Abstract
In immortalized cells of the erythroid lineage, the iron-regulatory protein (IRP) has been suggested to coregulate biosynthesis of the iron storage protein ferritin and the erythroid delta-aminolevulinate synthase (eALAS), a key enzyme in heme production. Under iron scarcity, IRP binds to an iron-responsive element (IRE) located in ferritin and eALAS mRNA leaders, causing a block of translation. In contrast, IRP-IRE interaction is reduced under high iron conditions, allowing efficient translation. We show here that primary chicken erythroblasts (ebls) proliferating or differentiating in culture use a drastically different regulation of iron metabolism. Independently of iron administration, ferritin H (ferH) chain mRNA translation was massively decreased, whereas eALAS transcripts remained constitutively associated with polyribosomes, indicating efficient translation. Variations in iron supply had minor but significant effects on eALAS mRNA polysome recruitment but failed to modulate IRP-affinity to the ferH-IRE in vitro. However, leukemic ebls transformed by the v-ErbA/v-ErbB–expressing avian erythroblastosis virus showed an iron-dependent reduction of IRP mRNA-binding activity, resulting in mobilization of ferH mRNA into polysomes. Hence, we analyzed a panel of ebls overexpressing v-ErbA and/or v-ErbB oncoproteins as well as the respective normal cellular homologues (c-ErbA/TR, c-ErbB/EGFR). It turned out that v-ErbA, a mutated class II nuclear hormone receptor that arrests erythroid differentiation, caused the change in ferH mRNA translation. Accordingly, inhibition of v-ErbA function in these leukemic ebls led to a switch from iron-responsive to iron-independent ferH expression.
IRON IS AN ESSENTIAL nutrient required by every proliferating cell for the function of multiple iron-containing enzymes and hemoproteins.1,2 Because free iron is a potential source for harmful oxygen radicals, rapid incorporation of intracellular iron into proteins is crucial to keep the element in a nontoxic form.3 Excess iron is detoxified by sequestration into the iron storage protein ferritin, a multimer composed of 24 ferritin heavy (ferH) and light (ferL) subunits.4 5
A generally accepted standard model has emerged to describe how different tissues and established erythroleukemic cell lines control homeostasis of iron internalized via transferrin receptor (TfR)-mediated endocytosis. This model supposes that the expression of TfR, ferritin, and the erythroid delta-aminolevulinate synthase (eALAS), a rate-limiting enzyme in heme biosynthesis exclusively present in erythroid cells, is coordinately regulated.6-8Control is mainly exerted by the iron-regulatory protein 1 (IRP1),9-12 a trans-acting factor binding to stem-loop structures in cognate mRNAs, termed iron-responsive elements (IREs).13 A second isoform of similar function, IRP2, has been described, which is ubiquitously expressed, with highest levels found in neuronal tissues.14 At low intracellular iron levels, IRPs interact with multiple IREs within the 3′-untranslated region (UTR) of TfR mRNA, thereby strongly increasing mRNA half-life and TfR protein expression at the cell surface.15,16 Concurrently, the binding of IRPs to IREs in the 5′-UTR of ferritin and eALAS transcripts17-20interferes with the assembly of functional 80S translation initiation complexes and prevents protein synthesis.21 22 In contrast, high iron abundance diminishes IRP-affinity to IREs, resulting in rapid TfR mRNA degradation and efficient translation of ferritin and eALAS transcripts. However, this model fails to explain how primary erythroblasts (ebls) undergoing terminal differentiation cope with the paradoxical situation that low IRP-activity, allowing efficient eALAS biosynthesis should also favor ferritin production, thereby forming a futile iron storage at times of massive iron demand for heme biosynthesis.
Until recently, studies on iron metabolism during erythropoiesis could only be performed in erythroleukemic cell lines.19,20,23All of these transformed cell types share common drawbacks with respect to erythroid differentiation: they (1) require nonphysiological stimuli to initiate differentiation, (2) mature incompletely or aberrantly (insufficient hemoglobin accumulation, abnormal morphology, lack of enucleation24), and (3) exhibit nontypical cell cycle control during differentiation.25 These erythroleukemic cells show regulation of iron metabolism according to the standard model.26 However, their abnormalities allow no reliable conclusions on the regulation of iron metabolism upon maturation of primary ebls.
Recently, culture conditions were established allowing the mass cultivation of erythroid progenitors derived from chick bone marrow. After a limited period of factor-dependent proliferation, termed self-renewal, these cells can be induced to highly synchronous differentiation, resulting in cells virtually indistinguishable from erythrocytes in peripheral blood.27,28 Hence, this system appeared ideally suited to study the regulation of iron utilization and storage under in vivo-like conditions. Moreover, transformation of these bone marrow-derived ebls with the avian erythroblastosis virus (AEV)-generated immortalized cell lines (eg, HD3)29 30 as well as mortal clones of AEV-infected ebls. These cell types allowed us to address the question of whether there are differences in the regulation of iron metabolism between normal versus transformed erythroid cells.
We show here that primary ebls exhibit a mode of iron-dependent gene regulation clearly distinct from the standard model. Under low as well as high exogenous iron supply, ferH mRNA remained essentially absent from polyribosomes and was translated at low levels, whereas the coexpressed eALAS transcripts were mainly bound to polysomes under both conditions. These differences in the mode of ferH versus eALAS mRNA translation could not be observed in HD3 leukemic cells. Extensive analysis showed that v-ErbA, 1 of the 2 oncoproteins expressed by AEV, induced and maintained regulation of iron metabolism according to the standard model in primary ebls. The idea that v-ErbA was sufficient for this molecular phenotype was supported by functional inactivation of this oncoprotein in AEV-transformed ebls: loss of v-ErbA activity resulted in a reversal to iron-independent ferritin production.
MATERIALS AND METHODS
Cell culture and retroviral infection.
The chicken hepatoma cell line LMH-2A was grown in Waymouth medium supplemented with 10% fetal calf serum (FCS), 2 mmol/L L-glutamine, and antibiotics at 37°C and 5% CO2. Tissue culture plates were coated with 0.1% gelatine 4 hours before use. Human HeLa cervical carcinoma cells were grown in Dulbecco's modified Eagle's medium (DMEM) plus 10% FCS and antibiotics.
Primary chicken erythroid progenitors were isolated and cultivated as described earlier,25,27,28,31 starting from 2 × 108 normal bone marrow cells of 3- to 7-day-old Spafas chicks. Briefly, 5 × 107 purified cells were plated into 10 mL modified colony-forming unit-erythrocyte (CFU-E) medium32 supplemented with 100 ng/mL recombinant avian stem cell factor (SCF)31 and 40 ng/mL recombinant human insulin-like growth factor (IGF-1; Sigma, St Louis, MO). These so-called SCF-progenitors were expanded at 37°C and 5% CO2 to numbers between 2 × 108and 5 × 108. Before harvesting or induction of differentiation, spontaneously maturing erythroid cells were removed by Percoll purification.
To induce terminal differentiation of 3- to 4-day-old SCF-progenitors, self-renewing cells were washed twice with serum-free DMEM and cultured in differentiation medium for 4 days, maintaining a density between 2 and 3 × 106 cells/mL. The differentiation medium represents a modified CFU-E medium without chicken serum (Epotest medium) supplemented with 1.4 nmol/L insulin and 3% anemic chicken serum as a source of avian erythropoietin (Epo).31
To infect primary ebls, retrovirus-producing chicken embryo fibroblasts (CEFs) were generated by cotransfection of the retroviral construct carrying the gene of interest with helper virus DNA and G418 selection.33 Subsequently, freshly prepared chick bone marrow cells were cocultivated with mitomycin-C–treated CEFs producing the particular recombinant retroviruses, essentially as described.34
Ebls expressing v-ErbA alone or in combination with the murine Epo receptor (EpoR)35 were expanded under self-renewing conditions in CFU-E medium supplemented with 100 ng/mL avian SCF, 40 ng/mL human IGF-1, 1 μmol/L estradiol (E2), and 3 μmol/L of the glucocorticoid-antagonist ZK 112993.36 In case of c-ErbA expression on its own or plus exogenous murine EpoR, the same factors were added; however, the CFU-E medium was stripped of endogenous thyroid hormone and retinoids.37 ts34A2-ebls, expressing a temperature-sensitive mutant of the v-ErbB oncoprotein at 37°C,38,39 were kept for proliferation in CFU-E medium containing 100 ng/mL avian SCF, 40 ng/mL human IGF-1, 1 μmol/L E2, and 1 μmol/L dexamethasone (Dex), whereas CER-2 ebls expressing c-ErbB40 were additionally supplemented with 20 ng/mL transforming growth factor α (TGFα; Promega, Madison, WI).
All AEV-transformed ebls, ie, the primary clone ts34A6,41the established cell line HD3E11,42 and the cell line HD3E11 ectopically expressing the murine EpoR, termed HD3E22,43 were cultured under proliferation conditions in either standard growth medium consisting of DMEM supplemented with 8% FCS, 2% chicken serum, 10 mmol/L HEPES, pH 7.2, and antibiotics or in Epotest medium plus 40 ng/mL human IGF-1. Differentiation of HD3E22 ebls was induced by incubation in differentiation medium supplemented with 2 U/mL recombinant human Epo, 1.4 nmol/L insulin, 1 μmol/L of the estrogen antagonist ICI 164384, 1 μmol/L ZK 112993, the ErbB inhibitor PD 153035,44 and, where appropriate, with the c-Kit antagonist EXBW 50 (a kind gift of Boehringer Ingelheim, Ingelheim, Germany; H. Beug, unpublished data). For all types of ebls, cell size and number were monitored daily in an electronic cell analyzer (CASY; Schärfe-System, Reutlingen, Germany).
To achieve iron deprivation, cells were incubated for 24 hours with 50 μmol/L of the specific iron chelator desferrioxamine (Des). High iron supply was accomplished by supplementing culture media with either 1 mg/mL iron-saturated chicken ovotransferrin (Tf, conalbumin; Sigma) or 50 μg/mL ferric ammonium citrate (FeCit).
Assays for differentiation parameters.
Cell populations induced to differentiate were daily analyzed for their stage of maturation by photometrical determination of hemoglobin concentrations as well as by cytocentrifugation onto slides and subsequent staining with neutral benzidine blue plus histological dyes as described previously.25
Cloning of chicken ferH IRE cDNA.
By using the forward primer 5′-GTC GAA TTC CAG AGC GCG TCG GCG AGG CTG-3′ and the reverse primer 5′-GGT TGG TCA CAT GGT CAC CCA GCT GC-3′, both derived from the genomic chicken ferritin H subunit sequence,45 a 597-bp DNA fragment (EcoRI-BstEII) was amplified from total chicken liver RNA by reverse transcription-polymerase chain reaction (RT-PCR). The sequence of the isolated PCR product was 100% identical to the cDNA sequence predicted from the chicken ferH gene, spanning a region from the transcription start site in exon I up to a single BstEII restriction site in exon IV. This EcoRI-BstEII ferH cDNA fragment was ligated into a pGEM-T vector (Promega) and termed pGEM-cferH597. Religation after removal of an Apa I fragment of pGEM-cferH597 (comprising the C-terminal part of exon I and exon II-IV) resulted in the plasmid pGEM-cferH-IRE, which carried 78 bp of the 5′-UTR (5′-CAGAGCGCGT CGGCGAGGCT GAGCGGAGCG GGTTCCTGCG TCAACAGTGC TTGGACGGAA CCGGCCGCGC TCGGGCCC-3′), including the entire IRE stem-loop structure; the characteristic terminal hexaloop-sequence and the unpaired bulged C-residue in the stem are underlined.
The 44 bp of the 5′-UTR of chicken eALAS cDNA (5′-ATTCCAGTGC GTTCGTCCTC AGCGCGGGGC AACGGCACAG GACG-3′) containing the entire IRE hairpin motif according to the published sequence46 was produced by oligonucleotide synthesis, inserted via BamHI linkers into pGEM3Zf(−) (Promega), resulting in the plasmid pGEM-eALAS-IRE, and verified by sequencing.
Sucrose gradient analysis.
The distribution of ferH chain and eALAS mRNA between ribosome-free and polyribosome-associated compartments was determined by separating cytoplasmic extracts from 2 × 107 LMH-2A hepatoma cells or 4 to 5 × 107 ebls in linear 15% to 40% sucrose gradients as described.47,48 Each gradient was harvested into 18 fractions and the extracted RNA analyzed by electrophoresis on denaturing 1% formal dehyde agarose gels and subsequent Northern blotting. After UV-cross-linking of RNA to the nylon membranes, the distribution of 18S and 28S rRNA was visualized by staining of filters with methylene blue. The membranes were sequentially hybridized with a random primed 32P-labeled 0.45-kb fragment (Nco I-BstEII) of the chicken ferH chain cDNA and a 1.4-kb fragment (Pst I-Pst I) of the chicken eALAS cDNA.49 Signals on the autoradiographs were scanned and quantified with a laser densitometer (Molecular Dynamics, Sunnyvale, CA).
Western blotting.
The harvested cells were washed in ice-cold phosphate-buffered saline and lysed in an isotonic buffer containing 10 mmol/L HEPES, pH 7.6, 40 mmol/L KCl, 5% glycerol, 1 mmol/L dithiotreitol, and 0.2% of the nonionic detergent NP40.48 Protein concentrations were determined by a Coomassie Brilliant Blue assay (Bio-Rad, Hercules, CA). Ten to 20 μg of cell extract from each sample was separated in sodium dodecyl sulfate (SDS) polyacrylamide gels and transferred to nitrocellulose (Protran; Schleicher & Schuell, Keene, NH). Reversible staining of the membranes with acidic Ponceau S solution (Merck, Darmstadt, Germany) was used to visualize proper protein transfer. After blocking with 5% low fat dry milk, the filters were probed with the respective antisera: antihorse spleen ferritin, raised in rabbit by using the affinity isolated antigen (Sigma); antirat IRP1 (a kind gift of R.S. Eisenstein)50; or antirat IRP2 (a kind gift of E.A. Leibold).12 Immunoreactive signals were detected by enhanced chemoluminescence (Amersham, Uppsala, Sweden) and quantified by laser densitometry.
Analysis of IRP protein abundance and mRNA binding activity.
The affinity of IRPs to the cis-acting palindromic IRE sequence in chicken ferH mRNA was analyzed by gel retardation assays essentially as described previously.48 51 The preparation of cytoplasmic extracts and quantitation of protein concentrations were performed as outlined above for Western blotting.32P-radiolabeled in vitro transcripts were produced by T7 RNA polymerase after linearization of the plasmid pGEM-cferH-IRE withApa I. A total of 1.3 × 106 dpm (∼1.0 ng) of labeled IRE-containing in vitro transcript was incubated with 2 μg of protein for 30 minutes at room temperature and subsequently treated with RNAse T1 and heparin. RNA-protein complexes were visualized after separation in 6% nondenaturing polyacrylamide gels that were fixed by drying onto a DEAE ion exchange paper and subjected to autoradiography. Signals corresponding to IRP-IRE complexes were quantified by phosphoimaging (Molecular Dynamics).
To analyze IRP1 protein abundance, extracts were incubated for 30 minutes with 2% 2-mercaptoethanol before the assay to reduce IRP1 for the determination of total RNA binding activity.51
For antibody-mediated supershift experiments, extracts were preincubated with 1 or 3 μL of the specific rat IRP1 or IRP2 antisera or an unspecific antibody (IgG) for 2 hours at 4°C before the binding reaction with 32P-labeled chicken ferH-IREs.
RESULTS
Iron-dependent translational control of ferritin and eALAS mRNAs in erythroleukemic and hepatoma cell lines.
Both ferH and eALAS mRNAs harbor single palindromic IREs in their leader region and thus are considered as targets for iron-dependent mRNA binding of IRP.6 However, previous studies focused either exclusively on the IRP-interaction with the mRNAs of ferritin subunits in different cell lines and tissues or on IRP-complex formation with eALAS mRNA in erythroleukemic cells.17-20Therefore, we examined the potential of IRP to actually coregulate translation initiation of ferH and eALAS transcripts. For this analysis, we initially used chicken erythroblasts (ebls) transformed by the cooperating AEV oncoproteins v-ErbB and v-ErbA and expressing the murine Epo receptor (EpoR) to allow terminal differentiation of these cells (termed HD3E22)43 with recombinant factors. Sucrose gradient analysis was performed with cells either iron-depleted by exposure to the specific chelator desferrioxamine (Des) or treated with physiologically high levels of iron-saturated transferrin (Tf; 1 mg/mL; L. Lobmayr, unpublished data), the physiological iron source.2 By this technique, untranslated mRNPs (<80S region) are separated from polyribosome-associated mRNAs, which are engaged in protein synthesis and sediment further down into the gradient.47
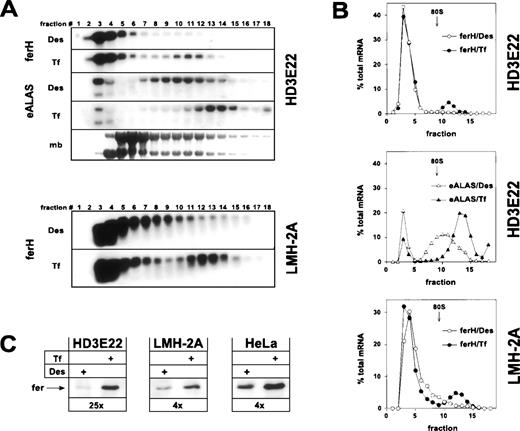
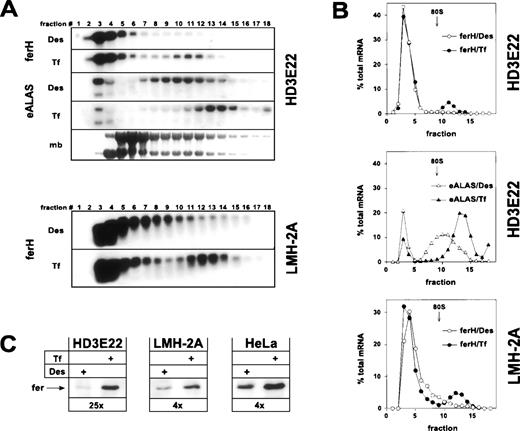
As expected, ferH mRNA was restricted to the ribosome-free pool (fractions 1-9) in iron-depleted HD3E22 ebls, whereas a significant mobilization into the polysome-bound compartment was observed under high Tf concentrations (Fig 1A and B). This shift was comparable to the one in exponentially growing chicken hepatoma cells (LMH-2A). It should be noted that differences in iron concentrations did not affect the overall ferH mRNA abundance in these cells. Similar results were obtained in primary chicken embryo fibroblasts (data not shown) and in previous studies on ferL mRNA translation in human HeLa cervical carcinoma cells.48 This indicated that the relative proportion of ferH mRNA bound to polysomes at high iron concentrations (10% to 20%) was comparable in these different cell types. To our surprise, already in iron-deprived HD3E22 ebls, a significant portion of eALAS transcripts was found in polysome fractions. However, the eALAS transcripts shifted onto even larger polysomes upon administration of high Tf levels, suggesting iron-dependent translational control to a certain extent (Fig 1A and B).
Iron-dependent translational control of ferH and eALAS mRNA in LMH-2A hepatoma cells and AEV-transformed ebls (HD3E22). Cells were incubated with the iron chelator desferrioxamine (DES; 50 μmol/L) or iron-loaded transferrin (TF; 1 mg/mL iron-saturated chicken ovotransferrin) for 24 hours before harvesting. (A) Cytoplasmic extracts were separated in linear 15% to 40% sucrose gradients47 and the RNA was isolated from 18 fractions analyzed by Northern blotting. Fraction 1 corresponds to the top and fraction 18 to the bottom of the gradient. The filters were hybridized with 32P-labeled probes specific for chicken ferH and, in case of HD3E22 ebls, subsequently with a chicken eALAS cDNA.49 The lowest panel displays the profile obtained by staining of the RNA with methylene blue (mb). The emergence of a constant molar ratio between 28S and 18S RNA (upper and lower band, respectively) around fraction 9 indicates the assembly of 80S initiation complexes and marks the boundary between the ribosome-free and polyribosome-bound compartment. Signals between gradients from the same cell type are directly comparable, because (1) the same aliquot of RNA from each fraction was blotted, (2) both filters were hybridized with the very same probe, and (3) exposed for the same time. Thus, the sum of signals over all the fractions corresponds to total cytoplasmic mRNA levels. (B) Quantitative analysis of hybridization signals from Northern blotting by laser densitometry. (Open symbols) Cells depleted of iron by Des; (solid symbols) cells under high Tf. To facilitate comparison between different experiments, the RNA content of each fraction was expressed as a percentage of the total amount of RNA contained in the gradient. (C) Ferritin protein expression in HD3E22, LMH-2A, and HeLa cells was determined by Western blotting as described in Materials and Methods. Numbers below each pair of lanes represent the factors of iron-dependent upregulation of ferritin expression (fold induction).
Iron-dependent translational control of ferH and eALAS mRNA in LMH-2A hepatoma cells and AEV-transformed ebls (HD3E22). Cells were incubated with the iron chelator desferrioxamine (DES; 50 μmol/L) or iron-loaded transferrin (TF; 1 mg/mL iron-saturated chicken ovotransferrin) for 24 hours before harvesting. (A) Cytoplasmic extracts were separated in linear 15% to 40% sucrose gradients47 and the RNA was isolated from 18 fractions analyzed by Northern blotting. Fraction 1 corresponds to the top and fraction 18 to the bottom of the gradient. The filters were hybridized with 32P-labeled probes specific for chicken ferH and, in case of HD3E22 ebls, subsequently with a chicken eALAS cDNA.49 The lowest panel displays the profile obtained by staining of the RNA with methylene blue (mb). The emergence of a constant molar ratio between 28S and 18S RNA (upper and lower band, respectively) around fraction 9 indicates the assembly of 80S initiation complexes and marks the boundary between the ribosome-free and polyribosome-bound compartment. Signals between gradients from the same cell type are directly comparable, because (1) the same aliquot of RNA from each fraction was blotted, (2) both filters were hybridized with the very same probe, and (3) exposed for the same time. Thus, the sum of signals over all the fractions corresponds to total cytoplasmic mRNA levels. (B) Quantitative analysis of hybridization signals from Northern blotting by laser densitometry. (Open symbols) Cells depleted of iron by Des; (solid symbols) cells under high Tf. To facilitate comparison between different experiments, the RNA content of each fraction was expressed as a percentage of the total amount of RNA contained in the gradient. (C) Ferritin protein expression in HD3E22, LMH-2A, and HeLa cells was determined by Western blotting as described in Materials and Methods. Numbers below each pair of lanes represent the factors of iron-dependent upregulation of ferritin expression (fold induction).
Concomitant with the iron-induced translocation of ferH mRNA from the ribosome-free to the polysome-associated compartment, we observed a regulation at the level of ferritin protein. This effect was strongest in HD3E22 ebls, where a 25-fold increase of ferritin abundance was found under high iron (Fig 1C; see also, Fig 3). A significant but more moderate change in ferritin levels (4×) occurred in LMH-2A (Fig1C), because these cells retained some more ferH mRNA in the ribosome-containing fractions (9-11) under iron depletion as compared with HD3E22 (Fig 1A and B). Two other AEV-transformed ebls, both lacking the murine EpoR, ie, the established cell line HD3E1142 and the clonal strain ts34A6,41yielded comparable results as obtained in HD3E22 cells (Table 1). These results are in line with the expected coregulation of ferH and eALAS mRNA, although the fraction of polysome-associated eALAS mRNA in the iron-deprived HD3E22 ebls was surprisingly high.
Translation of ferH mRNA is impaired at high iron concentrations in primary erythroblasts.
Erythroleukemic cell lines differentiate aberrantly and are defective in mature erythrocyte functions. Therefore, we became interested to investigate the translational control of ferH and eALAS transcripts in primary ebls, in which coregulation of these mRNAs would be functionally neutral or disadvantageous and thus should be futile. The erythroid progenitors used for this analysis were either maintained in the proliferative stage by stem cell factor (SCF; SCF-ebls) or induced to synchronous differentiation by Epo and insulin.30 During 96 hours of maturation, the cells increased their hemoglobin content to the level attained in peripheral blood erythrocytes, drastically reduced their cell volume, and showed a typical erythrocyte morphology after cytocentrifugation and staining to visualize hemoglobin (data not shown).25 As in the preceding experiments, cytoplasmic cell extracts were fractionated in sucrose gradients to monitor the ribosomal recruitment of iron-responsive target mRNAs.
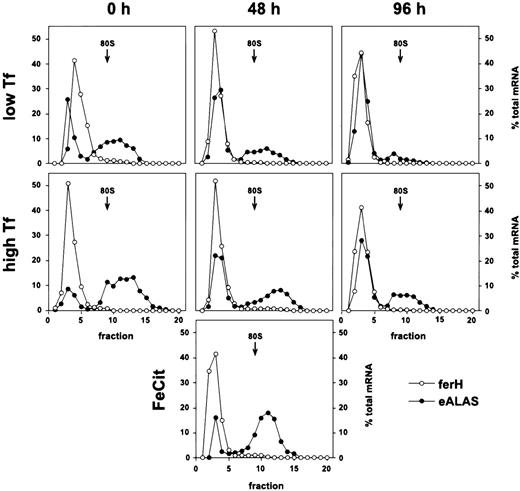
In sharp contrast to the standard model, ferH mRNA remained almost exclusively ribosome-free in these cells under low as well as high Tf supply, during self-renewal and after 48 or 96 hours of terminal differentiation (Fig 2). Moreover, this translational inhibition was independent of the iron source: even under high concentrations of ferric ammonium citrate, ferH transcripts were not translated more efficiently (48 hours differentiating SCF-ebls; Fig2, lowest panel). This phenotype suggested that the translation of ferH transcripts should be diminished, independently of iron abundance and progress into erythroid maturation.
Iron-independent reduction of ferH mRNA translation in proliferating and differentiating primary ebls. The SCF-dependent erythroid progenitors were grown from chicken bone marrow as described.25 Maturation was induced by removal of self-renewal factors (SCF and IGF-1) and addition of avian Epo (high-titer anemic serum) and insulin to the medium (see Materials and Methods). Self-renewing SCF-ebls and cells induced to differentiate for 48 or 96 hours were treated with low (0.03 mg/mL) or high levels of iron-saturated Tf (1 mg/mL) or high ferric ammonium citrate concentrations (FeCit; 50 μg/mL, lowest panel at 48 hours) for 24 hours. Sucrose gradient analysis and densitometrical quantitation were performed as described in the legend to Fig 1. (○) ferH mRNA; (•) eALAS mRNA.
Iron-independent reduction of ferH mRNA translation in proliferating and differentiating primary ebls. The SCF-dependent erythroid progenitors were grown from chicken bone marrow as described.25 Maturation was induced by removal of self-renewal factors (SCF and IGF-1) and addition of avian Epo (high-titer anemic serum) and insulin to the medium (see Materials and Methods). Self-renewing SCF-ebls and cells induced to differentiate for 48 or 96 hours were treated with low (0.03 mg/mL) or high levels of iron-saturated Tf (1 mg/mL) or high ferric ammonium citrate concentrations (FeCit; 50 μg/mL, lowest panel at 48 hours) for 24 hours. Sucrose gradient analysis and densitometrical quantitation were performed as described in the legend to Fig 1. (○) ferH mRNA; (•) eALAS mRNA.
E-ALAS mRNA was strongly bound to polyribosomes at low iron levels, and a moderate mobilization into larger polyribosomes was observed in response to high iron levels. This was true for proliferating cells as well as for those induced to differentiate for 48 and 96 hours. Thus, the ability for iron-dependent translational control of eALAS mRNA was maintained in primary SCF-ebls, comparable to the observations in transformed HD3 ebls (see Fig 2).
Taken together, these results indicated the existence of an iron-insensitive mechanism specific for the inhibition of ferH protein synthesis, which did not affect translation of eALAS transcripts. The restriction of ferH mRNA to the ribosome-free compartment correlated well with the lack of iron-dependent regulation of ferritin protein levels in normal SCF-ebls, although protein expression was not entirely eliminated (Fig 3B). Thus, ferH as well as eALAS mRNAs were differently used in committed erythroid cells, although their 5′-UTRs contain functional IREs, which in theory should have resulted in iron-responsive coregulation. In the following, the mechanism governing the differential translational control of ferH versus eALAS mRNA will be referred to as the erythroid mode of regulation of iron metabolism.
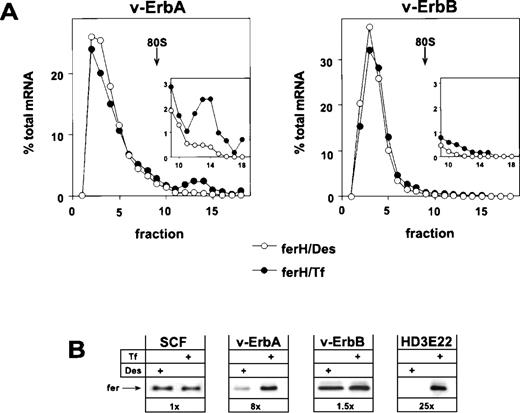
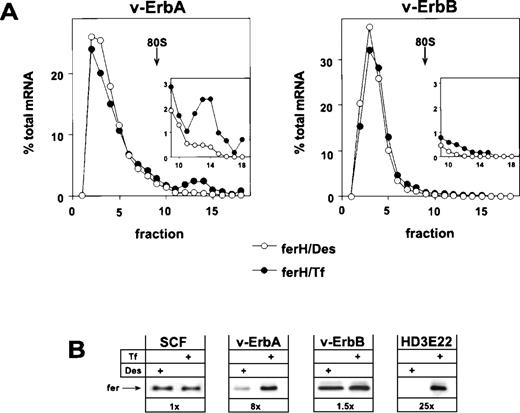
Retroviral expression of v-ErbA or v-ErbB in primary ebls and the consequences for ferritin biosynthesis. Bone marrow-derived, primary ebls expressing either the v-ErbA or the v-ErbB oncoprotein (see Materials and Methods) were kept for 24 hours under self-renewing conditions in the presence of 50 μmol/L Des to achieve iron depletion or were incubated with 1 mg/mL Tf to induce full iron saturation. (A) Distribution of ferH mRNA as determined by sucrose gradient analysis (for details, see legend to Fig 1). (Insets) The ribosome-bound mRNA compartment (fractions 9-18) is shown with an extended ordinate to highlight the difference between v-ErbA and v-ErbB expressing cells. (Open symbols) Des; (solid symbols) Tf. (B) Ferritin protein levels as detected by Western blotting in self-renewing primary ebls (SCF), v-ErbA– or v-ErbB–expressing ebls (v-ErbA, v-ErbB), and AEV-transformed red blood cells (HD3E22) under iron scarcity (Des) or high iron abundance (Tf). In case of SCF-progenitors, low Tf (0.03 mg/mL) instead of Des was used to induce IRP-activity. Numbers below each pair of lanes represent the factors of iron-dependent upregulation of ferritin expression (fold induction).
Retroviral expression of v-ErbA or v-ErbB in primary ebls and the consequences for ferritin biosynthesis. Bone marrow-derived, primary ebls expressing either the v-ErbA or the v-ErbB oncoprotein (see Materials and Methods) were kept for 24 hours under self-renewing conditions in the presence of 50 μmol/L Des to achieve iron depletion or were incubated with 1 mg/mL Tf to induce full iron saturation. (A) Distribution of ferH mRNA as determined by sucrose gradient analysis (for details, see legend to Fig 1). (Insets) The ribosome-bound mRNA compartment (fractions 9-18) is shown with an extended ordinate to highlight the difference between v-ErbA and v-ErbB expressing cells. (Open symbols) Des; (solid symbols) Tf. (B) Ferritin protein levels as detected by Western blotting in self-renewing primary ebls (SCF), v-ErbA– or v-ErbB–expressing ebls (v-ErbA, v-ErbB), and AEV-transformed red blood cells (HD3E22) under iron scarcity (Des) or high iron abundance (Tf). In case of SCF-progenitors, low Tf (0.03 mg/mL) instead of Des was used to induce IRP-activity. Numbers below each pair of lanes represent the factors of iron-dependent upregulation of ferritin expression (fold induction).
Translational inhibition of ferH mRNA in primary SCF-progenitors is abolished by the v-ErbA oncoprotein.
As shown above, AEV-transformed ebls conferred iron-dependent translational regulation according to the standard model (ie, coregulation of eALAS and ferH mRNA translation), whereas primary ebls exhibited the iron-insensitive erythroid mode. This raised the question of which of the 2 AEV oncogenes was responsible for this effect. In addition, we tested whether the induction of standard mode regulation was restricted to cells expressing the oncogenic ErbB and/or ErbA variant(s) or could also be induced by the respective, nonmutated cellular proto-oncogenes. Suitable retroviral vectors expressing these proteins were used to infect primary bone marrow ebls, either alone or in various combinations.31
The analysis showed that v-ErbA, a mutated version of the cellular thyroid hormone receptor α (TRα), on its own was capable to restore iron-dependent regulation of ferH protein synthesis in self-renewing ebls. Sucrose gradient analysis showed that v-ErbA–expressing erythroid cells exhibited a shift of ferH transcripts towards the polysomal compartment under high Tf concentrations. This mobilization was associated with an 8-fold upregulation in the level of ferritin protein (Fig 3A and B). Thus, v-ErbA–expressing primary ebls correspond to AEV-transformed ebls in their mode of translational regulation of ferH mRNA (see Figs 1 and 3B). Similar results were obtained using ebls coexpressing v-ErbA and the murine EpoR (Table 1).
In contrast to v-ErbA, the v-ErbB oncoprotein, a mutant of the cellular epidermal growth factor receptor (EGFR), failed to alter the erythroid mode of regulation of iron metabolism. Under conditions of iron depletion and excess, ferH mRNA was mainly restricted to the ribosome-free compartment (Fig 3A), and in consequence, ferritin protein levels were not modulated (Fig 3B). A comparable result was obtained in ebls overexpressing c-ErbB, the normal cellular EGFR (Table1). Similar to HD3E22 and SCF-ebls, the translational efficiency of eALAS mRNA moderately responded to depletion and excess of iron in both v-ErbA– and v-ErbB–expressing cells, as detected by sucrose gradient analysis (data not shown). This confirmed the data from SCF-ebls that translational control of eALAS mRNA is iron-dependent, whereas the utilization of ferH mRNA is differentially affected by the v-ErbA oncoprotein.
These results raised the question of whether the switch from the erythroid to the standard mode of ferritin biosynthesis by v-ErbA could also be obtained with c-ErbA/TRα. Upon overexpression, this nuclear class II hormone receptor promotes proliferation of ebls and differentiation arrest in a fashion indistinguishable to v-ErbA when completely withdrawn from ligand.37 To our surprise, c-ErbA/TRα had no effect on the inhibition of ferH mRNA translation in primary ebls, neither alone nor in combination with the murine EpoR (Table 1). This was true for both the inability of ferH mRNA to bind to polysomes in these cells and to produce increased levels of ferritin protein in response to high iron levels.
In conclusion, the v-ErbA oncoprotein was required to confer standard type regulation of iron metabolism in primary self renewing ebls. Neither its nonmutated form c-ErbA/TRα nor the EGFR in its normal (c-ErbB) or mutated oncogenic form (v-ErbB) were able to abrogate the translational inhibition of ferH mRNA, typical for the erythroid mode.
V-ErbA expression restores iron-dependent modulation of IRP-binding activity to the ferH-IRE.
To obtain first insights into the mechanism responsible for the erythroid mode of ferritin regulation, we studied the role of IRP in the interference with ferH mRNA translation. To this purpose, IRP abundance and IRE-affinity were determined in vitro from cytosolic extracts by gel retardation assays.47,48 To avoid potential artifacts arising from the binding of chicken IRP to mammalian IRE-templates, a chicken ferH cDNA containing the entire 5′-UTR and including the regulatory IRE stem-loop structure was cloned from total chicken liver RNA by RT-PCR as delineated from the published genomic sequence.45
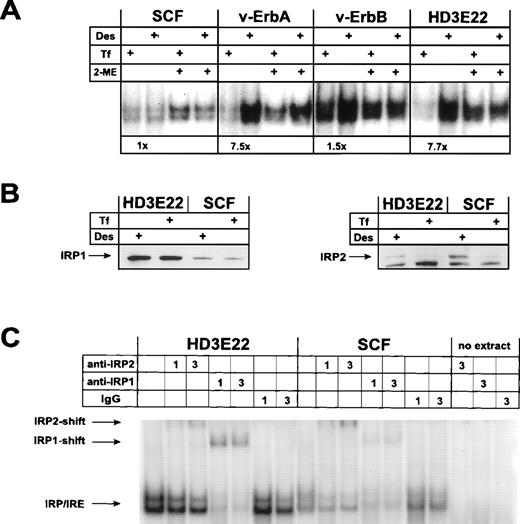
Surprisingly, IRP-mRNA binding activity could not be modulated in response to iron in primary SCF-progenitors (Fig 4 and L. Lobmayr, unpublished observation). In contrast, leukemic HD3E22 ebls displayed an 8-fold increase in IRP-IRE complex formation under iron depletion, as predicted by the standard model. Moreover, the total IRP abundance as monitored by the activation of nonbinding IRP molecules with 2-mercaptoethanol52 was low in the primary progenitors compared with their leukemic counterparts (Fig 4A). Similar results were obtained in 2 other strains of AEV-transformed ebls (HD3E11, ts34A6; see Table 1 and data not shown). In good agreement with the data obtained by sucrose gradient and Western blot analysis, we found that retrovirally expressed v-ErbA restored iron-dependent modulation of IRP-IRE binding affinity, as indicated by a greater than 7-fold increase in the formation of IRP-IRE complexes under conditions of iron deprivation. The abundance of IRP and the extent of its regulation closely corresponded to those observed in ebls expressing both AEV oncogenes (HD3E22; Fig 4A). Similar results were obtained with extracts of cells expressing v-ErbA in combination with murine EpoR (Table 1 and data not shown). On the contrary, exogenous v-ErbB or c-ErbB were unable to induce iron-dependent modulation of IRP-IRE complex formation in primary ebls. Cells overproducing c-ErbA/TRα, either alone or in combination with murine EpoR, were as iron-insensitive as the primary SCF-ebls (Table 1). From these data we concluded that the functional impact of the v-ErbA oncogene but not its cellular counterpart, c-ErbA, was sufficient to confer iron-dependent IRP modulation in SCF-progenitors.
Iron-dependent modulation of IRP-binding activity to the ferH-IRE in primary versus transformed ebls. (A) IRP-IRE-binding activity was determined in vitro by a gel retardation assay as outlined in Materials and Methods. Aliquots of the cell extracts from self-renewing ebls used for immunoblotting (see Fig 3) were incubated with an excess of radiolabeled ferH-IRE transcripts and the resulting RNA-protein complexes were separated in nondenaturing polyacrylamide gels. Where indicated, maximal IRP1-IRE binding activity was estimated by treatment of the extracts with 2-mercaptoethanol (2-ME) in vitro. The amount of radioactivity in the IRP-IRE complexes was quantified by phosphoimaging. Numbers below the pair of corresponding lanes represent the fold induction of IRP activity. (B) Levels of IRP1 (left panel) and IRP2 (right panel) in transformed versus primary ebls cultivated under different iron supply, as detected by immunoblotting. (C) IRP1 versus IRP2 binding activity to the ferH-IRE in extracts of transformed and primary ebls treated for 24 hours with Des. Gel retardation assays in combination with antibody-mediated supershifts were performed as described in detail under Materials and Methods. As a control (no extract), lysis buffer was incubated with 3 μL antibody and the radioactive ferH-IRE probe. -IRP1 and -IRP2, IRP1 and IRP2 specific antisera, respectively; IgG, nonspecific antibody.
Iron-dependent modulation of IRP-binding activity to the ferH-IRE in primary versus transformed ebls. (A) IRP-IRE-binding activity was determined in vitro by a gel retardation assay as outlined in Materials and Methods. Aliquots of the cell extracts from self-renewing ebls used for immunoblotting (see Fig 3) were incubated with an excess of radiolabeled ferH-IRE transcripts and the resulting RNA-protein complexes were separated in nondenaturing polyacrylamide gels. Where indicated, maximal IRP1-IRE binding activity was estimated by treatment of the extracts with 2-mercaptoethanol (2-ME) in vitro. The amount of radioactivity in the IRP-IRE complexes was quantified by phosphoimaging. Numbers below the pair of corresponding lanes represent the fold induction of IRP activity. (B) Levels of IRP1 (left panel) and IRP2 (right panel) in transformed versus primary ebls cultivated under different iron supply, as detected by immunoblotting. (C) IRP1 versus IRP2 binding activity to the ferH-IRE in extracts of transformed and primary ebls treated for 24 hours with Des. Gel retardation assays in combination with antibody-mediated supershifts were performed as described in detail under Materials and Methods. As a control (no extract), lysis buffer was incubated with 3 μL antibody and the radioactive ferH-IRE probe. -IRP1 and -IRP2, IRP1 and IRP2 specific antisera, respectively; IgG, nonspecific antibody.
To assess whether IRP1 and/or IRP2 was responsible for the observed mobility shift of the chicken ferH-IRE probe and the modulation of ferritin expression in the various erythroid cell types tested, Western blot analyses were performed. The amount of IRP1 expression was greatly increased in the AEV-transformed HD3E22 cells as compared with normal SCF erythroid cells (Fig 4B). This was not the case for IRP2 protein, whose abundance is even slightly decreased in the transformed cells. To our knowledge, this result also provided the first evidence for the existence of IRP2 in chicken. No iron-dependent variation in amounts of IRP1 protein was observed in the cell types tested, in accordance with previous reports,8 which showed that IRP1 activity is mainly governed by modifications of the FeS cluster near the RNA binding domain. In contrast, IRP2 expression was massively reduced by iron administration, again in agreement with data from the literature demonstrating a reduction of IRP2 protein stability in the presence of iron.53 54
The apparent coexpression of IRP1 and IRP2 in erythroid cells raised the question of which of these RNA binding proteins was responsible for the electrophoretic mobility shift described in Fig 4A. Moreover, the emergence of a double band might have suggested that each band corresponded to one of the IRPs. Hence, supershift assays were performed in which cellular extracts from HD3E22 as well as SCF-ebls were incubated with antisera specific for IRP1 or IRP2, respectively, before the IRP-IRE binding reaction. In the HD3E22 cells, the majority of both bands of the doublet could be supershifted with an antibody against IRP1; however, some supershifted complexes were produced by the IRP2-specific antibody (Fig 4C). In contrast, SCF cells appeared to contain less IRP1 activity and a slightly higher amount of IRP2 than the transformed cells. These data correlate well with the observations from the Western blot experiments (Fig 4B).
Taken together, these analyses strongly suggested that both IRP1 and IRP2 are responsible for both bands in the electrophoretic mobility shift with the chicken ferH-IRE. This was verified with nonerythroid chicken cells as well as mouse fibroblasts (data not shown), indicating that the double band is not cell type, but probe-specific. Thus, the lower intensity of the IRP-IRE complex formation in SCF cells seems to be a true reflection of reduced expression of IRP1 and IRP2 combined.
Reversion of standard to erythroid mode of ferritin expression by functional inactivation of v-ErbA in leukemic AEV-erythroblasts.
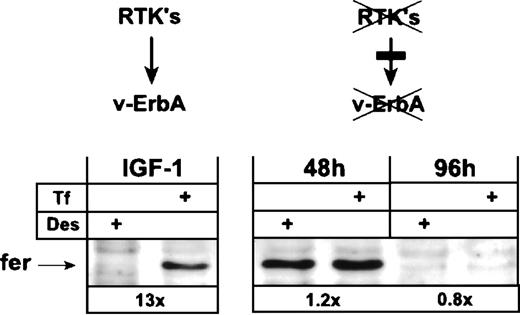
Earlier work has shown that the v-ErbA oncoprotein requires the cooperation of the receptor tyrosine kinase (RTK) c-Kit (or other tyrosine kinases such as v-ErbB) to display its biological activity, ie, continuous proliferation coupled with a tight differentiation arrest in primary erythroid progenitors.30 36 In the absence of cooperating RTK, v-ErbA is biologically inactive. We therefore asked the question of whether the standard-type regulation of ferH translation in leukemic HD3E22 ebls could be abolished by functional inactivation of v-ErbA via abrogation of RTK activity.
HD3E22 ebls express both v-ErbB and c-Kit. The constitutively active v-ErbB protein can be inhibited by the low molecular weight inhibitor PD 153035 specific for the EGFR.44 On the other hand, c-Kit is inactivated by withdrawal of its ligand, SCF, together with inhibition of its kinase activity by the low molecular weight c-Kit inhibitor EXBW 50. When HD3E22 cells are sub- jected to this combined treatment, they differentiate terminally in the presence of human Epo plus insulin (H. Beug, unpublished data).
After differentiation induction by the above-described treatment, HD3E22 cells drastically reduced their cell size, showed a greater than 10-fold increase in hemoglobin content, and displayed a typical erythrocyte morphology within 96 hours. In contrast, cells grown in the absence of Epo/insulin and inhibitors continued to proliferate and showed differentiation arrest (data not shown). Both cell types were analyzed for their expression of ferritin protein by Western blot after exposure to Des or high Tf levels. Interestingly, cells induced to differentiate for 48 to 96 hours by the above-described treatment failed to upregulate ferritin expression in response to iron, resembling the phenotype described for primary SCF-ebls (Fig 5; see also, Fig 3B). Ferritin was still expressed after 48 hours of differentiation, but the levels were similar under depletion and excess of iron. Ninety-six hours after differentiation induction, ferritin protein was downregulated under both conditions. In contrast, proliferating HD3E22 ebls that contained an active v-ErbA (ie, in the absence of inhibitors) exhibited the typical greater than 10-fold elevation in ferritin protein under high Tf conditions (Fig 5; see also, Fig 3B). HD3E22 ebls induced to differentiate for 48 hours by SCF withdrawal and v-ErbB inhibition by PD153035 only (ie, without adding the c-Kit inhibitor EXBW 50) showed iron-dependent modulation of ferritin expression to an extent comparable to proliferating cells (data not shown). This suggests that a residual c-Kit activity (or a closely related RTK) may have been sufficient to activate v-ErbA function with respect to alter regulation of ferH translation. These results support the hypothesis that the activity of the v-ErbA oncoprotein is sufficient to abolish the iron-independent inhibition of ferH translation in primary SCF-dependent erythroid progenitors.
Functional inactivation of the v-ErbA oncoprotein switches AEV-transformed ebls from iron-dependent to iron-independent ferritin expression. AEV-transformed ebls (HD3E22) were cultivated in Epotest medium either under proliferation conditions (IGF-1) or induced to terminal differentiation by supplementation with 2 U/mL human Epo, 1.4 nmol/L insulin, 1 μmol/L each of the steroid antagonists ICI 164384 and ZK 112993, the ErbB inhibitor PD 153035, and the c-Kit antagonist EXBW 50 for up to 96 hours (see Materials and Methods). Twenty-four hours before harvesting for Western blot analysis, ebls were switched to identical media containing either Des (50 μmol/L) or Tf (1 mg/mL). Numbers below each pair of lanes represent the fold induction of iron-dependent ferritin expression.
Functional inactivation of the v-ErbA oncoprotein switches AEV-transformed ebls from iron-dependent to iron-independent ferritin expression. AEV-transformed ebls (HD3E22) were cultivated in Epotest medium either under proliferation conditions (IGF-1) or induced to terminal differentiation by supplementation with 2 U/mL human Epo, 1.4 nmol/L insulin, 1 μmol/L each of the steroid antagonists ICI 164384 and ZK 112993, the ErbB inhibitor PD 153035, and the c-Kit antagonist EXBW 50 for up to 96 hours (see Materials and Methods). Twenty-four hours before harvesting for Western blot analysis, ebls were switched to identical media containing either Des (50 μmol/L) or Tf (1 mg/mL). Numbers below each pair of lanes represent the fold induction of iron-dependent ferritin expression.
DISCUSSION
The current investigation provides the first evidence for differential translational regulation of ferH versus eALAS mRNA during normal erythropoiesis. Using primary ebls, we show that ferritin mRNA translation is not subject to regulation by iron: binding of ferritin mRNA to polysomes is specifically impaired under low as well as high iron concentrations. In contrast, eALAS transcripts are already bound to polysomes under iron scarcity, but shift to larger polysomes in response to high Tf, corroborating previous evidence for translational control of eALAS. We further demonstrate that the hypothetical regulatory loop responsible for the inhibition of ferH protein synthesis in primary ebls can be efficiently shut off by the v-ErbA oncoprotein, restoring iron-responsive regulation of ferH mRNA translation, as observed in leukemic AEV-transformed ebls. The specificity of v-ErbA as a molecular switch between the standard and the erythroid mode of ferritin regulation could be confirmed by abolishing v-ErbA activity in erythroleukemic ebls, causing a change from iron-dependent to iron-independent ferritin expression typical for the erythroid mode of regulation of iron metabolism.
The moderate level of eALAS mRNA translational control might correspond to a variation in the hairpin motif of the chicken eALAS-IRE. As previously shown by sequence analysis of the chicken eALAS genomic DNA as well as mRNA,46 the consensus element 5′-CAGUGN-3′ representing the hypercritical loop region of the IRE is changed to 5′-CAGCGN-3′. A similarly aberrant sequence 5′-CAGGGN-3′ was independently reported.55 These naturally occurring variants as well as other point mutations generated in vitro have been described to impair the interaction with IRP.56 57 Preliminary results suggest that the alteration observed in the chicken eALAS IRE is indeed responsible for a 30- to 40-fold reduced affinity of IRP towards this particular element (data not shown).
In the chicken system, efficient retroviral vectors facilitate ectopical (over)expression of cellular proto-oncogenes and their corresponding viral counterparts.29,30 Our results from retroviral overexpression experiments clearly demonstrated that v-ErbA oncoprotein alone is sufficient to reverse the inhibition of ferH mRNA translation, resulting in standard-type regulation of iron storage as observed in AEV-transformed chicken ebls (Figs 3 and 4A). Similar to v-ErbA, overexpression of the cellular homologue c-ErbA/TRα has been shown to induce proliferation and to arrest differentiation of ebls, when allowed to cooperate with c-Kit and deprived of all ligands (T3 and retinoids activating the heterodimerization partner RXR). Hence, v-ErbA was proposed to represent a c-ErbA/TRα permanently frozen in the nonliganded conformation that is able to repress transcription around some or all of the c-ErbA/TRα-responsive target genes.37 However, as opposed to v-ErbA, exogenous c-ErbA/TRα did not influence the reduction of ferH mRNA translation efficiency (Table 1). This indicates that the mutations present in v-ErbA must endow it with additional features important for IRP-dependent translational regulation.
Leukemias are characterized by hyperproliferation of immature or partially mature hematopoietic progenitor cells. In vitro, normal bone marrow-derived SCF-progenitors undergo short-term self renewal for 8 to 10 generations until they differentiate or become apoptotic. However, overexpression of exogenous ErbA causes long-term self renewal (>25 generations), mimicking an erythroleukemic phenotype.30 Our data provide a potential correlation between the expression of ferritin under high Tf concentrations and the life span of erythroid progenitors. Whereas short-term proliferating SCF-ebls display low ferritin abundance at high iron levels, long-term self renewal is accompanied by an upregulation of ferritin expression in ebls overexpressing v-ErbA, reaching levels typical for the leukemic HD3 cells. These findings support the hypothesis that the ability to detoxify free iron via ferritin, to avoid the emergence of harmful radicals and reactive oxygen species, is essential for continuously growing cells. The idea is consistent with previous findings that rapidly proliferating tumor cells express high levels of ferritin.58 59 Thus, SCF-progenitors might not require high-level ferritin expression for iron detoxification, because these cells (1) use all available iron for hemoglobin production and (2) are eliminated after a relatively short time due to their limited number of cell divisions in which no long-term accumulation of damage caused by iron can occur.
At present, the molecular mechanisms governing the differential use of ferH and eALAS mRNAs in self-renewing as well as differentiating primary SCF-ebls remain essentially unknown. The phenotype cannot be explained by a differentiation-associated increase in the abundance of these mRNAs (exceeding the number of available IRP molecules), because differential translational control was already observed in self-renewing cells. In addition, analysis of the corresponding mRNA levels showed no massive alterations upon terminal erythropoiesis (data not shown).
Moreover, the inhibition of ferH mRNA translation cannot be due to alterations in IRP abundance. v-ErbA expressing transformed ebls with their relatively high IRP abundance (Fig 4A) allow translation of ferH mRNA under high iron, whereas normal SCF progenitors with their relatively low IRP expression reduce use of this mRNA independently of the iron status. What may cause the nonresponsiveness to iron? At least 2 possibilities are conceivable. First, in primary erythroid cells, IRP may be subjected to a posttranslational modification resulting in constitutive binding to ferH-IREs. Recent studies support the idea that phosphorylation of IRP can affect function.60 The translational control of eALAS transcripts, also presumably mediated by IRP, may well remain iron-sensitive in the same cells (SCF-progenitors) due to the alterations in the IRE consensus sequence mentioned above. Second, a putative factor X might specifically bind to ferH mRNA, freezing IRPs to their cognate ferH IREs by direct protein-protein interaction. Alternatively, complex formation between this putative factor X and ferH mRNA could inhibit ferH translation in an IRP-independent manner. Such an inhibitory factor should be either directly or indirectly inactivated by v-ErbA, in line with the ability of v-ErbA to transcriptionally repress distinct target genes in proliferating ebls.36,39 61
The possibility to compare 2 closely matched erythropoietic cell systems (AEV-transformed and primary chicken ebls) for their mode of iron use and storage enabled us to address the generally important question of how high ferritin levels can be compatible with high eALAS abundance in normal and aberrant ebls. In contrast to previous investigations in erythroleukemic cell lines,6 7 primary ebls displayed a differential translational control of ferH versus eALAS mRNA. This erythroid mode of iron metabolism offers a clear-cut rationale how a new balance is reached between (high) iron utilization and (low) storage capacity upon normal erythropoiesis: eALAS expression is on to ensure heme biosynthesis in the erythropoietic process of hemoglobinization, which requires tremendous amounts of iron, whereas de novo ferritin expression is off and iron storage capacity is low but still sufficient to avoid potential iron toxicity.
In conclusion, our work has shown that the standard model of iron metabolism represents just one of several possibilities particularly suitable for undifferentiated, rapidly proliferating cells. They require moderate Tf-iron levels for growth and survival, but need an effective mechanism to protect them from toxic radicals and reactive oxygen species generated by free iron. From a functional aspect, it is not surprising that a different mode of iron regulation is operative in erythroid cells that are transiently proliferating but exhibit a very high iron demand before and during differentiation. Here, regulation of iron-responsive genes according to the standard model would cause serious problems, which are circumvented by the erythroid mode of regulation.
ACKNOWLEDGMENT
The authors acknowledge the kind gift of IRP1 antiserum from Richard S. Eisenstein (University of Wisconsin, Madison, WI) and IRP2 antiserum from Elizabeth A. Leibold (University of Utah, Salt Lake City, UT). LMH-2A cells were kindly provided by Marcela Hermann and N. Erwin Ivessa. The authors thank Lukas C. Kühn for stimulating discussions and Eva Maria Deiner for expert technical assistance.
Supported by grants from the “Herzfeld Family Foundation” and the “Fonds zur Förderung der Wissenschaftlichen Forschung,” Austria.
The sequence data have been submitted to the EMBL/GenBank databases under accession no. Y14698.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Ernst W. Müllner, PhD, Institute of Molecular Biology, Vienna Biocenter, University of Vienna, Dr. Bohr-Gasse, A-1030 Vienna, Austria; e-mail: em@mol.univie.ac.at.