQTkgQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)