Abstract
The differentiation of osteoclasts is regulated by transcription factors expressed in cells of osteoclast lineage. We isolated here a potential transcription factor from a cDNA library of an enriched population of preosteoclasts and osteoclasts. The cDNA encodes a protein with N-terminal POZ domain and C-terminalKrüppel-like zinc fingers. We designate this protein as osteoclast-derived zinc finger (OCZF). OCZF was found to be rat homologue of mouse leukemia/lymphoma-related factor (LRF). Northern blot and in situ hybridization analysis showed OCZF mRNA at a high level in osteoclasts and kidney cells. OCZF had a nuclear targeting sequence and was localized in the nucleus of transfected cells. In addition, OCZF specifically bound to the guanine-rich consensus sequences of Egr-1 and c-Krox. Transient transfection assays indicate that OCZF can repress transcription activity like other POZ domain proteins. Furthermore, antisense but not sense phosphorothioate oligodeoxynucleotides (ODNs) for OCZF cDNA suppressed the formation of osteoclast-like multinucleated cells (MNCs) in bone marrow culture, whereas the same ODNs did not significantly affect the formation of macrophage polykaryons and mononuclear preosteoclast-like cells (POCs). These results suggest that OCZF is a unique transcription factor that plays an important role in the late stage of osteoclastogenesis.
OSTEOCLASTS ARE DERIVED from hematopoietic stem cells. They are multinucleated cells and are formed by the fusion of mononuclear preosteoclasts. Their differentiation is thought to be under the control of the bone microenvironment, which is supplied by stromal cells or osteoblasts and local factors.1 The detailed molecular mechanism underlying the differentiation of osteoclasts is not yet understood. Extracellular stimuli produced by the bone microenvironment induce an intracellular signal of the precursor cells for osteoclasts. The signal leads to the expression of target genes and differentiation toward osteoclasts. As in many developmental systems, the cell fate decision and promotion of differentiation are mediated by temporally expressed or activated transcription factors.
PU.1- and Fos-deficient mice have an osteopetrosis phenotype, suggesting that these transcription factors are involved in osteoclast differentiation.2-4 Fos protein is a major component of the AP-1 transcription factor complex. In c-fos mutant mice, the number of macrophages is not decreased, but the development of osteoclast is disordered. PU.1 mutation results in the elimination of the lineages for both osteoclasts and macrophages. PU.1 is thought to regulate the initial stages of myeloid differentiation, whereas Fos is thought to promote the differentiation of bipotential precursors into osteoclasts, indicating that these transcription factors are involved in the early stage of osteoclastogenesis. Conversely, there is yet little information regarding transcription factors expressed in the cells at the late stage of osteoclastogenesis.
In hematopoiesis, a number of transcription factors are known to participate in the proliferation and differentiation of each specified cell lineage. The transcription factors are classified into different types by specific motifs. The zinc finger domain, in particular, seems to comprise a major class of DNA-binding motifs, because many proteins possessing this domain have been described.5 Some zinc finger transcription factors are important, because they play a key role as a master switch at the branch point of differentiation in hematopoiesis. For example, GATA-1, which contains 2 Cys4-type zinc fingers, has an essential role in erythrocyte development.6,7 ErythroidKrüppel-like factor (EKLF), with Cys2His2-type zinc fingers, is an erythroid cell-specific transcription factor regulated by GATA-1.8Egr-1, another Krüppel-type zinc finger transcription factor with 3 Cys2His2-type zinc finger motifs, is expressed in macrophages and has been shown to play a critical role in the differentiation of the macrophage lineage.9 10 Such zinc finger transcription factors expressed in osteoclasts may have an important role in osteoclastogenesis.
In this study, we characterized a zinc finger protein with POZ domain that is preferentially expressed in osteoclasts. This protein, termed osteoclast-derived zinc finger (OCZF), showed an ability to bind specifically to a guanine-rich sequence and regulated transcriptional activity, suggesting that OCZF is a transcription factor. The functional role of OCZF in the differentiation of osteoclasts was investigated using specific antisense phosphorothionate oligodeoxynucleotides (ODNs). Interestingly, the blockage of the expression of OCZF results in a significant reduction of the formation of osteoclast-like multinucleated cells (MNCs) formed in rat bone marrow cultures. Our results suggest that OCZF may be one of several important regulators involved in the late stage of osteoclastogenesis.
MATERIALS AND METHODS
Materials.
Male Sprague-Dawley (SD) rats were purchased from SEAC Yoshitomi (Fukuoka, Japan). ROS17/2.8 and UMR106 cells (rat osteoblastic cells) were generously provided by Dr L. Bonewald (University of Texas Health Science Center, San Antonio, TX). L8 cells (rat myoblasts) and 293 (human kidney cells) cells were kindly provided by Drs S. Matsuhashi and K. Miyake (Saga Medical School, Saga, Japan). NRK cells (rat kidney cells) were obtained from the Riken Cell Bank (Tsukuba, Japan). 3Y1 (rat fibroblasts) and A11 (rat osteoblastic cells) cells were generously provided by Drs G. Kimura and T. Inai (University of Kyushu, Fukuoka, Japan), respectively. 293T cells (human kidney cells) were provided by Drs T. Watanabe and H. Kishi (University of Kyushu). WRT-7 P2 (P2) cells (rat leukemia cells) were kindly provided by Dr M. Kobayashi (University of Hokkaido, Sapporo, Japan). 1α,25dihydroxyvitamin D3[1α,25(OH)2D3] was purchased from Biomol Research Laboratories (Plymouth Meeting, PA).
Cell cultures.
A mouse hybridoma cell line for monoclonal antibody (MoAb) Kat6 and P2 cells were grown in Iscove’s modified Dulbecco’s medium (IMDM; GIBCO BRL, Gaithersburg, MD) containing 10% fetal calf serum (FCS). The differentiation of P2 cells into macrophages was induced by the addition of 10−7 mol/L phorbol myristate acetate (PMA; Sigma, St Louis, MO) for 3 days.11 The mouse hybridoma cell line for the MoAb Kat1 was grown in the same medium in the presence of 10 ng/mL human recombinant interleukin-6 (IL-6; R&D, Minneapolis, MN).12 The other cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO BRL) containing 10% FCS. Rat primary osteoblasts were isolated by sequential digestion from the calvariae of newborn rats according to the method of Takahashi et al13 and cultured in α minimum essential medium (αMEM; GIBCO BRL) containing 10% FCS.
For the formation of MNCs, bone marrow cells obtained from the tibiae and femurs of rats were cultured in the presence of 10−8 mol/L 1α,25(OH)2D3and 10% heat-treated conditioned medium derived from ROS17/2.8 cells (htROSCM), as described by Kukita et al.14For the formation of preosteoclast-like cells (POCs), nonadherent bone marrow cells from which adherent cells were eliminated using a Sephadex G-10 column were cultured in the presence of 10−8mol/L 1α,25(OH)2D3 and 10% (vol/vol) htROSCM, as described by Kukita et al.15 For the formation of macrophage polykaryons, bone marrow cells were cultured in the presence of 10−8 mol/L 1α,25(OH)2D3 and 10−7 mol/L PMA as described.16 These cells were cultured in αMEM containing 15% FCS (Biowittaker, Walkersville, MA) for 4 or 5 days. At the end of the culture, tartrate-resistant acid phosphatase (TRAP) staining was performed with a commercial kit (Sigma). TRAP-positive mononuclear cells as POCs or TRAP-positive multinucleated cells (>3 nucleus) as MNCs were counted. TRAP-negative multinucleated cells were counted as macrophage polykaryons.
Preparation of MoAb Kat6 and immunocytochemistry.
Immunization, hybridoma production, and screening were performed using MNCs as the antigen.12 The immunocytochemistry of bone marrow cells with MoAb Kat6 was performed as follows. Rat bone marrow cells were cultured in the presence of 10% (vol/vol) htROSCM and 10−8 mol/L 1α,25(OH)2D3. After 5 days of culture, the cells were detached from the plastic culture plates with 0.05% trypsin and 0.02% EDTA in phosphate-buffered saline (PBS) and centrifuged onto glass slides with the use of a cytospin. The cells were then fixed with 2% paraformaldehyde (PFA) in PBS for 5 minutes, followed by treatment with cold acetone for 2 minutes. The cells were stained by MoAb Kat 6 using fluorescein isothiocyanate (FITC)-conjugated antimouse IgM (μ-chain specific; Zymed, San Francisco, CA) as the second antibody.
Enrichment of osteoclasts and osteoclast-precursor cells by magnetic cell separator system (MACS).
Rat bone marrow cells were cultured in the presence of 10% (vol/vol) htROSCM and 10−8 mol/L 1α,25(OH)2D3. After 4 days of culture, the cells were detached from the plastic culture plates as described and incubated with MoAb Kat1 (500 μg/mL), which recognizes the surface antigen of osteoclasts, on ice for 20 minutes. After the cells were rinsed in ice-cold PBS, they were reacted with biotinylated antimouse IgM (μ-chain specific) on ice for 10 minutes. The cells were then rinsed in ice-chilled PBS and reacted with FITC-conjugated streptoavidin for 10 minutes, followed by the reaction with biotinylated MACS microbeads for 5 minutes according to the manufacturer’s protocol. The cells were then resuspended in PBS containing 2% bovine serum albumin (BSA) and applied to MACS (Miltenyi Biotec, Sunnyvale, CA), and the positive fraction (Kat1-positive cells) was collected.
Cloning of OCZF gene.
Poly(A)RNA was isolated from the Kat1-positive cell-enriched fraction with the use of an mRNA isolation kit (Pharmacia Biotech, Uppsala, Sweden), and cDNA synthesis was performed using a cDNA synthesis kit (Stratagene, La Jolla, CA). Briefly, the synthesis of double-stranded cDNA was primed with oligo dT linker-primer, and the cDNA was ligated to EcoRI adapter and then digested by Xho I. Synthesized cDNA was size-fractionated by electrophoresis on a 4% Nuisieve agarose gel (Takara, Tokyo, Japan). The cDNA larger than 1.0 kb was recovered and ligated to the EcoRI/Xho I site of λ Uni-ZAPII vector. A directional library was constructed with an in vitro phage packaging kit using a packaging extract (Stratagene). Screening of the library with MoAb Kat6 was performed by using an ABC-AP kit (Vector, Burlingame, CA). The positive clone was plaque-purified 3 times. The pBluscript plasmid containing the positive cloned DNA insert was excised by coinfection with helperphage, and a cDNA clone designated pK6-78 that encodes a partial sequence of theOCZF gene was obtained. To obtain the full-length sequence ofOCZF, the insert of pK6-78 (0.9 kb, 933 to 1,872 bp of pK6-78-4) was used as a probe to isolate overlapping clones from the Kat1-positive cell library. Plaque hybridization was performed using a32P-labeled insert of pK6-78 under high-stringency conditions. The screening of the library with this probe led to the isolation of several clones. The DNA sequence was determined using a DNA sequencer (Model 370A; Applied Biosystems, Foster City, CA) and a Taq DyeDeoxy cycle sequencing kit from Applied Biosystems. Sequencing of the full length of OCZF cDNA was performed using the longest clone, designated pK6-78-4.
In situ hybridization.
In situ hybridization was performed using sense and antisense RNA probes corresponding to about 0.9 kb (insert of pK6-78) of OCZFcDNA. The probes were labeled with digoxigenin with the use of an RNA synthesis kit (Nippon Gene, Toyama, Japan). Four-micrometer sections of rat mandible obtained from 6-day-old SD rats were prepared as described previously, except that fixation was performed by perfusion of the fixatives.17 Tissue sections were deparaffinized in xylene and rehydrated in ethanol series. After being rinsed in PBS, sections were treated with proteinase K (10 μg/mL; Sigma) for 10 minutes at 37°C, fixated in 4% PFA in PBS for 10 minutes, and then incubated with 0.2 mol/L HCl for 10 minutes. The sections were treated in PBS containing 0.2% glycine for 10 minutes 2 times, followed by rinsing in PBS. The sections were acetylated with 0.25% acetic anhydride in 0.1 mmol/L triethanolamine (pH 8.0) for 10 minutes. The prehybridization and hybridization were performed as previously described.17Hybridized digoxigenin-labeled probes were detected by use of a DIG Nucleic Acid Detection Kit (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer’s instructions. After color reaction, the sections were counterstained by 0.2% methylgreen.
Northern analysis of OCZF.
Poly(A)RNA was extracted from each of several tissues and cell lines with the use of a mRNA isolation kit (Pharmacia Biotech). Rat liver, testis, and brain poly(A)RNA were obtained from Clontech (Palo Alto, CA). Poly(A)RNA was also extracted from bone marrow cells treated with antisense, sense OCZF, and scramble ODNs after 4 days of culture. RNA was denatured at 65°C for 5 minutes in 50% formamide, electrophoresed through a 1.5% agarose gel containing 6.6% formaldehyde, and transferred to nylon membranes (Genescreen; NEN Research Products, Boston, MA). The insert of pK6-78 was labeled with [α-32P] cytosine triphosphate (CTP) using a random primer labeling kit (Nippon Gene) and used as a probe. Hybridization and washing were performed under stringent conditions at 60°C (the preparation was finally washed twice at 60°C in 0.1× saline sodium citrate plus 1% sodium dodecyl sulfate [SDS]).
DNA transfections.
For the expression of OCZF protein in cultured cells, full-lengthOCZF cDNA was cloned into the EcoRI and Xho I sites of the expression vector pME18S (provided by Dr K. Maruyama, Tokyo University, Tokyo, Japan), which has the SRα promoter. In some experiments, OCZF cDNA (nt 159-2817) and human Egr-1 cDNA (nt 331-3132)18 were cloned into expression vector pCDL-FLAG, which have the SRα promoter (M. Ouchida, unpublished results), restoring the open reading frame. Two types of pCDL-FLAG vectors, pCDL-FLAGa and pCDL-FLAGc, which differ only in the reading frame, were used for OCZF and Egr-1 cDNA, respectively. OCZF and Egr-1 proteins produced from these plasmids have S-tag and FLAG-tag epitope in its N-terminus and missed the first 10 and 21 amino acids, respectively. For the isolation of nuclear extract, 293T cells were plated in 10-mm dishes at a density of 4 × 105 cells before transfection. For immunocytochemical study, 293T cells were plated at a density of 2 × 104 cells on a glass chamber (Nunc, Roskilde, Denmark) before transfection. DNA transfection was performed by the calcium phosphate-DNA coprecipitation method as described.19 The expression of FLAG-tagged protein was confirmed by a Western analysis using anti-FLAG M2 MoAb (Sigma) as described.20
Electrophoretic mobility shift assays (EMSA).
Complementary oligonucleotides containing the Egr-1–binding sequence (underlined; 5′-ATCCCGGCGCGGGGGCGAGGGCGT-3′ and 5′-ACGCCCTCGCCCCCGCGCCGGGAT-3′) and the c-Krox–binding sequence (5′-CCA CGTCCCTCCCCCCTCGGCTCCCTCCCCTA-3′ and 5′-TAGG GGAGGGAGCCGAGGGGGGAGGGACGTGG-3′) were synthesized (Sawady Tec Co, Tokyo, Japan).21 The oligonucleotides were annealed by heating to 65°C for 10 minutes and then slowly cooled to room temperature. A consensus SP1 oligonucleotide (Promega, Madison, WI) was used as a competitor. Probes were prepared by end-labeling oligonucleotides with [γ-32P] adenosine triphosphate (ATP) and T4 polynucleotide kinase. Nuclear extracts were prepared from the cell lines 2 days after they were transfected with expression vector containing cDNA for OCZF by the calcium-phosphate precipitation method.22 The binding reaction was achieved by preincubating nuclear extracts with 2 μg poly(dI-dC)/poly(dI-dC) (Pharmacia Biotech) in a buffer containing 20 mmol/L HEPES, pH 7.9, 70 mmol/L KCl, 5 mmol/L MgCl2, 0.05% Nonidet P-40, 12% glycerol, 1 mg/mL BSA, 0.5 mmol/L dithiothreitol, 100 μmol/L ZnCl2, and 5 μmol/L p-Amidino phenylmethylsulphonyl fluoride (PMSF; Wakojunyaku, Osaka, Japan) at room temperature for 20 minutes as described.9 One nanogram of end-labeled probe (10,000cpm) was added to the reaction mixture containing the nuclear extract and incubated for 15 minutes at room temperature. For the competition experiments, excess unlabeled DNA was incubated with the reaction mixture for 15 minutes before the addition of the radiolabeled probe. In supershift assays, 3 μg of anti-FLAG M2 MoAb (Sigma) was incubated with the reaction mixture for 15 minutes before the addition of the radiolabeled probe. The samples were analyzed by a 4% polyacrylamide gel (PAGE) in 0.5× TBE (1× TBE consists of 0.089 mmol/L Tris-borate, 0.089 mmol/L boric acid, and 2 mmol/L EDTA) buffer at 150 V at room temperature.
S-protein pulldown experiments.
Nuclear extracts were prepared from 293T cells 2 days after they were transfected with pCDL-FLAG expression vectors (empty or encoding OCZF or Egr-1 protein). To purify S-tagged protein, S-protein agarose beads (Promega) were incubated with the nuclear extracts for 3 hours at 4°C and then washed 5 or 6 times in cold PBS. The bound proteins were then used for DNA binding reaction. The binding reaction was performed by incubating bound protein with 1 ng of end-labeled probe in same buffer used in EMSA for 30 minutes at room temperature. To remove unbound DNA, the bound DNA was then washed 3 times in the binding buffer and then the oligomers retained by the beads were released by the addition of deionizing water and boiling for 5 minutes. The resulting DNA was analyzed by a 10% PAGE.
Luciferase assays.
The reporter plasmid, pOA-Egr-TK-Luciferase (M. Ouchida, unpublished results), was constructed by inserting 4 tandem copies of double-stranded oligonucleotides containing Egr-1 binding sequence in upstream of the firefly luciferase gene of pOA-TK-Luciferase that contained 250 bp 5′ of the HSV thymidine kinase transcription initiation site. The internal control Renilla luciferase plasmid, pΔTK-RL, which has basal thymidine kinase promoter, was prepared from pRLTK (Promega). The expression vectors, pME18S (either empty or encoding OCZF protein), at 0.25 μg or 2.5 μg, were cotransfected with 1 μg of the reporter vector in 293 or 293T cells by calcium phosphate precipitation. The cells were harvested 48 hours later in the Promega Lysis Buffer. The activity of firefly and Renilla luciferase was measured using a reagent kit (Promega) and normalized to the Renilla luciferase activity of a cotransfected pΔTK-RL vector (0.25 μg) to correct for variation in transfection efficiency.
Antisense ODN experiments.
The phosphorothioate ODNs (20 bases in length) ODN-1 and ODN-2, complementary to the target sequence of OCZF mRNA, were designed and purified by high-performance liquid chromatography (HPLC) by Greiner Japan Co (Tokyo, Japan) and Toa Synthesis Co (Tokyo, Japan), respectively. Antisense OCZF ODN-1 located to a sequence starting at the ATG initiation codon of OCZF cDNA (5′-CCGTCCACGCCGCCAGCCAT-3′) and sense ODN-1 (5′-ATGCTGGCGGCGTGGGACGG-3′) were used in the experiments, the results of which are shown in Fig 8A (experiments no. 1 and 2). Antisense OCZF ODN-2 targeted against a sequence located 16 bases upstream of the initial ATG codon (5′-CATCTTCCGCGACACCTCT-3′), sense ODN-2 (5′-AGAGGTGTCGCGGAAGATG-3′), and scramble ODN-2 (5′-TGTCTCCTACC TGCCCAAC-3′) as a control were used in the experiments, the results of which are shown in Fig 8A (experiment no. 3), B, and C. Bone marrow cells or nonadherent bone marrow cells were cultured in 3 types of conditions to form MNCs, POCs, and macrophage polykaryons with ODNs at a final concentration of 1 μmol/L for 4 or 5 days at 37°C. Each culture was fed once every 3 days by replacing half of the culture medium with fresh medium, hormone, ODNs, and conditioned medium or PMA. Differentiation was assessed by staining the cells with a TRAP kit (Sigma).
RESULTS
Reactivity of MoAb Kat6 to a nuclear antigen in MNCs in bone marrow culture.
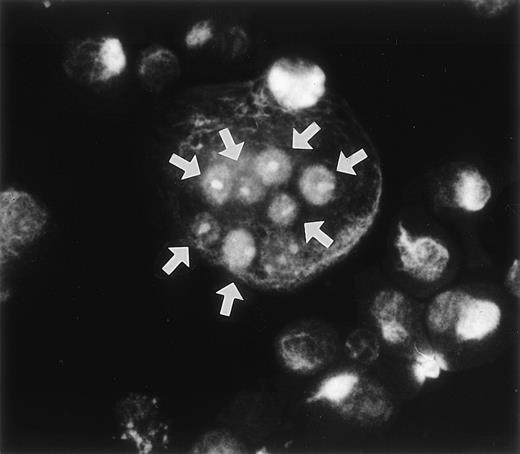
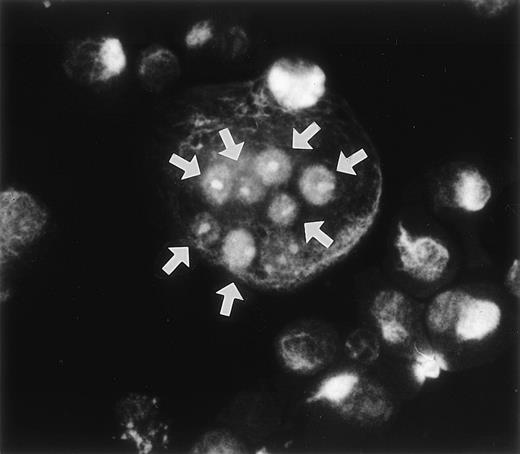
During the course of our search for the specific antigen expressed in osteoclast-like cells in rat bone marrow culture by making a panel of MoAbs,11 we found that MoAb designated Kat6 stained the nuclei of osteoclast-like cells. Figure 1demonstrates the positive staining of Kat6 in the nuclei of the multinucleated cells derived from bone marrow cultures. Similar positive staining with MoAb Kat6 was also seen in the mononuclear cells, but not in primary osteoblasts (data not shown). Because little information has been obtained regarding the nuclear proteins expressed in osteoclasts, we attempted cDNA cloning of the gene for Kat6 antigen.
Immunoreactivity of the MoAb Kat 6 to multinucleated cells formed in bone marrow culture. Rat bone marrow cells were cultured in the presence of htROSCM and 10−8 mol/L 1,25(OH)2D3 for 5 days. The detached cells were centrifuged onto a glass slide by using a cytospin, fixed with 2% PFA, and stained by MoAb Kat 6 using FITC-conjugated antimouse IgM as the second antibody. Positive signals in the nucleus of the multinucleated cells in the culture are indicated by arrows. (Original magnification × 443.)
Immunoreactivity of the MoAb Kat 6 to multinucleated cells formed in bone marrow culture. Rat bone marrow cells were cultured in the presence of htROSCM and 10−8 mol/L 1,25(OH)2D3 for 5 days. The detached cells were centrifuged onto a glass slide by using a cytospin, fixed with 2% PFA, and stained by MoAb Kat 6 using FITC-conjugated antimouse IgM as the second antibody. Positive signals in the nucleus of the multinucleated cells in the culture are indicated by arrows. (Original magnification × 443.)
Molecular cloning of OCZF cDNA.
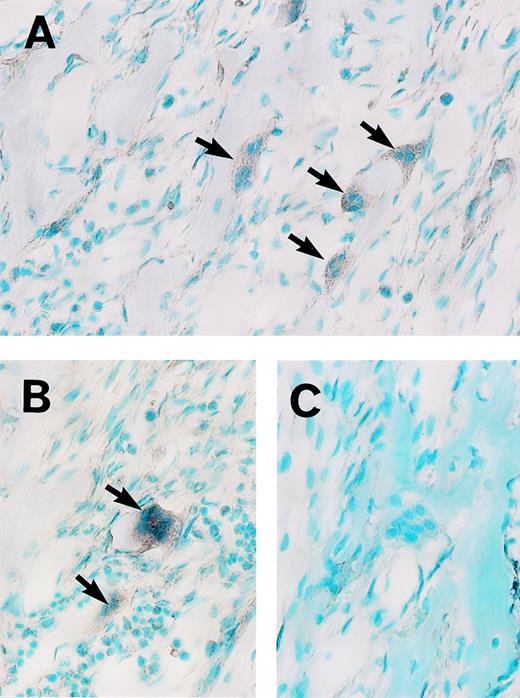
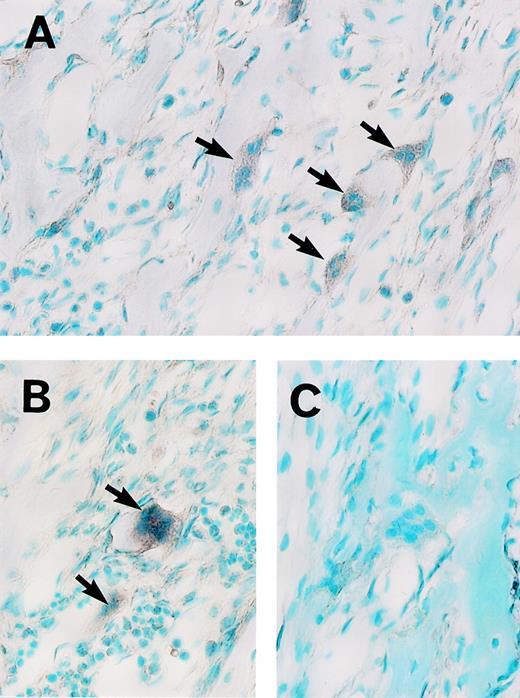
We recently isolated MoAb Kat1, which recognizes the surface antigen of osteoclasts.11 To perform the cDNA cloning efficiently, a population of the cells including preosteoclasts and osteoclasts was first enriched with this MoAb. A cDNA library (2.0 × 106 clones) in a phage vector (λ ZAPII) was constructed with mRNA from enriched cells. In the screening of cDNA library using the MoAb Kat6, we obtained a candidate clone (pK6-78). The nucleotide sequence of pK6-78 and its predicted amino acid sequence from one open reading frame (ORF) suggested that this protein encoded a partial sequence of a novel zinc finger protein. First, to determine the specificity of the expression of this cDNA, we performed an in situ hybridization analysis of rat mandible tissue sections with RNA probes prepared from pK6-78. As shown in Fig 2A and B, positive signals were detected with the antisense probe in the osteoclasts and some mononuclear cells. In contrast, similar staining was absent with the sense probe (Fig 2C). These results indicate that mRNA encoding a novel zinc finger protein is expressed in osteoclasts. We therefore designated this gene as an osteoclast-derived zinc finger gene,OCZF. We then screened again approximately 3 × 105 plaques of the Kat1-positive library with the insert of pK6-78 to obtain the full-length OCZF cDNA. The restriction enzyme mapping of the inserts (0.9 to 2.8 kb) of the 4 positive clones indicated that they overlapped. The longest clone (pK6-78-4) was selected and characterized.
Analysis of OCZF mRNA expression in osteoclasts from rat mandible by in situ hybridization. Sections were hybridized with a digoxygenin-labeled antisense (A and B) or sense (C) RNA probe. (A and B) The intense signal of OCZF mRNA is detectable in the osteoclasts with antisense RNA probe (the arrows). (C) No signal was observed in control experiment in which sense RNA was used in hybridization. (Original magnification × 239.)
Analysis of OCZF mRNA expression in osteoclasts from rat mandible by in situ hybridization. Sections were hybridized with a digoxygenin-labeled antisense (A and B) or sense (C) RNA probe. (A and B) The intense signal of OCZF mRNA is detectable in the osteoclasts with antisense RNA probe (the arrows). (C) No signal was observed in control experiment in which sense RNA was used in hybridization. (Original magnification × 239.)
Structure of OCZF cDNA.
The DNA sequence analysis of this cDNA showed an ORF encoding a 569 amino acid polypeptide of 60,539 Daltons (Fig 3). The putative ATG initial codon was preceded by an in-frame termination codon located 49 bases upstream. Because this terminal codon was confirmed by analyzing several independent clones, we concluded that we have cloned cDNA encoding the entire ORF. The predicted amino acid sequence contained 3 zinc fingers of the Cys2-His2 type near the C-terminus. A comparison of the OCZF sequence against Swiss Prot and GenBank/EMBL databases using the BLAST algorithms23 showed similarities with zinc finger proteins that are restricted to the region of the zinc finger and N-terminus. A murine cDNA lymphocyte/leukemia-related factor (LRF) that has been isolated very recently24 is 97% identical over their length, suggesting that OCZF is rat homologue of LRF. The high similarity between OCZF and LRF was also found in N-terminus and zinc finger regions of the proteins (Fig 3).
Deduced amino acid sequence of rat OCZF protein and comparison with that of mouse LRF. Dashes indicate shared identity between the mouse and rat proteins. The N-terminal POZ-like domain is underlined. The proline-rich sequence is double underlined. The domain of 3 zinc fingers is underlined. The nuclear target sequence corresponding to amino acids 480-496 is underlined with a dotted line. Amino acids are numbered at the end of each line. OCZF nucleotide sequence data are available from EMBL/GenBank/DDBJ under the accession no. D88450.
Deduced amino acid sequence of rat OCZF protein and comparison with that of mouse LRF. Dashes indicate shared identity between the mouse and rat proteins. The N-terminal POZ-like domain is underlined. The proline-rich sequence is double underlined. The domain of 3 zinc fingers is underlined. The nuclear target sequence corresponding to amino acids 480-496 is underlined with a dotted line. Amino acids are numbered at the end of each line. OCZF nucleotide sequence data are available from EMBL/GenBank/DDBJ under the accession no. D88450.
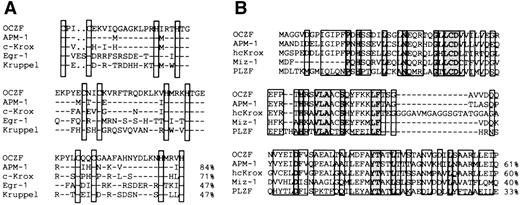
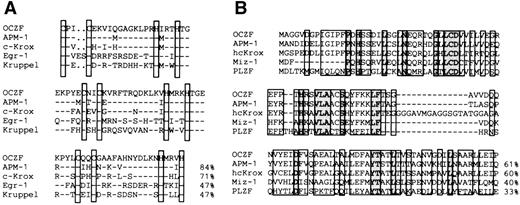
The comparison of the zinc finger sequence of OCZF showed features representative of the Cys2His2Krüppel-like family. The proteins that had the high matches in the zinc finger region were human APM-1,25 mouse c-Krox,26 and human hcKrox.27 The zinc finger sequences of c-Krox and hcKrox were identical. To analyze the zinc fingers in more detail, we aligned the zinc finger sequences of OCZF with those of human APM-1, mouse c-Krox, Egr-1,28and Krüppel,29 the segmentation gene inDrosophila (Fig 4A). The homology with APM-1, c-Krox, Egr-1, and Krüppel in the zinc finger domains was 84%, 71%, 47%, and 47%, respectively. The homology with other hematopoietic Krüppel-factors, such as EKLF8 and lung Krüppel-like factor (LKLF),30 was less than 30%. The N-terminal regions of OCZF had homology with the sequence of the so-called pox virus and zinc finger (POZ) domain seen in the N-terminal end of several proteins (Fig4B). The highest homology was found in the N-terminal region of APM-1 and hcKrox protein. This region of OCZF did have homology with the POZ domain of other zinc finger-transcription proteins, including the human c-myc–interacting Zn finger protein-1 (Miz-1)31 and promyelocytic leukemia zinc finger (PLZF; Fig 4B).32 The N-terminal regions also had homology with the regions ofBCL-633 involved in chromosome translocation in B-cell lymphoma, Drosophila kelch protein involved in nurse cell-oocyte interaction,34 as well as viral proteins (for example, VA55R) of the pox virus family (data not shown).35
Comparison of amino acid sequence of OCZF to other zinc finger proteins. (A) Homology of zinc finger domains of OCZF to those of other Krüppel zinc finger proteins. The cysteines and histidines of the zinc fingers are boxed. The homology with APM-1, c-Krox, Egr-1, and Krüppel in the zinc finger domains is shown on the right. The sequence of c-Krox is the same as that of hcKrox. (B) Homology of the N-terminal region of OCZF to those of other POZ domain proteins. The homology with APM-1, hcKrox, Miz-1, and PLZF in these regions is shown on the right. Identical residues are indicated with dashes.
Comparison of amino acid sequence of OCZF to other zinc finger proteins. (A) Homology of zinc finger domains of OCZF to those of other Krüppel zinc finger proteins. The cysteines and histidines of the zinc fingers are boxed. The homology with APM-1, c-Krox, Egr-1, and Krüppel in the zinc finger domains is shown on the right. The sequence of c-Krox is the same as that of hcKrox. (B) Homology of the N-terminal region of OCZF to those of other POZ domain proteins. The homology with APM-1, hcKrox, Miz-1, and PLZF in these regions is shown on the right. Identical residues are indicated with dashes.
The N-terminal POZ-like region of OCZF was followed by a proline-rich sequence that has been observed in the activator domains of other transcription factors.8 In the C-terminus, the consensus sequence (KKDGCNGVPSRRGRKPR) for the nuclear targeting sequence was found.36 This structural homology and characteristics suggest that OCZF functions as a transcription factor that regulates differentiation.
Expression of OCZF mRNA.
To investigate the tissue specificity of the expression ofOCZF, we prepared poly(A)RNA from various rat tissues and cell lines and performed an RNA blot analysis. OCZF mRNA of 2 different sizes (5.3 and 3.3 kb) were detected. The length of the smaller band (3.3 kb) was almost same as that of the OCZF ORF, whereas the signal of larger bands (5.3 kb) was higher than that of the smaller band (3.3 kb). The highest level of OCZF mRNA was detected in bone marrow cells, including MNCs that were stimulated with 1α,25(OH)2D3 and htROSCM for 4 days and NRK cells (kidney cells; Fig 5, lanes 6 and 12). OCZF mRNA was not seen in brain, unstimulated bone marrow cells, L8 (myoblasts), or UMR (osteosarcoma cells) cells (Fig 5, lanes 1, 5, 9, and 11). POCs were induced from nonadherent bone marrow cells (bone marrow cells depleted with stromal cells by using a Sephadex G-10 column) by treatment with 1α,25(OH)2D3 and htROSCM for 4 days. The OCZF mRNA level of the POCs (Fig 5, lane 15) was much lower than that of the bone marrow cells, including MNCs (Fig 5, lanes 6). Very low levels of the expression ofOCZF mRNA were seen in the testis, liver, spleen, primary osteoblasts, 3Y1 (fibroblasts), A11 (osteoblastic cells), and P2 (leukemia cells) cells. P2 cells differentiated into macrophages in the presence of 10−7 mol/L PMA. The expression ofOCZF mRNA was not changed when P2 cells were differentiated into macrophages in the presence of 10−7 mol/L PMA (Fig 5, lanes 13 and 14).
Expression of OCZF mRNA in various tissues and cell lines. Poly(A)RNA was prepared, and approximately 1 μg of each sample (indicated at the top of each lane) was separated on an agarose/formaldehyde gel and transferred to a nitrocellulose filter. The filter was hybridized at 60°C overnight with an OCZFcDNA probe and then washed. The same filter was rehybridized with a human β-actin probe as a control. Arrows indicate the bands ofOCZF and β-actin mRNA.
Expression of OCZF mRNA in various tissues and cell lines. Poly(A)RNA was prepared, and approximately 1 μg of each sample (indicated at the top of each lane) was separated on an agarose/formaldehyde gel and transferred to a nitrocellulose filter. The filter was hybridized at 60°C overnight with an OCZFcDNA probe and then washed. The same filter was rehybridized with a human β-actin probe as a control. Arrows indicate the bands ofOCZF and β-actin mRNA.
Detection and localization of OCZF expressed in the cells with MoAb Kat6.
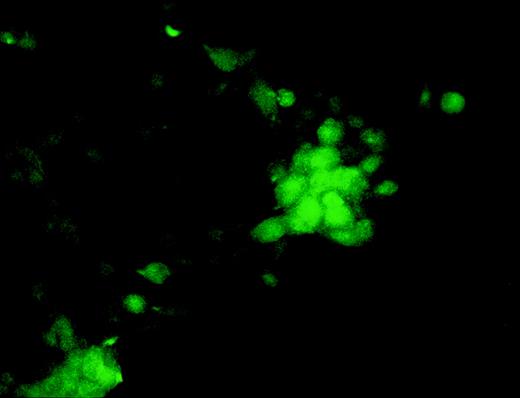
To confirm that MoAb Kat6 recognizes OCZF expressed in the cells and to examine the cellular localization of the protein, we transfected human kidney cells (293T) with an expression vector containing cDNA for OCZF and then stained with MoAb Kat6. As shown in Fig 6, OCZF was detected in the nuclei of some of the cells. A mock transfection (expression vector alone) was also performed and did not show any nuclear staining in these cells (data not shown).
Expression and detection of OCZF in human kidney cells.OCZF cDNA was inserted into an expression vector and transfected human kidney cells (293T) by the calcium phosphate-DNA coprecipitation method. After 48 hours, the cells were fixed and stained with MoAb Kat6 and FITC-labeled antimouse IgM. Dense staining was seen in the nuclei of some cells. (Original magnification × 202.)
Expression and detection of OCZF in human kidney cells.OCZF cDNA was inserted into an expression vector and transfected human kidney cells (293T) by the calcium phosphate-DNA coprecipitation method. After 48 hours, the cells were fixed and stained with MoAb Kat6 and FITC-labeled antimouse IgM. Dense staining was seen in the nuclei of some cells. (Original magnification × 202.)
Detection of OCZF binding to Egr-1 and c-Krox consensus sequence.
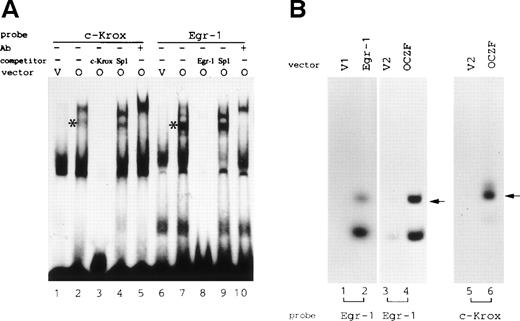
To examine the DNA-binding activity of OCZF, we performed an EMSA. We prepared nuclear extracts from 293T cells transfected with pCDL-FLAG expression vectors (empty or encoding OCZF protein). The nuclear extracts were then incubated with the probes. FLAG tagged OCZF did bind the consensus sequences of Egr-1 and c-Krox. A binding complex was detected in the EMSA using extract from 293T cells transfected with expression vector pCDL-FLAGc containing OCZF cDNA, but not with pCDL-FLAGc vector (Fig 7A, lanes 1, 2, 6, and 7). The formation of the complex was inhibited in the presence of a 100-fold molar excess of the unlabeled Egr-1 or c-Krox DNA but not in the presence of SP-1 DNA, indicating that OCZF specifically recognize these sequences (Fig 7A, lanes 3, 4, 8, and 9). Supershifting with FLAG M2 MoAb confirmed that the shifted band contained OCZF protein (Fig 7A, lanes 5 and 10).
(A) EMSA of nuclear extracts from 293T cells transfected with FLAG-tagged OCZF cDNA. c-Krox or Egr-1 DNA was incubated with the nuclear extract from 293T cells transfected with FLAG-OCZF cDNA (O; lanes 2 through 5 and 7 through 10) or vector cDNA (v; lanes 1 and 6), respectively. *The complex of OCZF and DNA. Competition experiments were performed in the presence of 100-fold molar excess of unlabeled c-Krox (lane 3), Egr-1 (lane 8), or SP1 (lanes 4 and 9) DNA. Binding reactions were also performed after preincubation of nuclear extracts with anti-FLAG MoAb (lanes 5 and 10). Thirty micrograms of nuclear extracts was analyzed. (B) Analysis of DNA binding activity with S-tagged Egr-1 and OCZF proteins. S-protein pulldown experiments were performed as described in Materials and Methods. The bound DNA was analyzed by a 10% PAGE. Samples purified on S-protein beads were prepared from 293T cells transfected with pCDL-FLAGa (v1; lane 1), pCDL-FLAGc (v2; lanes 3 and 5) or pCDL-FLAG containing Egr-1 (lane 2) or OCZF (lanes 4 and 6). They were analyzed for DNA binding activity using Egr-1 (lanes 1 through 4) or c-Krox (lanes 5 and 6) consensus sequences as probes. Arrows indicate the bound Egr-1 or c-Krox DNA. Lower bands indicate degraded products of Egr-1 DNA. Autoradiography was performed for 6 days at −80°C for OCZF protein or 3 days at room temperature for Egr-1 protein.
(A) EMSA of nuclear extracts from 293T cells transfected with FLAG-tagged OCZF cDNA. c-Krox or Egr-1 DNA was incubated with the nuclear extract from 293T cells transfected with FLAG-OCZF cDNA (O; lanes 2 through 5 and 7 through 10) or vector cDNA (v; lanes 1 and 6), respectively. *The complex of OCZF and DNA. Competition experiments were performed in the presence of 100-fold molar excess of unlabeled c-Krox (lane 3), Egr-1 (lane 8), or SP1 (lanes 4 and 9) DNA. Binding reactions were also performed after preincubation of nuclear extracts with anti-FLAG MoAb (lanes 5 and 10). Thirty micrograms of nuclear extracts was analyzed. (B) Analysis of DNA binding activity with S-tagged Egr-1 and OCZF proteins. S-protein pulldown experiments were performed as described in Materials and Methods. The bound DNA was analyzed by a 10% PAGE. Samples purified on S-protein beads were prepared from 293T cells transfected with pCDL-FLAGa (v1; lane 1), pCDL-FLAGc (v2; lanes 3 and 5) or pCDL-FLAG containing Egr-1 (lane 2) or OCZF (lanes 4 and 6). They were analyzed for DNA binding activity using Egr-1 (lanes 1 through 4) or c-Krox (lanes 5 and 6) consensus sequences as probes. Arrows indicate the bound Egr-1 or c-Krox DNA. Lower bands indicate degraded products of Egr-1 DNA. Autoradiography was performed for 6 days at −80°C for OCZF protein or 3 days at room temperature for Egr-1 protein.
To further confirm that OCZF has the DNA binding activity, we performed S-protein pulldown experiments using S-tagged OCZF protein produced from pCDL-FLAGc vector containing OCZF cDNA. In the similar experiment, S-tagged Eg-1 protein produced from pCD-FLAGa vector containing Egr-1 cDNA specifically bound its consensus sequence (Fig 7B, lane 2). We found that S-tagged OCZF protein also could specifically bind Egr-1 and c-Krox DNA (Fig 7B, lanes 4 and 6). In contrast, control samples prepared from the extracts transfected with empty vectors, pCDL-FLAGa, and pCDL-FLAGc did not show the band of bound DNA (Fig 7B, lanes 1, 3, and 5). This assay also indicated that the binding affinity of OCZF with these DNA seems to be weaker than that of Egr-1, because the time of autragiography for OCZF was longer than that for Egr-1. Taken together, these results indicate that OCZF could bind specifically to the consensus sequences of Egr-1 and c-Krox.
Transcriptional activity of OCZF.
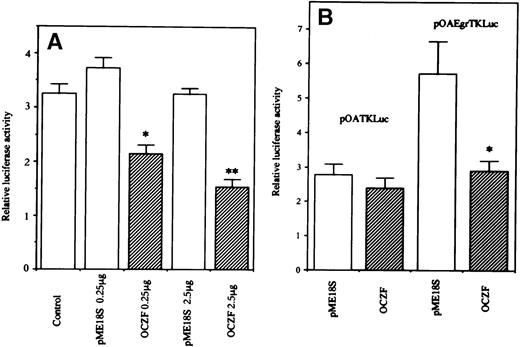
The transcriptional function of OCZF was studied by DNA transfection experiments with 293 and 293T cells using a reporter vector pOA-Egr-TK-Luciferase containing 4 copies of the Egr-1 binding element upstream of a basal TK promoter. When cotransfection of an expression vector containing Egr-1 cDNA with this reporter plasmid, Egr-1 increased the luciferase activity in 293 cells (data not shown). In 293T cells, the cotransfection of expression vector containing OCZF cDNA with a reporter vector pOA-Egr-TK-Luciferase induced a decrease in luciferase activity. The cotransfection of empty expression vector had no effect on the luciferase activity (Fig8A). The transcriptional effect was further studied in 293 cells. The cotransfection of expression vector containing OCZF cDNA with parental reporter plasmid pOA-TK-Luciferase had no effect on the levels of luciferase activity. But, cotransfection of expression vector with pOA-Egr-TK-Luciferase decreased the luciferase activity (Fig 8B). These results indicate that OCZF can act as a regulator of transcription through Egr-1 binding elements.
Transcriptional activity of OCZF. (A) 293T cells were transiently transfected with 1 μg of reporter plasmid pOAEgr-TKLuciferase, along with 0.25 or 2.5 μg of expression vector containing OCZF cDNA (OCZF) or empty vector (pME18S). (B) 293 cells were transiently transfected with 1 μg of reporter plasmid pOATKLuciferase, or pOA-Egr-TKLuciferase with 0.25 μg expression vector containing OCZF cDNA. Rennila luciferase expression vector, p▵TK-RL (0.25 μg), was used as an internal control for transfection. Bars represent the mean and SEM of 3 independent transfections. *P < .05, **P< .01 compared with control reporter vector.
Transcriptional activity of OCZF. (A) 293T cells were transiently transfected with 1 μg of reporter plasmid pOAEgr-TKLuciferase, along with 0.25 or 2.5 μg of expression vector containing OCZF cDNA (OCZF) or empty vector (pME18S). (B) 293 cells were transiently transfected with 1 μg of reporter plasmid pOATKLuciferase, or pOA-Egr-TKLuciferase with 0.25 μg expression vector containing OCZF cDNA. Rennila luciferase expression vector, p▵TK-RL (0.25 μg), was used as an internal control for transfection. Bars represent the mean and SEM of 3 independent transfections. *P < .05, **P< .01 compared with control reporter vector.
Effect of blockage of OCZF mRNA on osteoclast differentiation.
To determine the functional role of OCZF in the osteoclast differentiation, we added 2 types of OCZF sense and antisense ODNs (ODN-1 and ODN-2) to bone marrow culture that form MNCs for different periods. The addition of OCZF antisense ODN-1 for 1 to 5 days and 3 to 5 days partially inhibited the MNC formation (Fig 9A; experiments no. 1 and 2). To confirm the finding regarding the inhibitory effect of antisense OCZF ODN-1, we added OCZF sense, scrambled, and antisense ODN-2, which have sequences different from that of ODN-1, to 2 types of bone marrow cultures that form POCs or MNCs. As shown in Fig 9A (experiment no. 3), the treatment with OCZF antisense but not sense or scramble ODN-2 caused an inhibition of MNC formation. In contrast, the addition of antisense ODN-2 did not inhibit the formation of POCs in the stromal cell-deprived bone marrow culture (Fig 9B). These data suggest that OCZF is involved in the fusion process of mononuclear precursors in osteoclast differentiation. We therefore added the same ODN-2 to cultures that form macrophage polykaryons. In this culture system, macrophage polykaryons were formed that were negative for TRAP-staining. Interestingly, the antisense ODN-2 had no effect on the formation of the macrophage polykaryons (Fig 9C).
Effects of expressional blockage of OCZF cDNA with antisense ODN on the formation of MNCs, POCs, and macrophage polykaryons. (A) Inhibition of MNC formation with antisense ODN but not sense or scrambled ODN in bone marrow culture. Bone marrow cells were cultured with 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of MNCs. After 5 days of culture, TRAP-positive MNCs were counted. (B) Effect of ODN-2 on the formation of POCs. Nonadherent bone marrow cells were cultured in the presence of 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of POCs. After 5 days of culture, TRAP-positive mononuclear cells were counted. (C) Effect of ODN-2 on the formation of macrophage polykaryons. Bone marrow cells were cultured with 10−7mol/L PMA and 10−8 mol/L 1,25(OH)2D3. After 4 days of culture, TRAP-negative MNCs were counted. One micromole per liter ofOCZF antisense (A) or sense (S) ODN-1 ([A] experiments no. 1 and 2) or antisense (A), sense (S), or scrambled (Scr) ODN-2 ([A] experiment no. 3, [B], and [C]) was added to the culture. Data are the mean ± SEM of quadruplicate cultures. **P < .01 compared with control.
Effects of expressional blockage of OCZF cDNA with antisense ODN on the formation of MNCs, POCs, and macrophage polykaryons. (A) Inhibition of MNC formation with antisense ODN but not sense or scrambled ODN in bone marrow culture. Bone marrow cells were cultured with 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of MNCs. After 5 days of culture, TRAP-positive MNCs were counted. (B) Effect of ODN-2 on the formation of POCs. Nonadherent bone marrow cells were cultured in the presence of 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of POCs. After 5 days of culture, TRAP-positive mononuclear cells were counted. (C) Effect of ODN-2 on the formation of macrophage polykaryons. Bone marrow cells were cultured with 10−7mol/L PMA and 10−8 mol/L 1,25(OH)2D3. After 4 days of culture, TRAP-negative MNCs were counted. One micromole per liter ofOCZF antisense (A) or sense (S) ODN-1 ([A] experiments no. 1 and 2) or antisense (A), sense (S), or scrambled (Scr) ODN-2 ([A] experiment no. 3, [B], and [C]) was added to the culture. Data are the mean ± SEM of quadruplicate cultures. **P < .01 compared with control.
Northern analysis of OCZF mRNA from antisense, sense, and scrambled ODN-2–treated bone marrow cells. Bone marrow cells were cultured with 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of MNCs in the presence of 1 μmol/L OCZF ODN-2 for 4 days. RNA was isolated from the 4-day culture and approximately 1 μg of each sample was analyzed. Northern analysis was performed as described in Materials and Method. The same filter was rehybridized with a human β-actin probe as a control. Arrows indicate the 5.3- and 3.3-kb bands of OCZF and β-actin mRNA. Lanes correspond to RNA from scramble (lane 1), sense (lane 2), and antisense (lane 3) ODN-treated cells.
Northern analysis of OCZF mRNA from antisense, sense, and scrambled ODN-2–treated bone marrow cells. Bone marrow cells were cultured with 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of MNCs in the presence of 1 μmol/L OCZF ODN-2 for 4 days. RNA was isolated from the 4-day culture and approximately 1 μg of each sample was analyzed. Northern analysis was performed as described in Materials and Method. The same filter was rehybridized with a human β-actin probe as a control. Arrows indicate the 5.3- and 3.3-kb bands of OCZF and β-actin mRNA. Lanes correspond to RNA from scramble (lane 1), sense (lane 2), and antisense (lane 3) ODN-treated cells.
To confirm the antisense strategy, we next examined the effect of antisense ODN-2 on OCZF mRNA levels. Poly(A)RNA was isolated from the treated cells, and we performed a Northern analysis (Fig 10). The level of OCZF mRNA of the cells treated with antisense ODN-2 was significantly lower than those treated with sense or scrambled ODN-2. This result confirmed that antisense ODN-2 successfully blocked the expression of OCZF mRNA.
DISCUSSION
In the present study, we provide evidence that OCZF is a possible transcription factor that regulates the fusion process in osteoclastogenesis. Osteoclasts are highly specialized multinucleated bone-resorbing cells originating from hematopoietic stem cells. The process of osteoclast differentiation involves an early stage, the proliferation and differentiation of osteoclast progenitors into mononuclear preosteoclasts, and a late stage, the fusion of preosteoclast progenitors into multinucleated osteoclasts, which are under the control of the bone microenvironment. In the course of osteoclast differentiation, the late-stage multinucleation step is a critical step in bone resorption. Although data have accumulated related to the involvement of cytokines in osteoclastogenesis,37 the regulatory mechanism of this important process is poorly understood. Interestingly, our analysis of the functional role of OCZF with specific antisense ODN indicated that OCZF is involved in the formation of MNCs but not in that of POCs. In addition, OCZF antisense ODN did not inhibit the fusion of macrophages, suggesting that the inhibition of fusion by this ODN was specific for osteoclast differentiation. It was recently reported that surface molecules such as E-cadherin and CD98 expressed on osteoclast precursor cells are involved in the fusion process in osteoclast differentiation.38 39 The signal transduction of such molecules may lead to the expression of OCZF.
To elucidate the molecular mechanism of osteoclast differentiation, it is important to know the transcription factors specifically expressed in osteoclasts. However, there is as yet little information about nuclear transcription factors specifically expressed in mature osteoclasts. Jimi et al40 found that NFκB-like transcription factor is expressed in osteoclast-like cells. In addition, Iotsova et al41 recently reported that mice lacking both of the transcription factors NFκ-B1 and NFκ-B2 developed osteopetrosis. However, NFκB is a ubiquitous transcription factor expressed in various cells. Inoue et al42 recently reported that the expression of an osteoclast nuclear transcription factor (NFOC-1) that bound to the human T-cell leukemia virus type I-long terminal repeat enhancer element was found in osteoclast-like cells but not in macrophages. The detailed sequence of NFOC-1 is not known, but, interestingly, NFOC-1 seems not to be Fos, but had reactivity to anti-JunD antibody.42 In this study, we demonstrated by in situ hybridization that zinc finger protein OCZF is preferentially expressed in mature osteoclasts. In addition, the expression of OCZF mRNA is increased during the differentiation into multinucleated osteoclast-like cells, but is not increased during differentiation into macrophages.
OCZF encoded a protein with POZ domain and Krüppel-type 3 zinc fingers. OCZF was considered to be rat homologue of LRF that was recently reported as a potential target for BCL-6 oncogene. High conservation between rat OCZF and mouse LRF indicates that this factor has a very important role. Because it is not known whether BCL-6 is expressed in osteoclasts, further studies will be required to elucidate the function of the interaction of BCL-6 and OCZF in osteoclastogenesis. Five percent to 10% of Krüppel-type zinc finger proteins contain POZ domain in their N-terminus. This domain is known to form homomeric and heteromeric interactions with other POZ domains43,44 and is required for transcriptional repression of several proteins, including hcKrox, BCL-6, and PLZF.45-47 Punctuate nuclear staining patterns are commonly seen in these zinc-finger POZ proteins.48 49 In this study, we found that OCZF had the repressive activity of transcription like other POZ domain proteins. Interestingly, OCZF by immunofluorescence with Kat6 antibody showed the localization of this protein to discrete regions of the nucleus and did not show diffuse staining in osteoclasts (Fig 1).
The amino acid sequence of POZ domain of OCZF is very similar to those of human APM-1 and hcKrox. APM-1 is recently isolated human protein involved in human papillomavirus (HPV) integration region.24 APM-1 transcripts were detected in normal cervical keratinocytes, but not in the cervical carcinoma cell lines. Expression of APM-1 gene strongly reduced clonal cell growth, suggesting that APM-1 may function as a tumor-suppressor gene. The hcKrox gene was recently isolated as c-Krox homologue from a human fibroblast cDNA library.26 hcKrox has an ability to repress the transcription of extracellular matrix genes, including α1 type I procollagen and fibronectin mRNA. The POZ domain of OCZF protein had also homology with that of Miz-1 and PLZF, both of which were shown to have potent suppressive effect on cell growth.30 50 It is interesting that POZ domain of OCZF had homology with APM-1, Miz-1, and PLZF, which had cell growth inhibitory activity.
Recently, POZ domain proteins have been implicated in embryonic development and in hematopoiesis.32,51 PLZF and BCL-6 have important roles in the terminal differentiation of the cells. Loss of PLZF function blocks terminal differentiation of hematopoietic precursor cells.52 BCL-6 is expressed in many types of the cells but is expressed at a high level in muscle cells. During the differentiation of proliferating myoblasts into myotubes, BCL-6 expression is upregulated. BCL-6 was considered to be involved in muscle terminal differentiation.53 In terminal differentiation, the cells stop DNA synthesis. The terminal differentiation step into osteoclasts, the fusion process is not required for DNA synthesis.54 55 OCZF may be involved in the regulation of cell growth or cell cycle, which is possibly involved in the fusion process into osteoclasts.
The zinc finger region of OCZF had high homology with APM-1 and hcKrox. However, the detailed binding DNA sequences for these factors have been not shown. Because the zinc finger region of OCZF also had homology with c-Krox and Egr-1, we have analyzed the ability of DNA binding of OCZF to Egr-1 and c-Krox binding sequence. We have found that OCZF bound these sequences specifically. In this study, we also found that OCZF repressed the transcriptional activity through the Egr-1 binding site. These data suggest that some genes that are expressed in osteoclasts and have the Egr-1 binding sequence in the regulatory region may be regulated by OCZF. However, OCZF may bind similarly to another target sequence with high affinity, because the binding affinity of OCZF for Egr-1 binding sequence seems to be lower compared with that of Egr-1. To determine high-affinity binding sequences and elucidate target genes of OCZF, further studies will be required.
The developmental process of hematopoietic cells is controlled by the combination of several genes. The osteopetrotic phenotype of NFκ-B1 and NFκ-B2 double-knockout mouse model resembles that of c-fos–deficient animals; however, the induction of c-fos expression was detected in this mutant mouse. Because these 2 transcription factors expressed in wide types of the cells, these results suggest that some other common transcription factor expressed specifically in the cells of osteoclast lineages may interact with these factors to restrict their functions. In fact, some transcription factors are known to self-associate to form dimmers44 or interact with other transcription factors and regulate the differentiation in a combined fashion.51 56 Thus, the temporal expression of a set of several transcription factors is thought to be required for the ongoing process of differentiation into osteoclasts. The POZ domain of several transcription factors is a region interacting with other proteins, suggesting that POZ domain proteins have some role in the osteoclastogenesis. At present, the molecular mechanism of the involvement of OCZF in the multinucleation step in osteoclast differentiation is not entirely clear. Future studies will clarify the possible functions of the OCZF protein in the regulation of differentiation and transcription in osteoclast-specific lineage cells.
ACKNOWLEDGMENT
The authors thank Dr T. Watanabe (Kyushu University Medical Institute of Bioregulation) and Dr H. Kishi (Toyama University) for helpful suggestions regarding cDNA cloning and Dr S. Matsuhashi (Saga Medical School) for helpful suggestions regarding immunocytochemical study. We also thank Dr M. Kobayashi (Hokkaido University) for donating the WRT-7 P2 cell line. We also thank Dr K. Hori (Vice-President, Saga Medical School) for encouragement and Dr Lin Xin Xu for assistance.
Supported in part by a Grant for Scientific Research from the Japanese Ministry of Education, Science and Culture (Project No. 05671543).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Akiko Kukita, PhD, Department of Microbiology, Saga Medical School, 5-1-1, Nabeshima, Saga 849-8501, Japan; e-mail: kukita@smsnet.saga-med.ac.jp.



![Fig. 9. Effects of expressional blockage of OCZF cDNA with antisense ODN on the formation of MNCs, POCs, and macrophage polykaryons. (A) Inhibition of MNC formation with antisense ODN but not sense or scrambled ODN in bone marrow culture. Bone marrow cells were cultured with 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of MNCs. After 5 days of culture, TRAP-positive MNCs were counted. (B) Effect of ODN-2 on the formation of POCs. Nonadherent bone marrow cells were cultured in the presence of 10−8 mol/L 1,25(OH)2D3 and htROSCM for the formation of POCs. After 5 days of culture, TRAP-positive mononuclear cells were counted. (C) Effect of ODN-2 on the formation of macrophage polykaryons. Bone marrow cells were cultured with 10−7mol/L PMA and 10−8 mol/L 1,25(OH)2D3. After 4 days of culture, TRAP-negative MNCs were counted. One micromole per liter ofOCZF antisense (A) or sense (S) ODN-1 ([A] experiments no. 1 and 2) or antisense (A), sense (S), or scrambled (Scr) ODN-2 ([A] experiment no. 3, [B], and [C]) was added to the culture. Data are the mean ± SEM of quadruplicate cultures. **P < .01 compared with control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1987/5/m_blod41826009x.jpeg?Expires=1771166927&Signature=zkofeof7AY4xgVodbVOrzZZAxJE8k4xGXAZ-JzmDpc8qoFCJXUCjdbf1OleDCmr7yOZYsfkVG7-UaBsP6JOrE1zJvkMt6NhNPeHOL3HNgL1gJvpK1PfDeYOtTVf2JkvZNki7fmcQ5YBPwHu138ThWCM3ucAD2NO4J~vr1eCPan6OHkqlV3WILVN7JiMjpH99mnUldP6GyQCjKGJWFtNjHHFv-5NqXJRaW8Fqyl5wcbTu~a3mzOJkwc1CEt7F0PNa5zSmMvXGNcX6ob-nQV1G7QC-1sLK6SQN1vdVG5r6Bb4hsa6V88L3OjeEPA3NNDk5WLk61qnpIu-LpPH6ofj4qQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)