Abstract
Paired immunoglobulin-like receptors (PIR) are expressed on B cells and macrophages and include inhibitory and putative activating receptors referred to as PIR-B and PIR-A, respectively. Although PIR-B’s inhibitory pathway has been described, it is unknown whether PIR-A receptors can deliver activation signals to macrophages, and if so, through what mechanism. Here we use chimeric receptors to address the mechanisms of PIR-A signaling. Cotransfection of chimeric receptors comprised of the extracellular region of human CD4 and the transmembrane and cytoplasmic domains of murine PIR-A3 showed the ability of PIR-A3 to physically interact with the FcɛRIγ chain in 293T cells. This interaction is dependent on Arg632 within the PIR-A3 transmembrane domain. We also demonstrate PIR-A3 interaction with the endogenous FcɛRIγ of the ANA-1 macrophage cell line, again in an Arg632-dependent manner. Furthermore, we show that crosslinking of these chimeric receptors synergizes with IFN-γ in the production of nitric oxide. Our data are the first to show the potential of PIR-A3 to deliver activation signals to macrophages and establish its dependence on Arg632. These findings suggest that further study of the PIR-A receptors should be aggressively pursued toward a complete understanding of the intricate regulation of macrophage biology.
THE MODULATION of immune cell activation involves the coordinate regulation of inhibitory and stimulatory signaling pathways triggered by various cell surface receptors. The intracellular immunoreceptor tyrosine-based inhibitory motif (ITIM), defined as I/VxYxxL/I, is present in the cytoplasmic tails of many inhibitory receptors.1 ITIM-containing receptors described thus far include several members of the Ig superfamily (killer inhibitory receptor [KIR], paired Ig-like receptor [PIR], and immunoglobulin-like transcript [ILT] families2-9), as well as the c-type lectin superfamily (Ly49 and NKG2 families10 11).
Each of these families of inhibitory receptors (leukocyte inhibitory receptor [LIR], PIR, KIR, and Ly49) contain members that lack ITIMs in their cytoplasmic domains.2,5,7,10-12 Instead, these receptors contain a charged amino acid within their transmembrane domains in a fashion similar to the T-cell receptor (TCR) and some Fc receptors (FcR), suggesting that they may also transmit activation signals.13 The TCR and these FcR both lack intrinsic catalytic activity and therefore, transmit their activation signals via receptor-associated chains such as the CD3 invariant complex (including TCRζ) and FcεRIγ, respectively.14,15 Association of these signaling chains with the receptor complex is dependent on the presence of oppositely charged amino acid residues within the transmembrane region of the receptor and the signal transducing chain.16,17 Upon receptor ligation, the tyrosine residues within the immunoreceptor tyrosine-based activation motif (ITAM)s of the signaling chains are phosphorylated and initiate intracellular signaling events.17 18
The expression of the recently described PIR family is restricted to B and myeloid cells.7,8 This receptor family was discovered while searching for the murine homolog of the human Fcα receptor (FcαR) gene. Within the PIR family, two main isoforms have been described. The PIR-B receptor is an inhibitory receptor containing four cytoplasmic ITIMs. In contrast, the PIR-A receptors (PIR-A1-7) are putative activation receptors that lack both ITIMs and ITAMs and have a positively charged amino acid in their transmembrane domains.7
The PIR-A receptors of macrophages have a high degree of homology with FcRs, which have been well characterized as activating receptors. For example, stimulation of human monocytes through the FcαR activates a variety of biological responses, including tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), IL-8, and IL-1.19,20 Similarly, stimulation of murine macrophages through FcγR results in the production of nitric oxide (NO).21 22 These findings suggest that PIR-A may also be capable, through the interaction with a signal transduction chain, of activating macrophages to produce inflammatory cytokines and/or reactive nitrogen intermediates. However, the lack of reagents able to discriminate between the highly homologous PIR-A and PIR-B has hampered the effort to determine whether the PIR-A receptors might also regulate macrophage function. Here, we use a chimeric receptor approach to show that PIR-A3 physically interacts with FcεRIγ in murine macrophages via Arg632. Moreover, we show that PIR-A3 is capable of inducing the production of NO, a major mediator of macrophage tumoricidal and microbicidal activity, in a manner dependent on Arg632 and therefore, FcεRIγ. These findings show for the first time the potential impact of the PIR-A receptors on the regulation of immunity through their control of macrophage function.
MATERIALS AND METHODS
Cells.
The 293T human kidney embryonic cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The mouse macrophage cell line ANA-1 was established by infecting fresh bone marrow–derived cells from C57BL/6 mice with the J2 (v-raf/v-myc) recombinant retrovirus and has been extensively characterized.23 24 ANA-1 macrophages were cultured in DMEM containing 5% fetal bovine serum, glutamine, and antibiotics as above. Stably transfected ANA-1 cell lines were maintained as above with the addition of 200 μg/mL G418.
Antibodies.
The mouse monoclonal antibody recognizing human CD4 was purchased from Becton Dickinson Immunocytology Systems (San Jose, CA). Biotinylated 4G10 antibody, recognizing phosphotyrosine, and streptavidin conjugated with horseradish peroxidase were purchased from Upstate Biotechnologies, Inc (Lake Placid, NY). The polyclonal antibody recognizing murine FcεRIγ was a gift from Dr J.P. Kinet (Harvard University, Boston, MA). Goat F(ab′)2 against mouse IgG used in receptor crosslinking studies was purchased from Cappel (Durham, NC).
Plasmids.
The human CD4 cDNA was a gift from Dr Richard Axel via the AIDS (acquired immunodeficiency syndrome) Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).25 The murine PIR-A3 cDNA was identified by searching an Expressed Sequence Tag (EST) database using the National Cancer Institute (NCI) Blast program (Wisconsin Package Version 10.0, GCG, Madison, WI). The query sequence was the murine PIR-A1 sequence reported by Kubagawa et al.7 The EST (GenBank Accession NumberAA184287) was purchased from ATCC (Rockville, MD). The CD4/PIR-A3 chimeric construct was prepared by polymerase chain reaction (PCR) amplification of cDNA with the addition of aVspI linker. The PCR primers used to amplify the extracellular domain of human CD4 cDNA were 5′ ATGCGGTACCATTTCTGTGGGCTCAG 3′ (forward primer, adding a 5′ Kpn I linker) and the reverse primer 5′ CTAGAATTAATGCCATTGGCTGCACCGG 3′ (adding a 3′ VspI linker). The PCR primers used to amplify the cDNA representing the transmembrane and intracellular regions of murine PIR-A were 5′ GATCATTAATCAGGATGGGGATGGCA 3′ (adding a 5′ VspI linker) and 5′ CGTAGAATTCAGAGTGTAGAACATTGA 3′ (reverse primer, adding a 3′EcoRI linker). The cDNAs were subcloned into the KpnI/EcoRI site of the pcDNA3 expression vector (Promega, Madison, WI) and sequenced using the T7 Sequenase version 2.0 DNA sequencing kit (United States Biochemical, Cleveland, OH). The resulting receptor carries the extracellular domain of human CD4 and the transmembrane and cytoplasmic domain of PIR-A3 with the exception of a conservative substitution of valine for isoleucine within the transmembrane domain. The mutant chimeric receptor construct was generated using the Transformer Site-Directed Mutagenesis Kit (Clontech, Palo Alto, CA), in which the arginine corresponding to Arg632 in the PIR-A3 receptor sequence was mutated to Leu (CD4/PIR-A3R632L). The murine FcεRIγ expression construct was a gift from Dr J.P. Kinet (Harvard University).26 27
Transfections.
One day before transfection, 293T cells were seeded in six-well plates at a density of 3 × 105/well and cultured overnight. Cells were then transfected with the indicated amounts and combinations of constructs using the FuGENE 6 Transfection reagent as directed by the manufacturer (Boehringer Mannheim, Indianapolis, IN). The total amount of cDNA per transfection was 1 μg and was normalized by the addition of pcDNA3 plasmid. ANA-1 macrophages (10 × 106) were transfected with 10 μg of the indicated constructs using a Gene Pulser electroporator (BioRad, Hercules, CA). After electroporation, the cells were allowed to recover for 3 days in complete media. Cells were then selected in 1 mg/mL G418 for approximately 2 weeks, followed by subcloning. Expression of the chimeric receptors was monitored by flow cytometry.
Biotinylation, stimulation, immunoprecipitation, electrophoresis, and blotting.
Where specified, transfected 293T cells were biotinylated using the Cellular Labeling and Immunoprecipitation Kit from Boehringer Mannheim, as directed. Cells were stimulated with 1 mmol/L pervanadate for 15 minutes at 37°C as described.28 After stimulation, the cells were pelleted, the supernatant was removed, and the cells were lysed in 1 mL lysis buffer (1% Triton X-100 [TTX-100] or 1% Brij-96, and 50 mmol/L Tris, pH 7.4, 300 mmol/L NaCl, 2 mmol/L EDTA, 0.4 mmol/L Na3VO4, 2.5 mmol/L leupeptin, 2.5 mmol/L aprotinin, and 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF]).28 Lysates were clarified by centrifugation and were immunoprecipitated with the specified antibody bound to Protein G or Protein A agarose beads for 2 to 3 hours at 4°C. Immune complexes were washed in lysis buffer containing 0.1% TTX-100 or 0.1% Brij-96. Proteins were eluted in either 2X nonreducing or reducing Laemmli sample buffer as indicated and separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to Immobilon-P membrane (Millipore, Bedford, MA) and were blocked in phosphate-buffered saline (PBS) containing 5% bovine serum albumin (BSA) and 0.1% Tween-20 (Sigma, St Louis, MO). Biotinylated 4G10 antibody followed by streptavidin-conjugated horseradish peroxidase (SA-HRP) was used in the detection of phosphoproteins. Western blot analysis using FcεRIγ antisera followed by a horseradish peroxidase–linked goat antirabbit Ig (Boehringer Mannheim) was performed using Tris-buffered saline containing 5% nonfat milk and 0.1% Tween-20. Blots were developed using an enhanced chemiluminescence kit (ECL; Amersham, Arlington Heights, IL) and were exposed to XAR-5 film (Kodak, Rochester, NY).
Cell stimulation with CD4 antibody.
In experiments using anti-CD4 as a stimulus, 1 μg/mL goat antimouse IgG in Hanks’ Balanced Salt Solution (HBSS) was used to coat the wells of a 24-well plate at 4°C overnight. The wells were washed two times with HBSS before the addition of reagents and cells. The following day, cells were washed in 0.1% fetal bovine serum (FBS) in Dulbecco’s phosphate buffered saline (DPBS), resuspended at 1 × 106/mL, and incubated on ice for 1 hour with 40 μL anti-CD4 per 1 × 106 cells. After the incubation period, the cells were washed with cold DPBS and cultured in complete media at 1 × 106/mL overnight with any additional reagents as described below.
NO2− assay.
The accumulation of nitrite (NO2−) in macrophage culture supernatants was used as a relative measurement of NO production.29 Transfected ANA-1 macrophages were cultured at a concentration of 1 × 106 cells/well in a 24-well microtiter plate containing a total volume of 1 mL. After the indicated stimulations, 50 μL of cell-free culture supernatant was incubated with 50 μL Griess reagent (1% sulfanilimide/0.1% naphthylenediamine dihydrochloride/2.5% H3PO4) at room temperature for 5 minutes, and the absorbance at 550 nm was determined in a MR500 microplate reader (Dynex Technologies, Inc, Chantilly, VA). The concentration of NO2− was determined from a least squares linear regression analysis of a sodium nitrite standard curve generated with each experiment.
RESULTS AND DISCUSSION
CD4/PIR-A3 associates with FcεRIγ in 293T cells.
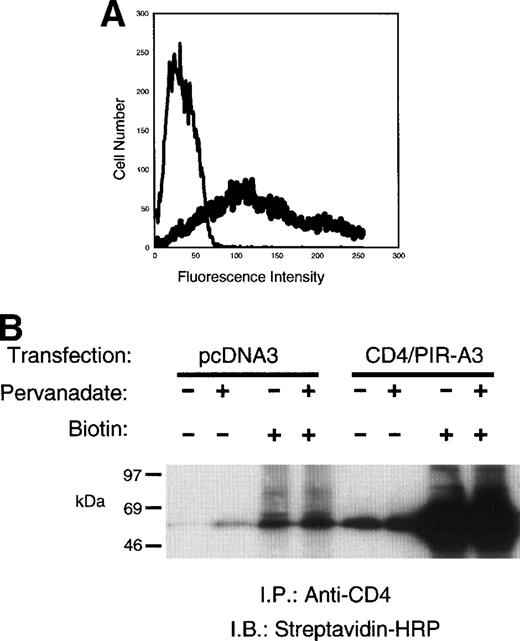
The presence of a charged transmembrane arginine residue and the lack of an ITAM in the cytoplasmic domain in the PIR-A receptors suggest the ability to associate with a signal transducing chain.7 The signal transduction chains associated with multichain immune recognition receptors such as the TCR, B-cell receptor (BCR), FcR (reviewed in Keegan et al18), and, most recently, the noninhibitory KIR30 and Ly4931 32 proteins, have been described. However, the lack of specific reagents for the putative activating PIR-A versus the inhibitory PIR-B has precluded the study of possible signaling chains that might interact with these new receptors. In fact, there are no data to show the ability of PIR-A receptors to lead to a functional outcome in any cell, suggesting that they may be incapable of delivering enough of an intracellular signal to result in gene transcription, cytokine production, or cytotoxicity. Therefore, to directly examine the ability of the PIR-A receptors to form a signal-transducing complex in macrophages, we constructed a chimeric receptor (CD4/PIR-A3) that contains the extracellular domain of human CD4 linked to the transmembrane and cytoplasmic domains of PIR-A3 (Fig 1). The construct (0.5 μg) was transiently transfected into 293T cells, and the expression level of the CD4/PIR-A3 was assessed by flow cytometry using phycoerythrin (PE)-conjugated anti-human CD4 monoclonal antibody (MoAb) (Fig 2A). Transfection with these concentrations of plasmid routinely results in more than 70% of the cells expressing the chimera. To confirm the recognition of CD4/PIR-A3 by the human CD4 MoAb for biochemical studies, 293T cells transiently transfected with the receptor were surface labeled with biotin, lysed, and immunoprecipitated with anti-CD4. The immune complexes were separated by SDS-PAGE and detected with SA-HRP. Figure 2B shows the immunoprecipitation of a labeled protein at approximately 48 kD consistent with the predicted mass of the CD4/PIR-A3 chimeric receptor. Moreover, these data show that treatment of the cells with pervanadate before lysis and immunoprecipitation does not affect the ability of anti-CD4 to immunoprecipitate chimeric receptors.
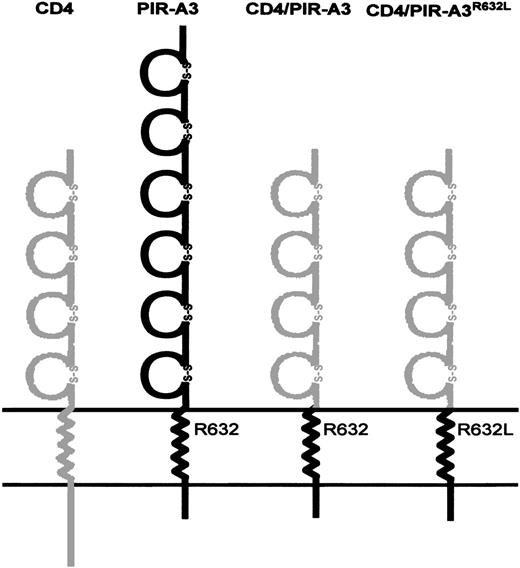
Schematic diagram of CD4/PIR-A3 chimeric receptors. The four Ig-like extracellular domains of human CD4 were linked to the transmembrane and cytoplasmic domains of murine PIR-A3. The transmembrane region of PIR-A3 contains a positively charged arginine at position 632. The gray and black areas represent human CD4 and murine PIR-A3, respectively.
Schematic diagram of CD4/PIR-A3 chimeric receptors. The four Ig-like extracellular domains of human CD4 were linked to the transmembrane and cytoplasmic domains of murine PIR-A3. The transmembrane region of PIR-A3 contains a positively charged arginine at position 632. The gray and black areas represent human CD4 and murine PIR-A3, respectively.
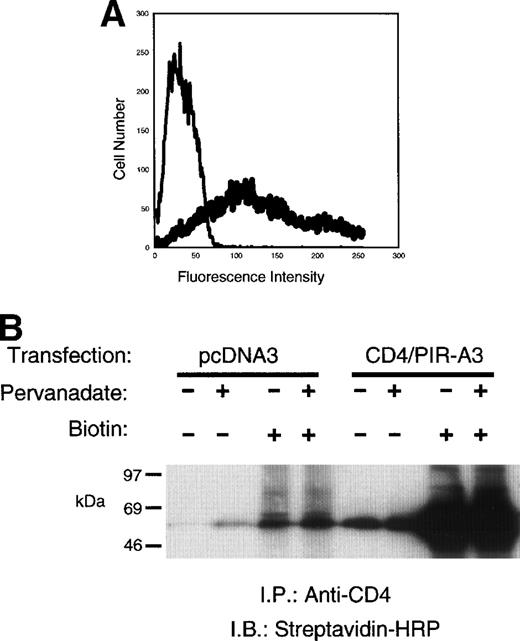
CD4/PIR-A3 is expressed on 293T cells and is recognized by anti-human CD4. (A) Expression of CD4/PIR-A3 in 293T cells. 293T cells were transfected with 0.5 μg of either pcDNA3 (thin trace) or CD4/PIR-A3 (thick trace) cDNA. Twenty-four hours after transfection, the cells were labeled with PE-conjugated anti-human CD4 for flow cytometric analysis. (B) Anti-CD4 immunoprecipitates CD4/PIR-A3. Twenty-four hours after transfection, 293T cells were surface labeled with biotin followed by pervandate stimulation, as indicated. The cells were lysed in a buffer containing 1% TTX-100, and lysates were immunoprecipitated with anti-CD4. Immune complexes were eluted in reducing Laemmli sample buffer, separated by 8% to 16% SDS-PAGE, and detected with SA-HRP and ECL.
CD4/PIR-A3 is expressed on 293T cells and is recognized by anti-human CD4. (A) Expression of CD4/PIR-A3 in 293T cells. 293T cells were transfected with 0.5 μg of either pcDNA3 (thin trace) or CD4/PIR-A3 (thick trace) cDNA. Twenty-four hours after transfection, the cells were labeled with PE-conjugated anti-human CD4 for flow cytometric analysis. (B) Anti-CD4 immunoprecipitates CD4/PIR-A3. Twenty-four hours after transfection, 293T cells were surface labeled with biotin followed by pervandate stimulation, as indicated. The cells were lysed in a buffer containing 1% TTX-100, and lysates were immunoprecipitated with anti-CD4. Immune complexes were eluted in reducing Laemmli sample buffer, separated by 8% to 16% SDS-PAGE, and detected with SA-HRP and ECL.
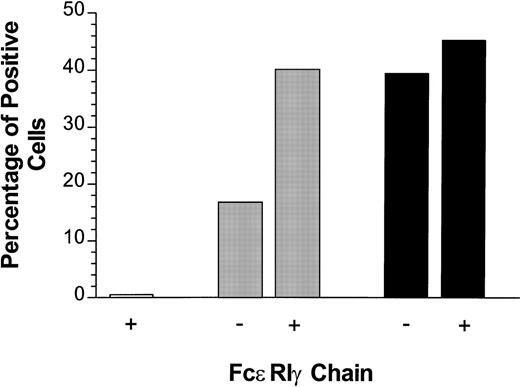
The PIR family was originally identified by screening mouse genomic and splenic cDNA libraries with the α chain of FcαR.7,8 As expected, the resulting receptors show high homology to several Ig-superfamily receptors, including human FcαR, bovine Fcγ, and the human KIRs. The transmembrane domain of PIR-A receptors is most homologous to those of the human FcαR, suggesting that the signaling mechanisms of these two receptors may be similar.7,32Recently, FcαR has been shown to use FcεRIγ.33,34Transfected FcαR is poorly expressed without cotransfection with FcεRIγ,33 and ligation of FcαR results in phosphorylation of FcεRIγ.34 Mutations of an arginine residue within the FcαR transmembrane domain prevented its association with FcεRIγ and ablated its signaling capability.33 Like FcαR, PIR-A3 contains an arginine in its transmembrane domain, and this residue is in the same relative position within the membrane-spanning segment of both receptors.7 In fact, they exist within the conserved sequence, LIRM. This suggested that PIR-A might use this arginine (Arg632) to couple physically and/or functionally to FcεRIγ. To test this hypothesis directly, we mutated Arg632 in CD4/PIR-A3 to Leu. The mutant chimeric receptor (CD4/PIR-A3R632L), when transiently transfected (0.5 μg) into 293T cells, is expressed at levels similar to CD4/PIR-A3, as assessed by flow cytometry (data not shown). Although both chimeric receptors can be expressed on the surface of 293T cells after transfection of large amounts of plasmid DNA (0.5 μg) in the absence of FcεRIγ, we considered the possibility that cotransfection of a signaling chain might enhance the cell surface expression of the receptors at lower plasmid DNA concentrations. To address this possibility, 50 ng of chimeric receptor cDNA was transfected in combination with 500 ng murine FcεRIγ cDNA into 293T cells. Expression of the chimeric receptors was monitored using anti-CD4. As shown in Fig 3, cotransfection of FcεRIγ increased the surface expression of CD4/PIR-A3 approximately threefold, while the expression of CD4/PIR-A3R632L was virtually unchanged. Interestingly, CD4/PIR-A3 expression in the presence of FcεRIγ was increased to approximately the same level as that of the mutated receptor. It is unclear whether the lower expression of FcαR and our chimera in the absence of FcεRIγ is the result of receptor being sequestered in the cytoplasm caused by an inability to integrate into the plasma membrane, or caused by degradation of uncoupled receptor similar to that seen with the chains of the TCR.16 Regardless, our results show that FcεRIγ likely interacts with the transmembrane domain of PIR-A3. Moreover, they suggest that Arg632 within the LIRM sequence may mediate both the interaction with FcεRIγ as well as the sequestration and/or degradation of receptor in the absence of coexpressed signal transduction chains.
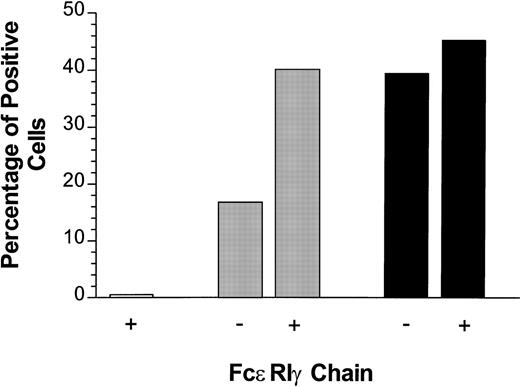
Cotransfection of FcɛRIγ with CD4/PIR-A3 enhances cell surface expression in 293T cells. Fifty nanograms of pcDNA3 (□), CD4/PIR-A3 (▩), or CD4/PIR-A3R632L (▪) cDNA were transiently transfected in the absence or presence of 500 ng murine FcɛRIγ cDNA. Twenty-four hours later, the cultures were labeled with PE-conjugated anti-human CD4 for flow cytometric analysis.
Cotransfection of FcɛRIγ with CD4/PIR-A3 enhances cell surface expression in 293T cells. Fifty nanograms of pcDNA3 (□), CD4/PIR-A3 (▩), or CD4/PIR-A3R632L (▪) cDNA were transiently transfected in the absence or presence of 500 ng murine FcɛRIγ cDNA. Twenty-four hours later, the cultures were labeled with PE-conjugated anti-human CD4 for flow cytometric analysis.
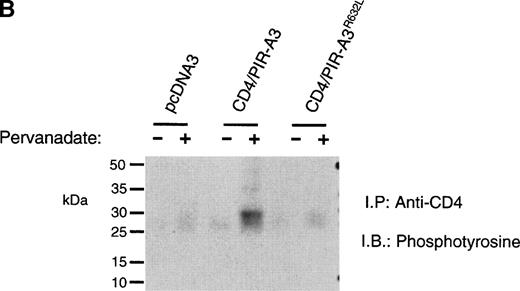
To show a physical interaction of FcεRIγ with CD4/PIR-A3, 293T cells were transiently transfected with the chimeric receptors and FcεRIγ, using 500 ng of each DNA to ensure equivalent surface expression. After stimulation with pervanadate, the transfected cells were lysed in Brij-96 buffer followed by immunoprecipitation with anti-CD4. Nonreduced immunoblot analysis with antiphosphotyrosine (Fig 4A) revealed an associated, phosphorylated protein approximately the size of FcεRIγ from the CD4/PIR-A3 sample specifically. The apparent 50-kD band in lanes 4 and 5 is not a consistent finding and likely represents nonspecific reactivity with the antiphosphotyrosine. Mutation of Arg632completely abolished the chimeric receptor’s ability to interact with FcεRIγ. Immunoblotting with antiphosphotyrosine is extremely sensitive, but does not allow the definitive identification of the phosphoprotein as FcεRIγ. In addition, this result leaves open the possibility that because of the induction of tyrosine phosphorylation, FcεRIγ is interacting with an intermediary protein via an src homology 2 domain interaction. Therefore, to definitively identify the associated polypeptide as FcεRIγ and rule out phosphotyrosine-mediated interactions, similar immunoprecipitations were performed without pervanadate stimulation followed by immunoblotting with a polyclonal antibody recognizing FcεRIγ. Figure 4B unequivocally reveals the coimmunoprecipitation of the γ chain with CD4/PIR-A3 and not with CD4/PIR-A3R632L, showing the importance of Arg632 within the LIRM sequence for receptor complex formation. Several other recently identified receptors, including the ILT receptors,4 the likely human counterparts of the PIR, and putative activating LIR,5 also contain the sequence LIRM near the outer leaflet of the plasma membrane, suggesting that they may also interact with FcεRIγ. Notably, the charged residues of the activating human KIR and murine Ly49D are not within this context, and they are spaced so as to be nearly exactly in the middle of their respective transmembrane domains; this even though KIR are type 1 Ig superfamily receptors and Ly49D are type 2 C-type lectin receptors. Both the activating KIR and activating Ly49s have now been shown to use the newly described signaling chain, DAP12, suggesting that relative position of a receptor’s transmembrane arginine residue may be critical in determining the signal transduction chain used by that receptor.30-32
FcɛRIγ associates with CD4/PIR-A3 via Arg632 in 293T cells. 293T cells were transfected with 500 ng of pcDNA3, CD4/PIR-A3, or CD4/PIR-A3R632L cDNA in the absence or presence of 500 ng of FcɛRIγ DNA. (A) A tyrosine phosphoprotein is associated with CD4/PIR-A3 after pervanadate stimulation. The transfected cells were stimulated with pervanadate and lysed in 1% Brij-96 buffer, and whole cell lysates were immunoprecipitated with anti-CD4. Immune complexes were eluted in nonreducing Laemmli sample buffer and separated by 8% SDS-PAGE followed by immunoblotting with antiphosphotyrosine. (B) FcɛRIγ associates with CD4/PIR-A3 via Arg632 in 293T cells. The transfected cells were lysed and processed as above, using reducing Laemmli sample buffer followed by separation with 4% to 20% SDS-PAGE and immunoblotting with antimurine FcɛRIγ.
FcɛRIγ associates with CD4/PIR-A3 via Arg632 in 293T cells. 293T cells were transfected with 500 ng of pcDNA3, CD4/PIR-A3, or CD4/PIR-A3R632L cDNA in the absence or presence of 500 ng of FcɛRIγ DNA. (A) A tyrosine phosphoprotein is associated with CD4/PIR-A3 after pervanadate stimulation. The transfected cells were stimulated with pervanadate and lysed in 1% Brij-96 buffer, and whole cell lysates were immunoprecipitated with anti-CD4. Immune complexes were eluted in nonreducing Laemmli sample buffer and separated by 8% SDS-PAGE followed by immunoblotting with antiphosphotyrosine. (B) FcɛRIγ associates with CD4/PIR-A3 via Arg632 in 293T cells. The transfected cells were lysed and processed as above, using reducing Laemmli sample buffer followed by separation with 4% to 20% SDS-PAGE and immunoblotting with antimurine FcɛRIγ.
CD4/PIR-A3 associates with FcεRIγ in murine macrophages.
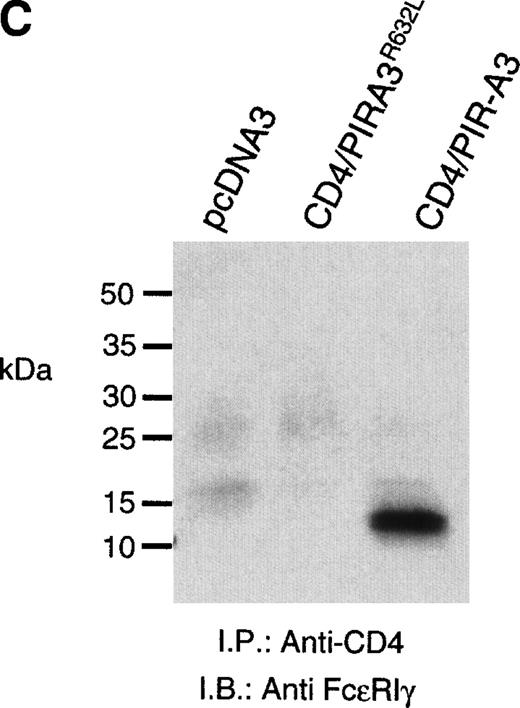
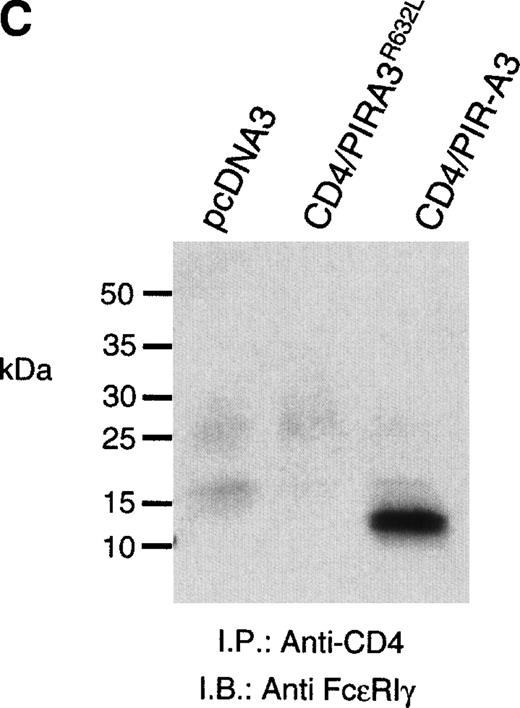
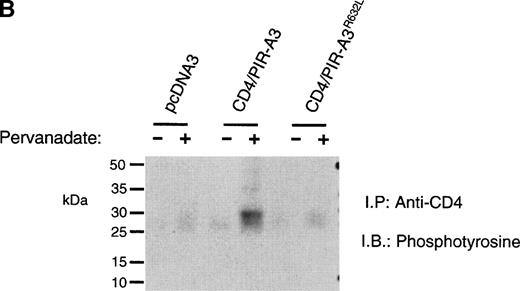
Because the expression of the native PIR proteins seems to be restricted to B cells and myeloid cells,7 8 we transfected the chimeric receptors into the murine macrophage cell line ANA-1 to ask whether this receptor would associate with endogenous FcεRIγ. ANA-1 cells were transfected with the pcDNA3 vector, the CD4/PIR-A3 chimera, or the chimera carrying the R632L mutation. After transfection, the cells were selected in G418 and cloned by limiting dilution. Cell surface expression of the receptors on the clones was determined by flow cytometry using anti-CD4 MoAb. Although the expressed levels of receptor on the clones were weak, the surface expression was consistent and easily detectable. As shown in Fig 5A, one CD4/PIR-A3 clone and one CD4/PIR-A3R632L clone with comparable cell surface expression were selected for further study. After stimulation with pervanadate, the clones were lysed in Brij-96 buffer and immunoprecipitated with anti-CD4. Immunoblot analysis with antiphosphotyrosine under nonreducing conditions revealed a tyrosine phosphoprotein of approximately 28 kD in anti-CD4 immunoprecipitates of the CD4/PIR-A3 clone only (Fig 5B). To confirm the identity of this protein as FcεRIγ, a similar experiment was performed under reducing conditions without pervanadate treatment followed by immunoblotting with anti-FcεRIγ. As shown in Fig 5C, coimmunoprecipitation of FcεRIγ was detectable from the CD4/PIR-A3 clone and not from CD4/PIR-A3R632L or the vector control clones. These data show the ability of PIR-A3 transmembrane domains to physically associate with the endogenous FcεRIγ of murine macrophages. Again, this interaction was found to be dependent on Arg632 of the PIR-A3 sequence.
CD4/PIR-A3 is expressed and associates with FcɛRIγ in ANA-1 stable transfectants. (A) ANA-1 express CD4/PIR-A3 and CD4/PIR-A3R632L. ANA-1 macrophages were transfected with pcDNA3 (thin traces), CD4/PIR-A3 (left panel, thick trace), or CD4/PIR-A3R632L (right panel, thick trace) expression vectors, as described in Materials and Methods. Clones were screened for expression using PE-conjugated anti-human CD4. (B) CD4/PIR-A3 is associated with a tyrosine phosphoprotein after pervanadate stimulation in ANA-1. ANA-1 clones (5 × 106 cells/point) were stimulated in the absence or presence of pervanadate followed by lysis in 1% Brij-96 buffer. Whole cell lysates were immunoprecipitated with anti-human CD4 and eluted in nonreducing Laemmli sample buffer. Immune complexes were separated by 4% to 20% SDS-PAGE and immunoblotted with antiphosphotyrosine. (C) FcɛRIγ associates with CD4/PIR-A3 via Arg632. ANA-1 clones (25 × 106 cells/point) were lysed and processed as above, using reducing Laemmli sample buffer followed by separation with 4% to 20% SDS-PAGE and immunoblotting with antimurine FcɛRIγ.
CD4/PIR-A3 is expressed and associates with FcɛRIγ in ANA-1 stable transfectants. (A) ANA-1 express CD4/PIR-A3 and CD4/PIR-A3R632L. ANA-1 macrophages were transfected with pcDNA3 (thin traces), CD4/PIR-A3 (left panel, thick trace), or CD4/PIR-A3R632L (right panel, thick trace) expression vectors, as described in Materials and Methods. Clones were screened for expression using PE-conjugated anti-human CD4. (B) CD4/PIR-A3 is associated with a tyrosine phosphoprotein after pervanadate stimulation in ANA-1. ANA-1 clones (5 × 106 cells/point) were stimulated in the absence or presence of pervanadate followed by lysis in 1% Brij-96 buffer. Whole cell lysates were immunoprecipitated with anti-human CD4 and eluted in nonreducing Laemmli sample buffer. Immune complexes were separated by 4% to 20% SDS-PAGE and immunoblotted with antiphosphotyrosine. (C) FcɛRIγ associates with CD4/PIR-A3 via Arg632. ANA-1 clones (25 × 106 cells/point) were lysed and processed as above, using reducing Laemmli sample buffer followed by separation with 4% to 20% SDS-PAGE and immunoblotting with antimurine FcɛRIγ.
CD4/PIR-A3 activates murine macrophages.
In addition to PIR-A3, macrophages express Fc receptors that physically associate with FcεRIγ.17 Aggregation of these receptors with antibody or immune complexes can deliver potent activation signals to macrophages, resulting in production of cytokines and reactive nitrogen intermediates.21,22,35 Having established that PIR-A3 physically interact with FcεRIγ, we next asked whether ligation of our PIR-A3 chimera would result in macrophage activation. Others have shown an interaction between a PIR-A receptor and the FcεRIγ chain in mast cells.36 37 Whereas the exact PIR-A receptor used in their study is unclear, crosslinking resulted in calcium mobilization, although there was no indication of cellular activation. Therefore, we next tested whether PIR-A3 activation could lead to NO production by analyzing NO (measured as accumulated NO2−) release from cells after ligation of chimeric receptors. As shown in Fig 6, NO production (20 to 30 μmol/L) was easily detectable from CD4/PIR-A3, CD4/PIR-A3R632L, as well as the vector control, when the cells were stimulated with interferon γ (IFN-γ) plus lipopolysaccharide (LPS). However, only CD4/PIR-A3 was able to produce NO in response to IFN-γ plus anti-CD4. The failure of CD4/PIR-A3R632L to produce NO is reflective of its inability to form a signaling complex because of the mutation of its transmembrane arginine. In addition, the lack of NO production by CD4/PIR-A3R632L definitively rules out Fc receptor effects in these assays.
CD4/PIR-A3 induces NO production in ANA-1 macrophages. ANA-1 clones (1 × 106 cells/well) were stimulated with medium, 100 U/mL IFN-γ, 1 μg/mL goat antimouse IgG, 10 ng/mL LPS, and 40 μL anti-CD4, as indicated for 24 hours, as described in Materials and Methods. Cell-free culture supernatants were then assayed for NO production. (□), pcDNA3; (▩), CD4/PIR-A3; (▪), CD4/PIR-A3R632L.
CD4/PIR-A3 induces NO production in ANA-1 macrophages. ANA-1 clones (1 × 106 cells/well) were stimulated with medium, 100 U/mL IFN-γ, 1 μg/mL goat antimouse IgG, 10 ng/mL LPS, and 40 μL anti-CD4, as indicated for 24 hours, as described in Materials and Methods. Cell-free culture supernatants were then assayed for NO production. (□), pcDNA3; (▩), CD4/PIR-A3; (▪), CD4/PIR-A3R632L.
Macrophages are a key element in host defense. They are an important source of a wide variety of secretory products, including inflammatory cytokines and NO, and are endowed with antitumoral and antimicrobicidal activities. The ability of PIR-A3 to induce NO suggests that the PIR-A family of receptors will play an important role in the modulation of the macrophage-mediated immune response. Unfortunately, the ligands for the PIR have not yet been defined. The existence of multiple genes for PIR-A proteins would, however, suggest that these receptors will function to regulate macrophage NO production in response to various highly homologous ligands.7 Macrophages are found in different tissues where they are likely to encounter various ligand configurations. The ability of the macrophage to express multiple PIR-A receptors might allow recognition of a variety of ligands and therefore, complex regulation of their responses. Our data clearly show that even low levels of PIR-A3 expression are sufficient for macrophage activation in the presence of IFN-γ, but further study of PIR-A effects on macrophage biology will be required to address whether these receptors function independently or as coreceptors. Regardless, our findings show the capability of this new family of receptors to activate macrophages and suggest that the signal transduction pathways of this activation will be similar to those defined for other FcεRIγ-coupled receptors, specifically the Fc receptors themselves. Continued study of this emerging family of receptors should shed considerable light on the role of macrophage activation in regulation of the immune response.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
This is a US government work. There are no restrictions on its use.
REFERENCES
Author notes
Address reprint requests to Daniel W. McVicar, PhD, NCI-FCRDC Building 560/Room 31-93, Frederick, MD 21702; e-mail: MCVICAR@NIH.GOV.