Abstract
We have identified specific iron (Fe) chelators of the pyridoxal isonicotinoyl hydrazone (PIH) class that are far more effective ligands than desferrioxamine (DFO; Richardson et al, Blood 86:4295, 1995; Richardson and Milnes, Blood 89:3025, 1997). In the present study, we have compared the effect of DFO and one of the most active chelators (2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone; 311) on molecular targets involved in proliferation. This was performed to further understand the mechanisms involved in the antitumor activity of Fe chelators. Ligand 311 was far more active than DFO at increasing Fe release from SK-N-MC neuroepithelioma and BE-2 neuroblastoma cells and preventing Fe uptake from transferrin. Like DFO, 311 increased the RNA-binding activity of the iron-regulatory proteins (IRPs). However, despite the far greater Fe chelation efficacy of 311 compared with DFO, a similar increase in IRP-RNA binding activity occurred after 2 to 4 hours of incubation with either chelator, and the binding activity was not inhibited by cycloheximide. These results suggest that, irrespective of the Fe chelation efficacy of a ligand, an increase IRP-RNA binding activity occurred via a time-dependent step that did not require protein synthesis. Further studies examined the effect of 311 and DFO on the expression of p53-transactivated genes that are crucial for cell cycle control and DNA repair, namely WAF1,GADD45, and mdm-2. Incubation of 3 different cell lines with DFO or 311 caused a pronounced concentration- and time-dependent increase in the expression of WAF1 and GADD45 mRNA, but not mdm-2 mRNA. In accordance with the distinct differences in Fe chelation efficacy and antiproliferative activity of DFO and 311, much higher concentrations of DFO (150 μmol/L) than 311 (2.5 to 5 μmol/L) were required to markedly increase GADD45 and WAF1 mRNA levels. The increase in GADD45 and WAF1 mRNA expression was seen only after 20 hours of incubation with the chelators and was reversible after removal of the ligands. In contrast to the chelators, the Fe(III) complexes of DFO and 311 had no effect on increasing GADD45 and WAF1 mRNA levels, suggesting that Fe chelation was required. Finally, the increase in GADD45 and WAF1 mRNAs appeared to occur by a p53-independent pathway in SK-N-MC and K562 cells, because these cell lines lack functional p53. Our results suggest that GADD45 and WAF1 may play important roles in the cell cycle arrest observed after exposure to these chelators.
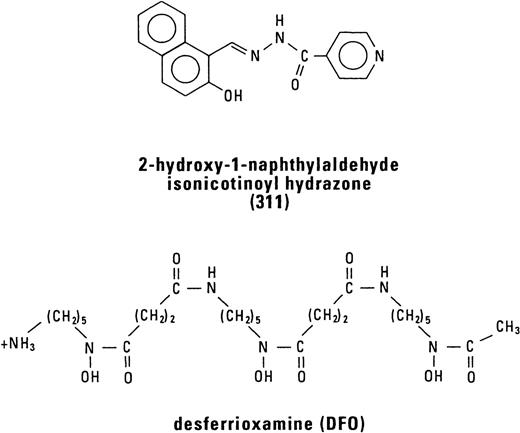
A WIDE VARIETY OF studies have suggested that cancer cell proliferation can be inhibited using a range of specific iron (Fe) chelating agents.1-6 The ability of these ligands to inhibit growth reflects the well-known importance of Fe in a variety of crucial metabolic pathways, including DNA synthesis and ATP production.6 The most well-characterized Fe chelator in terms of its antiproliferative activity is desferrioxamine (DFO; Fig 1), the drug used to treat Fe overload disease. DFO has been shown to be effective in vitro at inhibiting the growth of a number of neoplastic cell types, with the most well-studied tumors being neuroblastoma7-10 and leukemia.11These in vitro studies have encouraged limited clinical trials in which DFO has shown promise both as a single agent12,13 or in combination with other cytotoxic drugs.14
The antiproliferative effect of DFO prompted us to investigate the antineoplastic activity of Fe chelators of the pyridoxal isonicotinoyl hydrazone (PIH) class. These compounds have a high affinity and specificity for Fe that is similar to that found for DFO and much greater than that of EDTA.15,16 Moreover, PIH and some of its analogues have been shown to possess marked Fe chelation efficacy in a wide variety of biological systems both in vitro and in vivo.17-20 Considering their high Fe chelation efficacy, in recent studies we have screened the antineoplastic activity of 3 groups of ligands based on PIH.21,22 From these investigations, one of the most active Fe chelators identified was 2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311; Fig1).21 22
At present, very little is known concerning the molecular mechanisms by which Fe chelators inhibit cellular proliferation and the cell cycle. It has been well documented that some Fe chelators can inhibit the activity of ribonucleotide reductase, the critical enzyme involved in the conversion of ribonucleotides to deoxyribonucleotides.23,24 The reduction in deoxyribonucleotide concentrations is thought to prevent DNA synthesis, which then results in G1/S arrest.3,10,22,25 26Apart from this, nothing is known concerning why some Fe chelators are more effective antiproliferative agents than others.
In an initial attempt to further elucidate the molecular targets of Fe chelators, we have compared the effect of DFO, which shows relatively low antiproliferative efficacy, with that of 311.21,22 In these studies, we have compared the Fe chelation efficacy of these ligands and their effect on the RNA-binding activity of the iron-regulatory proteins (IRPs). The IRPs are key regulators of intracellular Fe metabolism and control the expression of a number of proteins, including those that are involved in Fe uptake (the transferrin receptor [TfR]) and Fe storage (ferritin-H and -L subunits).27-29 We have also examined the effect of DFO and 311 on the expression of a number of genes that are thought to play key roles in cellular proliferation and the regulation of the cell cycle. These include the p53-responsive genes WAF1 (wild-type p53 activating fragment 1 gene), GADD-45 (growth arrest and DNA damage gene), and human mdm-2 (murine double minute gene). WAF-1 is a potent universal inhibitor of cyclin-dependent kinases30 and can induce a G1/S arrest31-33 and possibly a G2/M arrest.34 GADD-45 is induced upon DNA damage, can arrest the cell cycle, and is also involved in DNA nucleotide excision repair.33,35 On the other hand, p53-mediated transactivation of mdm-2 results in a feedback control mechanism of p53 activity.33,36,37 Interestingly, both the expression of WAF1 and GADD45 can also be controlled by p53-independent pathways.38 39
Our present studies clearly demonstrate that 311 rapidly chelates intracellular Fe and prevents Fe uptake from transferrin (Tf) at very low concentrations, being far more effective than DFO. Despite the far greater Fe chelation efficacy of 311 compared with DFO, the increase in IRP-RNA binding activity of the IRPs occurred by similar kinetics and was not inhibited by cycloheximide. These latter data suggest that, irrespective of Fe chelation efficacy, the increase in IRP-RNA binding activity was a time-dependent event that did not require protein synthesis. In agreement with the marked differences in Fe chelation efficacy and antiproliferative activity of 311 compared with DFO, a pronounced increase in WAF1 and GADD45 mRNA levels was observed at a DFO concentration of 150 μmol/L, whereas 311 was effective at concentrations as low as 2.5 to 5 μmol/L. The results suggest that the increase in GADD45 and WAF1 mRNA expression may be important in the cell cycle arrest observed after exposure to potent Fe chelators.
MATERIALS AND METHODS
Preparation of 311 and its iron(III) complex.
Chelator 311 was synthesized by Schiff base condensation between 2-hydroxy-1-naphthylaldehyde and isonicotinic acid hydrazide using standard procedures.40 DFO was purchased from Ciba-Geigy Pharmaceutical Co (Summit, NJ). Chelator 311 and its Fe(III) complex ([Fe(311)2] (NO3)·5H2O) were characterized by a combination of elemental analysis, infrared spectroscopy, NMR spectroscopy, and x-ray crystallography.41 The Fe complex of DFO (ferrioxamine) was prepared as reported in previous studies.42 Chelator 311 was dissolved in dimethyl sulphoxide (DMSO) as a 10 mmol/L stock solution immediately before an experiment and then diluted in 10% fetal calf serum (FCS) so that the final concentration of DMSO was equal to or less than 0.5% (vol/vol).21 All incubations of cells with the chelators were performed in 10% FCS.
Cell culture.
The human BE-2 neuroblastoma cell line was kindly provided by Dr Greg Anderson (Queensland Institute for Medical Research, Herston, Brisbane, Queensland). The SK-N-MC cell line (American Type Culture Collection [ATCC], Rockville, MD) was originally classified as a neuroblastoma, but it has been recently reclassified as a neuroepithelioma, a closely related, primitive neuroectodermal malignancy. The human K562 erythroleukemia cell line was also from the ATCC. The SK-N-MC cell line was grown in Eagle’s modified minimum essential medium (MEM; GIBCO BRL, Sydney, Australia) containing 10% FCS (Commonwealth Serum Laboratories, Melbourne, Australia), 1% (vol/vol) nonessential amino acids (GIBCO), 2 mmol/L L-glutamine (Sigma Chemical Co, St Louis, MO), 100 μg/mL of streptomycin (GIBCO), 100 U/mL penicillin (GIBCO), and 0.28 μg/mL of fungizone (Squibb Pharmaceuticals, Montréal, Quebec, Canada). The BE-2 and K562 cell lines were grown in RPMI (GIBCO) containing 10% FCS and the supplements described above for MEM. Cells were grown in an incubator (Forma Scientific, Marietta, OH) at 37°C in an humidified atmosphere of 5% CO2/95% air and subcultured as described previously.43 Cellular growth and viability were monitored using phase-contrast microscopy and trypan blue staining.
Protein labeling.
Apotransferrin (Sigma Chemical Co) was prepared and labeled with59Fe (as ferric chloride in 0.1 mol/L HCl; Dupont NEN, Boston, MA) to produce59Fe2-transferrin (59Fe-Tf) using standard procedures.43
Metabolic labeling.
Labeling of cellular proteins with 3H-leucine (52 Ci/mmol; Dupont NEN) was estimated after precipitation with trichloroacetic acid (TCA). Briefly, after the required exposure period to cycloheximide,3H-leucine (1 μCi/mL) was added and the cells were incubated for 1 hour at 37°C. The petri dishes were then placed on ice and washed four times with ice-cold Hanks’ balanced salt solution (BSS; GIBCO BRL), and the cells were detached from the plate using 1 mmol/L EDTA in Ca/Mg-free saline. The cells were then pelleted by centrifugation, the supernatant was removed, and the pellet was frozen at −70°C. After thawing the cells on ice, 1 mL of ice-cold 20% TCA was added and the solution was then vortexed and kept on ice for 1 hour with periodic vortex mixing. This solution was then centrifuged at 15,000 rpm for 15 minutes at 4°C and the supernatant was removed. The pellet was then washed twice with 1 mL of ice-cold 10% TCA and dissolved in 0.5 mL of 1 mol/L NaOH. After transfering this solution to scintillation tubes, 3 mL of scintillant was added and the radioactivity was measured on a β-scintillation counter (LKB Wallace, Turku, Finland).
Iron uptake and iron efflux experiments.
The effect of chelators on 59Fe uptake from59Fe-Tf and 59Fe release from prelabeled cells was studied using standard methods reported previously.21
IRP gel-retardation assay.
The gel-retardation assay was used to measure the interaction between the IRPs and iron-responsive element (IRE) using established techniques.44 45 Briefly, after incubation with medium alone (control) or medium containing ferric ammonium citrate (100 μg/mL; Aldrich Ltd., Sydney, Australia) or the chelators, 2 to 5 × 106 cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed at 4°C in ice-cold Munro extraction buffer (10 mmol/L HEPES, pH 7.6, 3 mmol/L MgCl2, 40 mmol/L KCl, 5% glycerol, 1 mmol/L dithiothreitol, and 0.5% Nonidet P-40). After lysis, the samples were then centrifuged at 10,000g for 3 minutes at 4°C to remove nuclei and the supernatant was stored at −70°C. Frozen cytoplasmic extracts were thawed on ice and then centrifuged at 15,000 rpm for 10 minutes at 4°C. The protein concentration of the supernatant was determined using the Bio-Rad protein assay (Bio-Rad Ltd, Hercules, CA). Samples of cytoplasmic extracts were diluted to a protein concentration of 100 μg/mL in Munro buffer without Nonidet P-40, and 1-μg aliquots were analyzed for IRP by incubation with 0.1 ng (∼1 × 105 cpm) of 32P-labeled pGL66 RNA transcript (pGL66 was kindly provided by Dr Elizabeth Leibold, University of Utah, Salt Lake City, UT). The riboprobe was transcribed in vitro from linearized plasmid templates with SP6 RNA polymerase in the presence of α-32P UTP (Dupont, NEN) using the Promega Riboprobe In Vitro Transcription Kit (Promega, Madison, WI). The probe was subsequently purified on a 6% urea/polyacrylamide gel electrophoresis (PAGE) gel. To form RNA-protein complexes, cytoplasmic extracts containing 1 μg of protein were incubated for 10 minutes at room temperature with the 32P-labeled riboprobe. Unprotected probe was degraded by incubation with 1 U of RNAse T1 for 10 minutes at room temperature. Heparin (Sigma) at a final concentration of 5 mg/mL was then added and incubated with the extract for another 10 minutes at room temperature to exclude nonspecific binding. RNA-protein complexes were analyzed in 6% nondenaturing polyacrylamide gels at 4°C. Gels were dried, covered in plastic film, and exposed to Kodak XAR films (Eastman Kodak, Rochester, NY) at −70°C with an intensifying screen.
Northern blot analysis.
Northern blot analysis was performed by isolating total RNA using the Total RNA Isolation Reagent from Advanced Biotechnologies Ltd (Surrey, UK). The RNA (15 μg) was heat-denatured at 90°C for 2 minutes in RNA-loading buffer and then loaded onto a 1.2% agarose-formaldehyde gel. After electrophoresis, RNA was transferred to a nylon membrane (GeneScreen; New England Nuclear, Boston, MA) in 10× SSC using the capillary blotting method. The RNA was then cross-linked to the membrane using a UV-crosslinker (UV Stratalinker 1800; Stratagene Ltd, La Jolla, CA).
The membranes were hybridized with probes specific for the human TfR, β-actin, N-myc, WAF1, GADD-45, and mdm-2. The TfR probe consisted of a 2.8-kb coding region from the human TfR cDNA cloned into pCD-TR1 (kindly supplied by Dr L.C. Kühn, Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland). The β-actin probe consisted of a 1.4-kb fragment from human β-actin cDNA cloned into pBluescript SK- (ATCC; catalogue no. 37997). The N-myc probe was a 1-kb fragment from human N-myc cDNA cloned into pBR322 (ATCC; catalogue no. 41011). The WAF1 probe consisted of a 1-kb fragment from pSXV (ATCC; catalogue no. 79928). The GADD45 probe consisted of a 760-bp fragment from human GADD45 cDNA cloned into pHu145B2 (kindly supplied by Dr Albert Fornace, National Cancer Institute, National Institutes of Health, Bethesda, MD). The human mdm-2 probe was a 1.5-kb fragment derived from the full-length human mdm-2 cDNA cloned into the pGEM vector (kindly supplied by Dr Andreas Evdokiou, Royal Adelaide Hospital, Adelaide, South Australia).
Hybridization of probes to the membranes and their subsequent washing were performed as described by Mahmoudi and Lin46 using a Hybaid Shake and Stack Hybridization oven (Hybaid Ltd, Middlesex, UK). Membranes were then wrapped in plastic film and exposed to Kodak XAR films at −70°C with an intensifying screen. Probes were stripped from the nylon membrane by boiling in a solution containing 10 mmol/L Tris-HCl (pH 7.0), 1 mmol/L EDTA (pH 8.0), and 1% sodium dodecyl sulfate (SDS) for 15 to 30 minutes, as described by the membrane manufacturer. Densitometric data were collected with a Laser Densitometer and analyzed by Kodak Biomax I Software (Kodak Ltd, Rochester, NY).
RESULTS
The effect of DFO and 311 on iron release from the SK-N-MC and BE-2 cell lines.
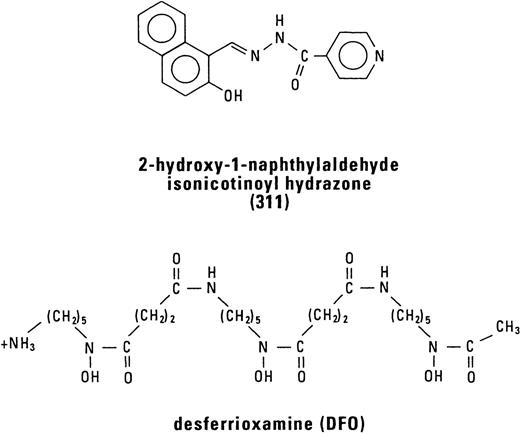
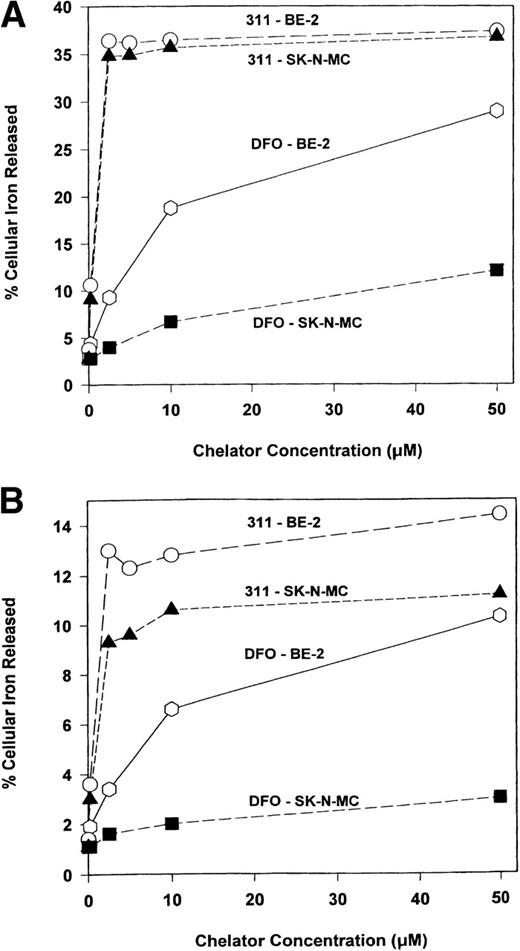
The effect of DFO and 311 concentration on Fe mobilization from the SK-N-MC and BE-2 cell lines were examined after a 3-hour (Fig 2A) and 24-hour (Fig 2B) labeling period with 59Fe-Tf (0.75 μmol/L), followed by 3 hours of reincubation in the presence and absence of the chelators (0.2 to 50 μmol/L). Chelator 311 was far more effective than DFO and showed similar activity in both cell types markedly increasing59Fe release (Fig 2A). Increasing the concentration of 311 from 2.5 μmol/L up to 50 μmol/L did not increase 59Fe release (Fig 2A), suggesting that the chelator depleted the59Fe pool that it had targeted. However, increasing the concentration of DFO from 2.5 μmol/L up to 50 μmol/L resulted in additional 59Fe release, but even at a DFO concentration of 50 μmol/L, the percentage of 59Fe released from SK-N-MC and BE-2 cells (12% and 29%, respectively) was less than that observed at a 311 concentration of 2.5 μmol/L (Fig 2A).
The effect of the concentration of DFO or 311 on59Fe efflux from the SK-N-MC and BE-2 cell lines that have been labeled with 59Fe-transferrin (0.75 μmol/L) for (A) 3 hours or (B) 24 hours and then reincubated in the presence of the chelators (0.2 to 50 μmol/L) for 3 hours at 37°C. Results are means of duplicate determinations in a typical experiment of two experiments performed.
The effect of the concentration of DFO or 311 on59Fe efflux from the SK-N-MC and BE-2 cell lines that have been labeled with 59Fe-transferrin (0.75 μmol/L) for (A) 3 hours or (B) 24 hours and then reincubated in the presence of the chelators (0.2 to 50 μmol/L) for 3 hours at 37°C. Results are means of duplicate determinations in a typical experiment of two experiments performed.
Comparing DFO and 311-mediated 59Fe release after a 3-hour (Fig 2A) and 24-hour (Fig 2B) labeling time, it is obvious that, after a 24-hour label, the percentage of cellular 59Fe released was far less (Fig 2B). For example, even at a 311 concentration of 50 μmol/L, 59Fe efflux from SK-N-MC and BE-2 cells was equal to 11% and 14%, respectively (Fig 2B). In contrast to the effect of 311, which showed similar activity in each cell type, DFO was far more effective at increasing 59Fe release from BE-2 cells compared with the SK-N-MC cell line after both a 3-hour (Fig 2A) and 24-hour (Fig 2B) labeling period.
The effect of DFO and 311 on iron uptake from Tf by the SK-N-MC and BE-2 cell lines.
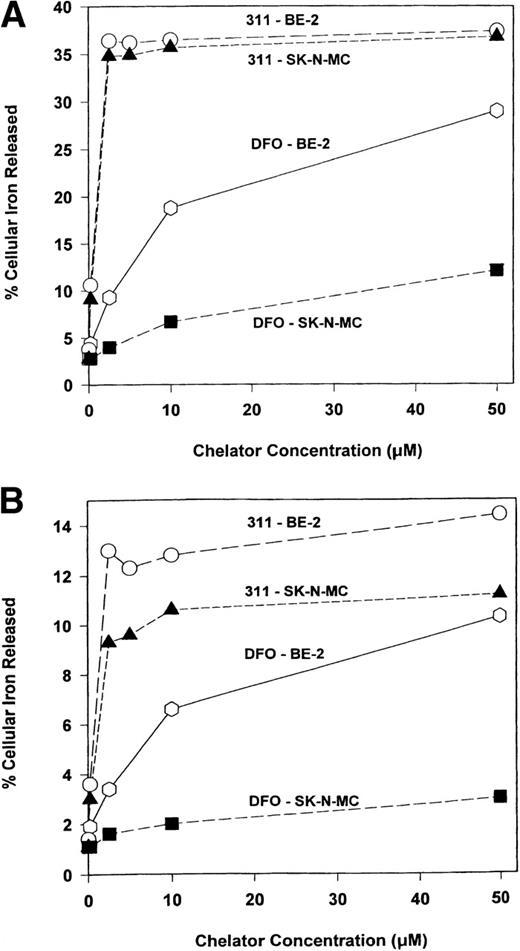
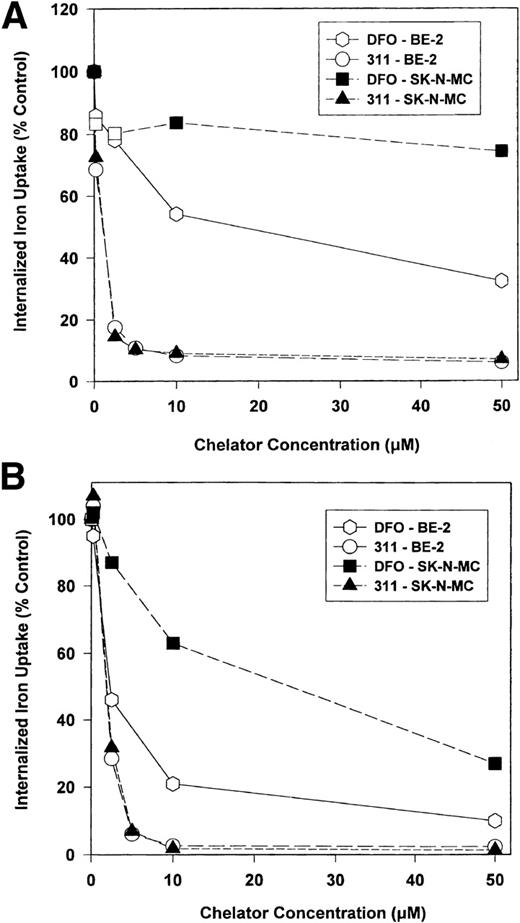
To determine the ability of DFO and 311 to inhibit 59Fe uptake from 59Fe-Tf (0.75 μmol/L), cells were incubated with the chelators (0.2 to 50 μmol/L) and 59Fe-Tf (0.75 μmol/L) for 3 hours (Fig 3A) or 24 hours at 37°C (Fig 3B). Chelator 311 markedly decreased 59Fe uptake from 59Fe-Tf in a similar way for both the SK-N-MC and BE-2 cell lines after the 3-hour and 24-hour labeling times (Fig 3A and B). In contrast, DFO was far less effective than 311 at preventing59Fe uptake from 59Fe-Tf, especially in SK-N-MC cells after both 3 and 24 hours of incubation (Fig 3A and B).
The effect of the concentration of DFO or 311 on iron uptake from 59Fe-transferrin (0.75 μmol/L) by BE-2 and SK-N-MC cells over (A) 3 hours and (B) 24 hours of incubation at 37°C. After this incubation, the cells were washed four times with ice-cold BSS and incubated with pronase (1 mg/mL) at 30 minutes for 4°C.21 43 Results are means of duplicate determinations in a typical experiment of two experiments performed.
The effect of the concentration of DFO or 311 on iron uptake from 59Fe-transferrin (0.75 μmol/L) by BE-2 and SK-N-MC cells over (A) 3 hours and (B) 24 hours of incubation at 37°C. After this incubation, the cells were washed four times with ice-cold BSS and incubated with pronase (1 mg/mL) at 30 minutes for 4°C.21 43 Results are means of duplicate determinations in a typical experiment of two experiments performed.
The effect of DFO and 311 on the RNA-binding activity of the IRPs.
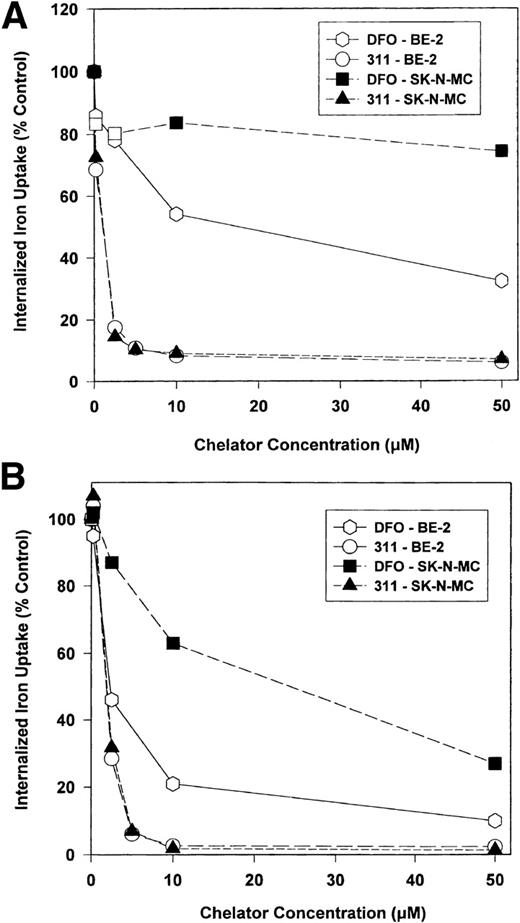
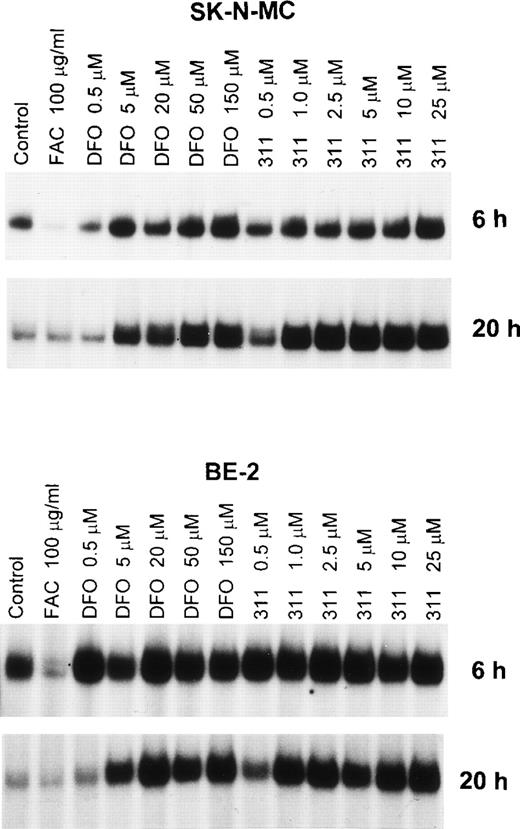
An important effect of intracellular Fe depletion using DFO is the activation of RNA-binding activity of the IRPs. Although the effect of DFO on the RNA-binding activity of this protein has been well characterized,28 the effect of other Fe chelators such as 311 have not been examined. In initial experiments, we examined the effect of range concentrations of DFO (0.5 to 150 μmol/L) and 311 (0.5 to 25 μmol/L) on IRP activation over 6 and 20 hours of incubation using the SK-N-MC and BE-2 cell lines (Fig 4). From Fig 4 it is obvious that one major IRP-IRE band is present, because human IRP1-IRE and IRP2-IRE complexes comigrate in nondenaturing polyacrylamide gels.47The IRP-RNA binding activity after exposure to DFO or 311 was far less pronounced after 6 hours compared with 20 hours of incubation in SK-N-MC cells, whereas in the BE-2 cell line it was similar (Fig 4). In the SK-N-MC cell line after 6 and 20 hours of incubation, and in the BE-2 cell line after 20 hours of incubation, 311 was more effective on a molar basis than DFO at increasing RNA-binding activity of the IRPs. For example, after 20 hours of incubation of SK-N-MC cells with 1 μmol/L 311, IRP-RNA binding activity was very similar to that found after incubation with 50 μmol/L DFO (Fig 4). These results agree with our studies showing marked differences in Fe chelation efficacy between DFO and 311 (Figs 2 and 3). However, after 6 hours of incubation of BE-2 cells with DFO or 311, there was no difference in their ability to increase IRP-RNA binding activity, which was only slightly increased over the control at all chelator concentrations (Fig 4).
The effect of the concentration of DFO (0.5 to 150 μmol/L) or 311 (0.5 to 25 μmol/L) on the RNA-binding activity of the IRPs from the SK-N-MC and BE-2 cell lines after 6 and 20 hours of incubation at 37°C. This result is a typical experiment from three performed for each time point.
The effect of the concentration of DFO (0.5 to 150 μmol/L) or 311 (0.5 to 25 μmol/L) on the RNA-binding activity of the IRPs from the SK-N-MC and BE-2 cell lines after 6 and 20 hours of incubation at 37°C. This result is a typical experiment from three performed for each time point.
As expected, after 6 hours of incubation with the Fe donor ferric ammonium citrate (FAC), there was a significant decrease in IRP-RNA binding activity in both SK-N-MC and BE-2 cells compared with the controls (Fig 4). In contrast, after 20 hours of incubation with FAC, the decrease in IRP-RNA binding activity was not so obvious when compared with the relevant control (Fig 4). This may be because the IRP-RNA binding activity of the controls is far less after 20 hours compared with 6 hours of incubation, suggesting that the control cells are Fe-replete after the longer incubation.
Further studies examined the dependence of incubation time with 311 and DFO on the RNA-binding activity of the IRPs (data not shown). Despite marked differences in the Fe chelation efficacy between 311 and DFO (Figs 2 and 3), activation of the IRPs was observed after 2 to 4 hours of incubation with both these chelators in BE-2 and SK-N-MC cells (for example, see Fig 6).
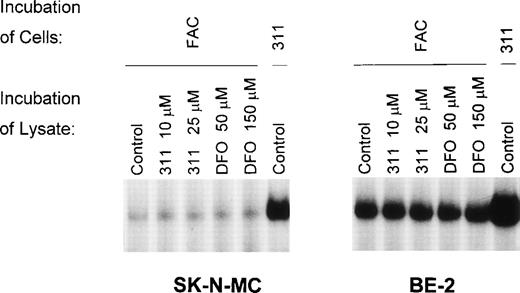
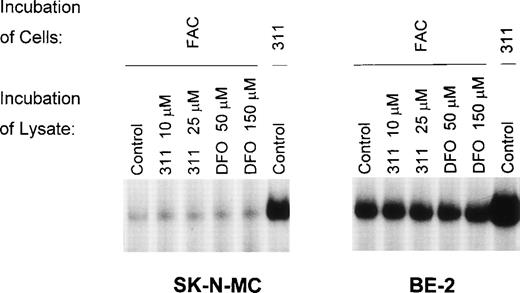
To determine if 311 (10 or 25 μmol/L) or DFO (50 or 150 μmol/L) could remove Fe from the FeS cluster of IRP1, we examined the ability of the chelators to increase IRP-RNA binding activity by incubating DFO or 311 with lysates prepared from cells pretreated with FAC (100 μg/mL) or 311 (25 μmol/L) for 20 hours (Fig 5). Using lysates prepared from cells preincubated with FAC for 20 hours, the IRPs only weakly bind the32P-labeled IRE compared with lysates derived from cells that had been preincubated with 311 (25 μmol/L; Fig 5). These data suggest that, as observed previously,28 29 after preincubation of cells with FAC, much of the IRP1 present has an FeS cluster. Incubation of these lysates for 30 minutes at 4°C with 311 or DFO had no effect at increasing the RNA-binding activity of the IRPs (Fig 5), suggesting that neither chelator could directly remove Fe from the Fe-S cluster of IRP1. Longer incubation periods with the chelators (up to 20 hours) also did not increase IRP-RNA binding activity (data not shown).
The effect on IRP-RNA-binding activity of incubating cell lysates with either Munro buffer (control) or Munro buffer containing either 311 (10 or 25 μmol/L) or DFO (50 or 150 μmol/L) for 30 minutes at 4°C. The lysates were derived from cells pretreated for 20 hours at 37°C with either FAC (100 μg/mL) or 311 (25 μmol/L). The result is a typical experiment from two experiments performed.
The effect on IRP-RNA-binding activity of incubating cell lysates with either Munro buffer (control) or Munro buffer containing either 311 (10 or 25 μmol/L) or DFO (50 or 150 μmol/L) for 30 minutes at 4°C. The lysates were derived from cells pretreated for 20 hours at 37°C with either FAC (100 μg/mL) or 311 (25 μmol/L). The result is a typical experiment from two experiments performed.
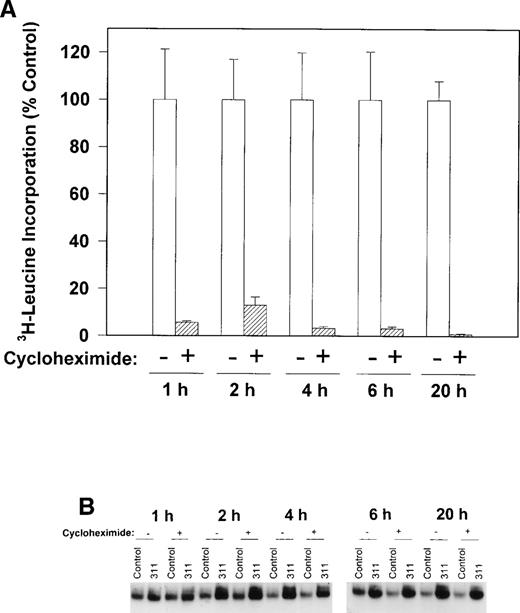
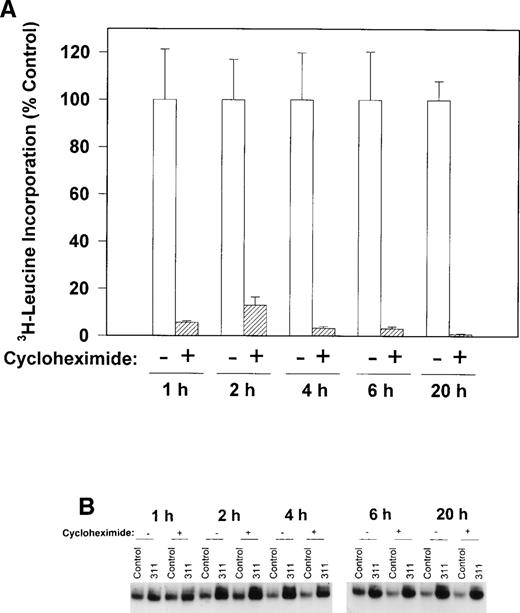
To determine if protein synthesis was necessary to increase IRP-RNA binding activity, we examined the ability of cycloheximide to inhibit3H-leucine incorporation into protein and prevent the increase in the RNA-binding activity of the IRPs during incubation with 311 in human SK-N-MC cells (Fig 6A and B). Cycloheximide (40 μg/mL) markedly reduced the incorporation of3H-leucine into protein after all incubation periods from 1 to 20 hours (Fig 6A) without having any effect on cellular viability. However, this agent had little to no appreciable effect on the increase in RNA-binding activity of the IRPs in the presence of 311 in this cell line (Fig 6B). These results suggest that protein synthesis was not responsible for the increase in RNA-binding activity of the IRPs after exposure to 311. Very similar results were also seen for the BE-2 cell line, and cycloheximide also had little effect on IRP-RNA binding activity during incubation of cells with DFO (data not shown).
The effect of the protein synthesis inhibitor, cycloheximide (40 μg/mL), on (A) 3H-leucine incorporation into protein over 1 to 20 hours and (B) on the RNA-binding activity of the IRPs during 1 to 20 hours of incubation with control medium (control) or 311 (25 μmol/L). The result in (A) is the mean ± SD of 5 replicates in a typical experiment from two experiments performed. The result in (B) is a typical experiment from four experiments performed.
The effect of the protein synthesis inhibitor, cycloheximide (40 μg/mL), on (A) 3H-leucine incorporation into protein over 1 to 20 hours and (B) on the RNA-binding activity of the IRPs during 1 to 20 hours of incubation with control medium (control) or 311 (25 μmol/L). The result in (A) is the mean ± SD of 5 replicates in a typical experiment from two experiments performed. The result in (B) is a typical experiment from four experiments performed.
Effect of DFO and 311 on the expression of genes involved in iron metabolism, cell proliferation, and the cell cycle.
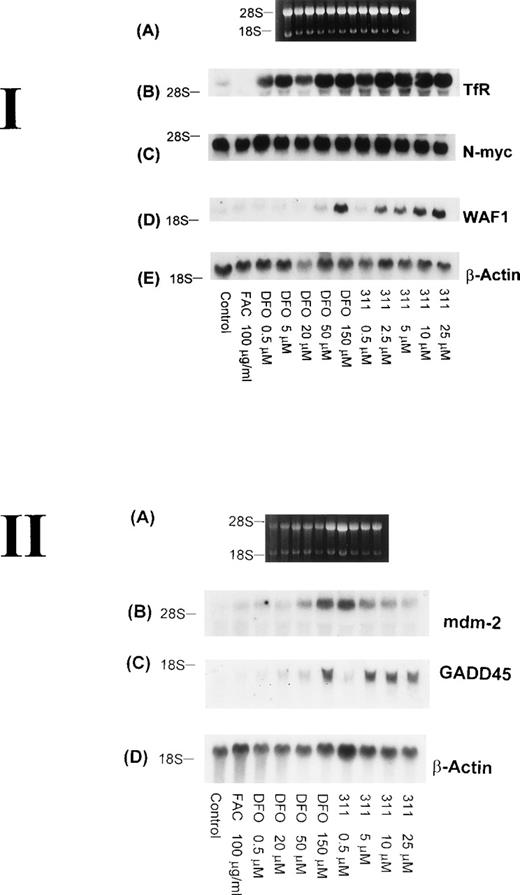
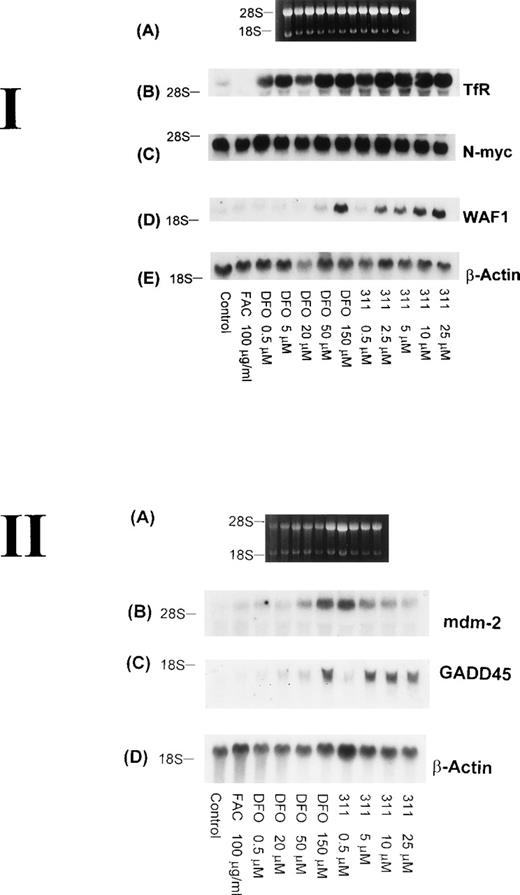
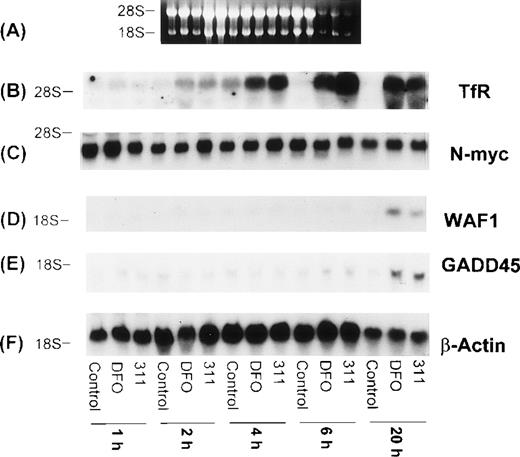
The TfR is necessary for the proliferation of most cells, because this molecule is involved in the uptake of Fe from Tf.29Although it is well known that TfR mRNA expression can be markedly upregulated by DFO, very little is understood concerning the effect of 311, which has antiproliferative activity. Hence, it was important to understand the effect of 311 on TfR mRNA levels. After 20 hours of incubation with increasing concentrations of DFO (0.5 to 150 μmol/L) or 311 (0.5 to 25 μmol/L), TfR mRNA levels increased in a concentration-dependent manner, with 311 being more effective than DFO on a molar basis (Fig 7 I-B). In contrast, incubation of cells with FAC (100 μg/mL) decreased TfR expression compared with the control. The increase in TfR mRNA levels after exposure to DFO and 311 was also time-dependent, with increased expression being observed only after 2 hours of incubation with the chelators (Fig 8B).
The effect of the concentration of DFO or 311 on mRNA levels of the TfR, N-myc, WAF1, β-actin, mdm-2, and GADD-45 in BE-2 neuroblastoma cells. (I-A) Ethidium bromide staining of the agarose gel; (I-B) TfR; (I-C) N-myc; (I-D) WAF1; (I-E) β-actin. (II-A) Ethidium bromide staining of the agarose gel; (II-B) mdm-2; (II-C) GADD45; (II-D) β-actin. The result shown is a typical experiment from three experiments performed.
The effect of the concentration of DFO or 311 on mRNA levels of the TfR, N-myc, WAF1, β-actin, mdm-2, and GADD-45 in BE-2 neuroblastoma cells. (I-A) Ethidium bromide staining of the agarose gel; (I-B) TfR; (I-C) N-myc; (I-D) WAF1; (I-E) β-actin. (II-A) Ethidium bromide staining of the agarose gel; (II-B) mdm-2; (II-C) GADD45; (II-D) β-actin. The result shown is a typical experiment from three experiments performed.
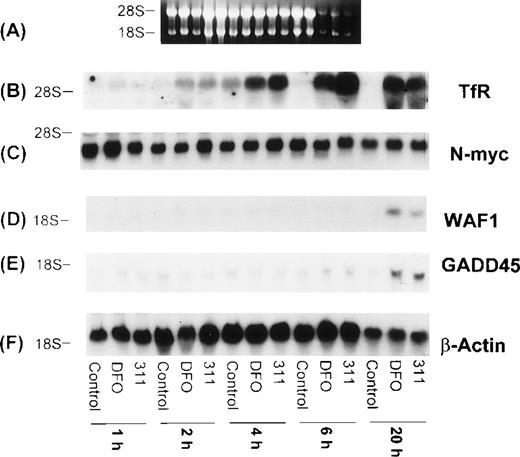
The effect of incubation time (1 to 20 hours) with DFO (150 μmol/L) or 311 (25 μmol/L) on mRNA levels of the TfR, N-myc, WAF1, GADD45, mdm-2, and β-actin in BE-2 neuroblastoma cells. (I-A) Ethidium bromide staining of the agarose gel; (I-B) TfR; (I-C) N-myc; (I-D) WAF1; (I-E) GADD45; (I-F) β-actin. The result illustrated is a typical experiment from three experiments performed.
The effect of incubation time (1 to 20 hours) with DFO (150 μmol/L) or 311 (25 μmol/L) on mRNA levels of the TfR, N-myc, WAF1, GADD45, mdm-2, and β-actin in BE-2 neuroblastoma cells. (I-A) Ethidium bromide staining of the agarose gel; (I-B) TfR; (I-C) N-myc; (I-D) WAF1; (I-E) GADD45; (I-F) β-actin. The result illustrated is a typical experiment from three experiments performed.
Increased expression of the proto-oncogene N-myc has been suggested to play a role in the malignant behavior of neuroblastoma.48 In several reports using neuroblastoma cells, treatment of a number of cell lines with DFO resulted in a decrease in the expression of N-myc mRNA, and this was thought to be due to a decrease in N-myctranscription.49 50 However, in our experiments using the BE-2 neuroblastoma cell line, neither DFO nor 311 had any effect onN-myc mRNA levels (Figs 7 I-C and 8C).
Although it is well known that DFO, 311, and other Fe chelators can cause cell cycle arrest,3,10,22,25 very little is understood concerning the mechanisms responsible for this activity. As part of our initial investigation into the molecular events involved in the antiproliferative effects of Fe chelators, we have examined the effects of 311 and DFO concentration on WAF1, GADD45, and human mdm-2 mRNA levels. These latter genes play crucial roles in cell cycle control and the repair of DNA damage.33 Interestingly, both Fe chelators caused an obvious concentration-dependent increase in the levels of WAF1 and GADD45 mRNA in BE-2 cells (Fig 7 I-D and II-C). Examining WAF1 and GADD45 mRNA levels, it is important to note that DFO only caused a marked increase in expression at a concentration of 150 μmol/L, whereas 311 increased the levels of these transcripts at concentrations as low as 2.5 to 5 μmol/L (Fig 7 I-D and II-C).
The increase in the levels of both the WAF1 and GADD45 transcripts in the presence of the chelators was time-dependent and only became appreciable after 20 hours of incubation of BE-2 cells with DFO (150 μmol/L) or 311 (25 μmol/L; Fig 8D and E). In contrast to the clear concentration- and time-dependent increase in WAF1 and GADD45 transcripts after exposure to DFO and 311, no such relationships were obvious for mdm-2 mRNA levels (Fig 7 II-B and data not shown). The elevation in the levels of WAF1 and GADD45 transcripts may be very important in terms of explaining the arrest in the cell cycle, which is known to occur after exposure to both DFO and 311.9,10 22Exactly comparable results to those observed for the BE-2 cell line in Figs 7 and 8 were also observed for the SK-N-MC and K562 cell lines (data not shown).
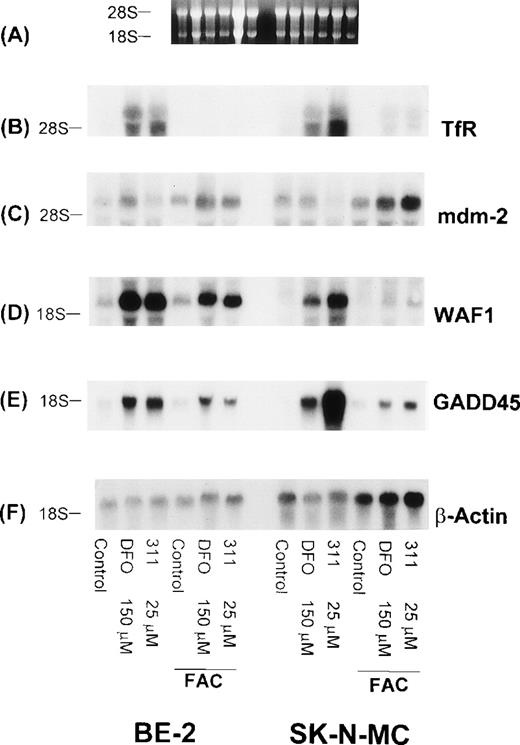
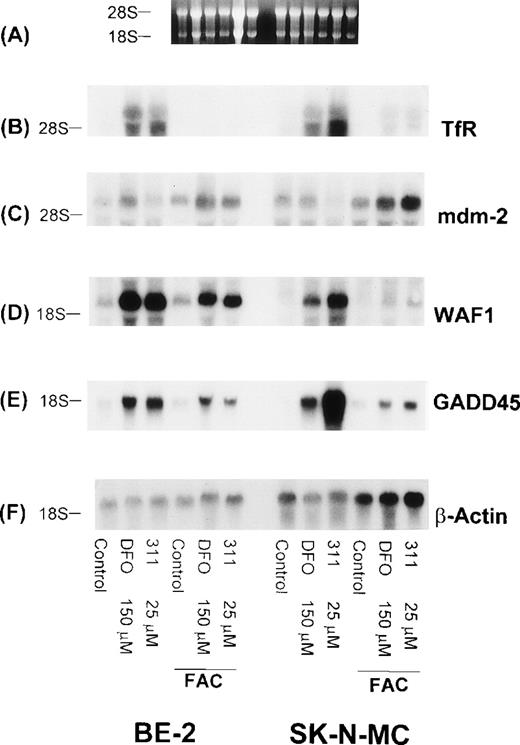
Further studies were designed to determine if the increase in the levels of WAF1 and GADD45 transcripts observed after 20 hours of exposure to chelators could be reversed by removing DFO (150 μmol/L) and 311 (25 μmol/L), extensively washing the cells (3 washes of the monolayer followed by two 30-minute incubations in MEM), and then reincubating them for 20 hours in the presence of medium containing 100 μg/mL of FAC as an Fe source (Fig 9). As shown above, 20 hours of incubation with DFO or 311 caused a marked increase in the levels of TfR, WAF1, and GADD45 mRNAs when compared with the relevant controls (Fig 9). After removal of the chelators and reincubation in the presence of FAC (100 μg/mL), there was a decrease in the levels of WAF1, GADD45, and TfR mRNA transcripts in both the BE-2 and SK-N-MC cell lines, whereas no such change was observed in the level of mdm-2 mRNA (Fig 9). It is of interest to note that reincubation of SK-N-MC cells in the presence of FAC caused a much larger decrease in WAF1 and GADD45 mRNA levels than that found for the BE-2 cell line (Fig 9).
The effect on mRNA levels of the TfR, mdm-2, WAF1, GADD45, and β-actin after exposure of SK-N-MC and BE-2 cells to DFO (150 μmol/L) or 311 (25 μmol/L) for 20 hours, and also cells exposed to these agents for 20 hours followed by reincubation for 20 hours in medium containing FAC (100 μg/mL). (A) Ethidium bromide staining of the agarose gel; (B) TfR; (C) mdm-2; (D) WAF1; (E) GADD45; (F) β-actin. The result shown is a typical experiment from two experiments performed.
The effect on mRNA levels of the TfR, mdm-2, WAF1, GADD45, and β-actin after exposure of SK-N-MC and BE-2 cells to DFO (150 μmol/L) or 311 (25 μmol/L) for 20 hours, and also cells exposed to these agents for 20 hours followed by reincubation for 20 hours in medium containing FAC (100 μg/mL). (A) Ethidium bromide staining of the agarose gel; (B) TfR; (C) mdm-2; (D) WAF1; (E) GADD45; (F) β-actin. The result shown is a typical experiment from two experiments performed.
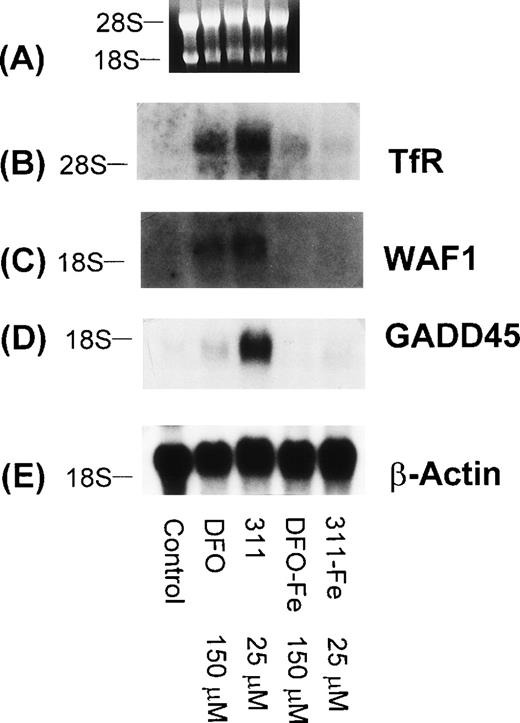
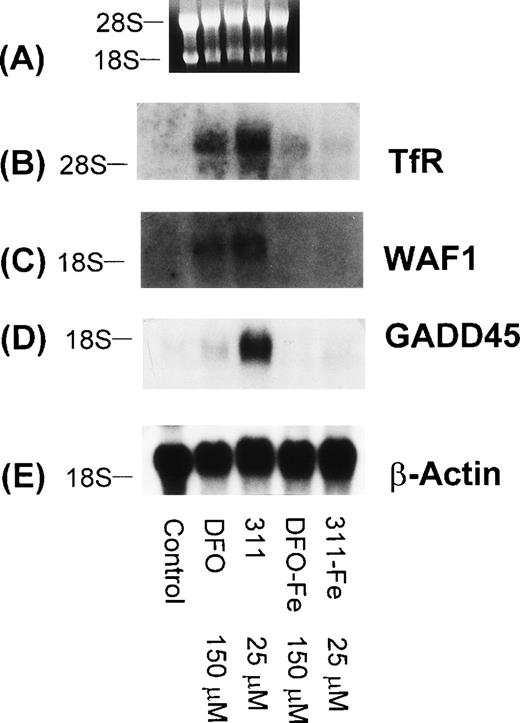
To determine that the increase in WAF1 and GADD45 mRNA levels were due to the ability of DFO and 311 to bind Fe, the Fe(III) complexes of these ligands (DFO-Fe, 311-Fe) were prepared and then incubated with SK-N-MC cells for 20 hours (Fig 10). In contrast to DFO (150 μmol/L) and 311 (25 μmol/L) that increased the levels of the TfR, GADD45, and WAF1 mRNAs when compared with the controls, the Fe complexes of these ligands at the same concentration had no appreciable effect (Fig 10). Very similar results were also seen for the BE-2 cell line (data not shown).
The effect of DFO, 311, and their iron(III) complexes (DFO-Fe or 311-Fe) on the levels of TfR, WAF1, GADD45, and β-actin mRNAs in SK-N-MC cells. (A) Ethidium bromide staining of the agarose gel; (B) TfR; (C) WAF1; (D) GADD45; (E) β-actin. The result shown is a typical experiment from two experiments performed.
The effect of DFO, 311, and their iron(III) complexes (DFO-Fe or 311-Fe) on the levels of TfR, WAF1, GADD45, and β-actin mRNAs in SK-N-MC cells. (A) Ethidium bromide staining of the agarose gel; (B) TfR; (C) WAF1; (D) GADD45; (E) β-actin. The result shown is a typical experiment from two experiments performed.
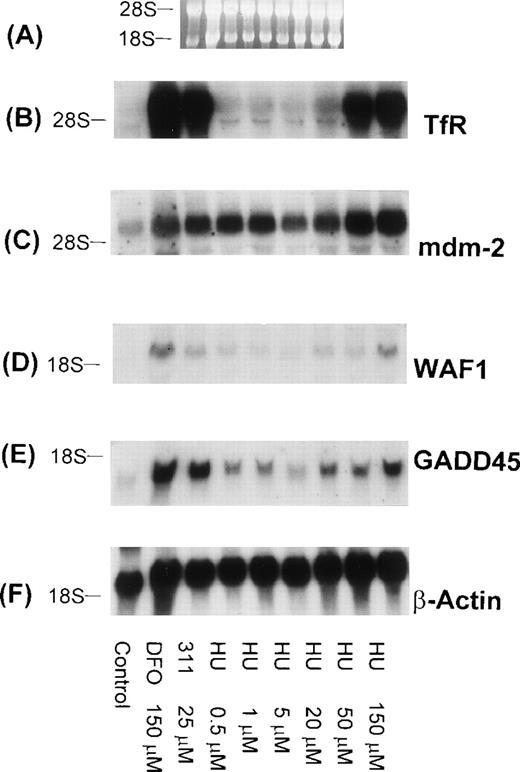
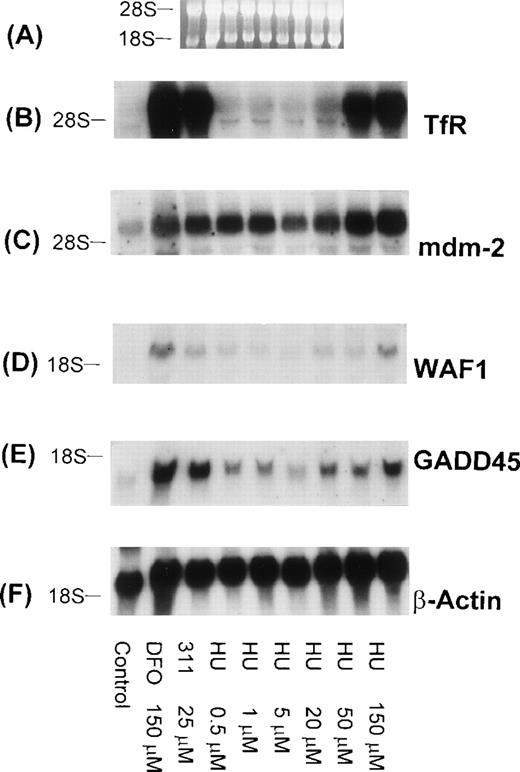
It is well known that DFO is an inhibitor of ribonucleotide reductase activity, and our previous studies have demonstrated that 311 can markedly inhibit 3H-thymidine incorporation, being far more effective than DFO.22 Considering that both GADD45 and WAF1 can be transactivated by p53 and that the levels of p53 can be increased by a decline in deoxyribonucleotide levels,51 we examined the effect of the potent ribonucleotide reductase inhibitor, hydroxyurea (HU), on TfR, mdm-2, WAF1, GADD45, and β-actin mRNA levels in BE-2 cells (Fig 11). As found for 311 (25 μmol/L) and DFO (150 μmol/L), both WAF1 and GADD45 mRNA levels were increased after exposure to HU, whereas little effect was observed on the level of mdm-2 mRNA (after normalization to β-actin; Fig 11C). Interestingly, as found in a lymphoblastic leukemic cell line,47 high concentrations of HU (50 and 150 μmol/L) were found to increase TfR mRNA levels (Fig 11B).
The effect of the concentration of HU on mRNA levels of the TfR, mdm-2, WAF1, GADD45, and β-actin in BE-2 neuroblastoma cells. (A) Ethidium bromide staining of the agarose gel; (B) TfR; (C) mdm-2; (D) WAF1; (E) GADD45; (F) β-actin. The result shown is a typical experiment from two experiments performed.
The effect of the concentration of HU on mRNA levels of the TfR, mdm-2, WAF1, GADD45, and β-actin in BE-2 neuroblastoma cells. (A) Ethidium bromide staining of the agarose gel; (B) TfR; (C) mdm-2; (D) WAF1; (E) GADD45; (F) β-actin. The result shown is a typical experiment from two experiments performed.
DISCUSSION
Although treatment of cells with Fe chelators is known to inhibit ribonucleotide reductase activity26 and result in a G1/S block,3,10,22 very little is known about the changes in gene expression that may play an important role in cell cycle arrest. WAF1 is a universal inhibitor of cyclin-dependent kinases and has been shown to be crucial in terms of mediating cell cycle arrest at G1/S.30-32 In addition, GADD45 is induced upon DNA damage and can also arrest the cell cycle.33,35 In the present study, we clearly demonstrate a concentration-dependent increase in WAF1 and GADD45 mRNA expression after exposure to both DFO and 311 (Figs 7 I-D and II-C and 9D and E), which was reversible after these chelators were removed (Fig 9). Importantly, there was a marked difference in the concentration of DFO and 311 that were required to increase WAF1 and GADD45 mRNA levels (Fig7 I-D and II-C). For example, the concentrations of DFO and 311 required to obtain a marked increase in WAF1 and GADD45 transcripts were 150 μmol/L and 2.5 to 5 μmol/L, respectively. This variation in activity corresponds to the differences in Fe chelation efficacy (Figs 2 and 3) and antiproliferative effects of these chelators.21 22
The effect of the ligands at increasing GADD45 and WAF1 mRNA levels were very similar in 3 different human cell lines, namely SK-N-MC neuroepithelioma cells, BE-2 neuroblastoma cells, and K562 erythroleukemia cells. Previous studies have demonstrated that SK-N-MC cells do not have functional p53, probably due to a deletion in the gene,52,53 and it is unknown whether the BE-2 cell line has wild-type p53. In addition, it has been demonstrated that the K562 cell line does not express wild-type p53 at the mRNA or protein level,54,55 whereas we have observed a marked increase in GADD45 and WAF1 mRNA levels in K562 cells after exposure to 311 and DFO (data not shown). Hence, at least for the SK-N-MC and K562 cell lines, the increase in GADD45 and WAF1 mRNA levels after exposure to chelators may be via a p53-independent pathway. Indeed, whereas it is well known that p53 activates transcription of WAF1 and GADD45 by its binding to the p53-binding site in their respective promoters, transcriptional activation of these genes can also occur by a p53-independent process.38,39,56,57 However, the mechanisms involved in p53-independent regulation of WAF1 and GADD45 expression are not well understood and may occur by multiple pathways, depending on the stimulus.39
Complexation of DFO or 311 with Fe prevented the effect of these chelators at increasing the expression of GADD45 and WAF1 mRNAs (Fig10). These results suggest that the effect of 311 and DFO at increasing the levels of these latter transcripts were due to their ability to chelate Fe from cells. In addition, the Fe complexes of both 311 and DFO did not inhibit cellular proliferation, in marked contrast to the ligands.41 It can be speculated that the effect of these chelators at increasing the levels of GADD45 and WAF1 mRNAs may be due to intracellular Fe chelation, which inhibits ribonucleotide reductase activity and decreases deoxyribonucleotide levels. Our previous studies with 311 and DFO have shown that both chelators inhibited the incorporation of 3H-thymidine into DNA, but 311 was far more effective.22 It has been demonstrated that a decline in the intracellular concentration of deoxyribonucleotides increases p53 levels that can transactivate WAF1 and GADD45.51However, whether a similar stimulus can increase WAF1 and GADD45 mRNA levels by p53-independent pathways remains unclear. The fact that treatment of cells with the potent ribonucleotide reductase inhibitor HU also increased WAF1 and GADD45 mRNA levels (Fig 11) lends support to our suggestion that inhibition of ribonucleotide reductase activity may be a regulatory step. As a working hypothesis that will be tested in future studies, we suggest that the ligands examined in the present investigation may act by chelating intracellular Fe, which then results in a decrease in ribonucleotide reductase activity. This decrease in enzyme activity causes a decline in deoxyribonucleotide levels that leads to an increase in the expression of WAF1 and GADD45 that act to inhibit the cell cycle. Obviously, other mechanisms of regulation are also possible, and further studies are essential to establish the precise molecular processes involved.
The Fe chelation experiments performed in the present study directly demonstrate the much greater efficacy of 311 compared with DFO in terms of its ability to increase Fe release and inhibit Fe uptake from Tf (Figs 2 and 3). Our studies have shown that both 311 and DFO are far less effective at mobilizing 59Fe from cells that have been labeled with 59Fe-Tf for 24 hours compared with 3 hours (Fig 2A and B). Because a greater proportion of Fe is found in ferritin after long labeling periods with Tf than short incubation times,22 these results suggest that both DFO and 311 chelate a labile form of 59Fe. We have also found that there was variation in the ability of DFO to chelate Fe from the SK-N-MC and BE-2 cell lines, in contrast to 311, which was equally efficient in both (Figs 2 and 3). This may be due to differences in the ability of DFO or its Fe complex to permeate the cell membrane or may suggest a difference in the Fe metabolism between these cell types.
Because chelator 311 was more effective than DFO in terms of its ability to chelate Fe from BE-2 and especially SK-N-MC cells (see Figs2 and 3), we compared the efficacy of each chelator at activating the RNA-binding activity of the IRPs. Both DFO and 311 took 2 to 4 hours to increase the RNA-binding activity of the IRPs (Fig 6 and data not shown). These results suggest that, irrespective of Fe chelation efficacy of the ligands, there was a time-dependent step that was required before IRP-RNA binding activity can increase. The protein synthesis inhibitor cycloheximide had little to no effect on the increase in the IRP-RNA binding activity in the presence of 311, suggesting that de novo protein synthesis was not required to increase RNA-binding activity. At present, the evidence regarding how an increase in IRP-RNA binding activity occurs in the presence of DFO is controversial, with several studies being performed in mouse Ltk− cells.45,58 Using cycloheximide as the protein synthesis inhibitor, one group of investigators suggested that protein synthesis was necessary to increase IRP-RNA binding activity in the presence of DFO.45In contrast, Pantopolous et al,58 using the same cell line, have shown that, in the presence of DFO, cycloheximide did not prevent activation of IRP-1 but completely blocked the appearance of IRP2-RNA binding activity. These latter results have led to the suggestion that the Fe-S cluster in IRP1 can be removed by some uncharacterized mechanism to generate an active RNA-binding protein.28Further studies are obviously essential to determine the process responsible for the increase in IRP-RNA binding activity in the presence of Fe chelators.
Neither DFO nor 311 when added to lysates from Fe-replete cells increased the RNA-binding activity of the IRPs (Fig 5). This result indicated that neither chelator acted directly on the cluster to remove Fe and increase RNA-binding activity. Therefore, the activation of RNA-binding activity of the IRPs by these chelators may be due to an indirect effect caused by the depletion of Fe in a labile intracellular pool. Indeed, we have shown that both 311 and DFO act on the same or a similar labile pool of Fe that markedly decreases as the incubation time with Tf increased (Fig 2A and B). Our present results using 311 and DFO are in agreement with previous investigations in which very high concentrations of DFO (10 mmol/L) added to purified IRP1 only resulted in a slight increase in RNA-binding activity.59Moreover, the results concur with studies showing that the Fe-S cluster of IRP1 is highly stable, requiring high concentrations of ferricyanide and EDTA to remove it.60
In summary, we have demonstrated that 311 is a highly effective Fe chelator compared with DFO. Despite the far greater Fe chelation efficacy of 311 compared with DFO, the increase in IRP-RNA binding activity in the presence of these chelators occurred by similar kinetics and was not inhibited by cycloheximide. Our studies demonstrate that DFO and 311 markedly increase WAF1 and GADD45 mRNA levels in a concentration- and time-dependent manner. However, only very low concentrations of 311 (2.5 to 5 μmol/L) compared with DFO (150 μmol/L) were required to induce this effect, in direct accordance with the difference in the Fe chelation efficacy and antiproliferative activity of these ligands. We suggest that, because both WAF1 and GADD45 are critical in cell cycle arrest, the increased expression of these molecules may play an important role in the antiproliferative activity of these chelators.21 22
ACKNOWLEDGMENT
Dr Greg Anderson and Lex Cowley are thanked for their generous assistance in setting up the IRP gel retardation assay.
Supported by a project grant from the National Health and Medical Research Council of Australia (Grant No. 970360), the Kathleen Cuningham Breast Cancer Research Foundation, and a Research Fellowship/Senior Research Fellowship from the Department of Medicine, University of Queensland (to D.R.R.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to D.R. Richardson, PhD, Department of Medicine, Clinical Sciences Building Floor C, Royal Brisbane Hospital, Brisbane, Australia 4029; e-mail:D.Richardson@medicine.herston.uq.edu.au.