Abstract
Nitric oxide (NO) regulates production of vascular endothelial growth factor (VEGF) by normal and transformed cells. We demonstrate that NO donors may up-regulate the activity of the human VEGF promoter in normoxic human glioblastoma and hepatoma cells independent of a cyclic guanosine monophosphate–mediated pathway. Deletion and mutation analysis of the VEGF promoter indicates that the NO-responsive cis-elements are the hypoxia-inducible factor-1 (HIF-1) binding site and an adjacent ancillary sequence that is located immediately downstream within the hypoxia-response element (HRE). This work demonstrates that the HRE of this promoter is the primary target of NO. In addition, VEGF gene regulation by NO, as well as by hypoxia, is potentiated by the AP-1 element of the gene. Our study also reveals that NO and hypoxia induce an increase in HIF-1 binding activity and HIF-1 protein levels, both in the nucleus and the whole cell. These results suggest that there are common features of the NO and hypoxic pathways of VEGF induction, while in part, NO mediates gene transcription by a mechanism distinct from hypoxia. This is demonstrated by a difference in sensitivity to guanylate cyclase inhibitors and a different pattern of HIF-1 binding. These results show that there is a primary role for NO in the control of VEGF synthesis and in cell adaptations to hypoxia. (Blood. 2000;95:189-197)
Angiogenesis, the sprouting of new capillaries from preexisting blood vessels, is a multistep process that involves migration and proliferation of endothelial cells, remodeling of the extracellular matrix, and functional maturation of the newly assembled vessels.1,2 Physiologically, angiogenesis is a tightly regulated process, resulting from the balance of angiogenic and angiostatic stimuli. These stimuli are regulated temporally and spatially, as for example during early embryonic development, organogenesis, and wound healing. At other times, angiogenesis is completely inhibited.3 Unregulated angiogenesis is the cause of severe tissue dysfunction, and has been directly implicated in the pathogenesis of various diseases including retinopathies, psoriasis, rheumatoid arthritis, and other chronic inflammatory diseases.4 Moreover, angiogenesis is essential for solid tumor outgrowth.5
The endothelial cell-specific vascular endothelial growth factor (VEGF) exerts a pivotal role in normal and pathological angiogenesis.6 Its production by stromal or epithelial cells is sufficient to trigger angiogenesis, and inactivation of the corresponding gene results in abnormal blood vessel development and embryonic lethality in mice.7 Indeed, synthesis of VEGF, followed by its secretion into the extracellular environment, is 1 of the primary steps in the angiogenic cascade and controls the onset, extent, and duration of this process. A number of angiogenic stimuli have been found to induce VEGF expression including several growth factors and cytokines, hormones, phorbol esters, oncogenes, nitric oxide (NO), and hypoxia.8 VEGF gene expression in hypoxic cells is characterized by its transcriptional activation,9-12 primarily through the hypoxia-response element (HRE) that includes cis-acting DNA elements recognized by multiple transactivators.9 12-14
Hypoxia-inducible factor-1 (HIF-1) is the best-characterized regulator of the VEGF gene transcription. In its active form, it is a dimer composed of 2 distinct subunits, both of which belong to the basic helix-loop-helix–per-arnt-sim (bHLH-PAS) protein family: HIF-1α and HIF-1β, the aryl hydrocarbon receptor nuclear translocator (ARNT).15 Under hypoxic conditions, active HIF-1 complexes accumulate in the cell nucleus. They bind to the target DNA sequence (HIF-1 binding site) within the HRE and enhance the hypoxia-inducible gene transcription rate.15
Nitric oxide is an intracellular and intercellular signaling molecule, generated in eukaryotic cells from L-arginine by a reaction catalyzed by NO synthases.16 A wide range of biological effects are attributed to this molecule.17 Some effects are linked to its intracellular second messenger nature, while others result from its paracrine actions, mediated by activation of the guanylate cyclase/3′,5′-cyclic guanosine monophosphate (GC/cGMP) pathway.18,19 Indeed, although NO is highly reactive and believed to be quite unstable in vivo, once produced in sufficient amounts it can travel significant distances in the tissue to reach multiple cellular targets.20
There is a considerable body of evidence that NO downregulates the expression of VEGF gene.21-25 In spite of these observations, production of angiogenic activity by human monocytes has been found to depend on NO,26 and NO-generating compounds have been shown to stimulate the VEGF gene transcription in human glioblastoma and hepatoma cells in culture.27 Furthermore, a strong positive correlation between NO synthase (NOS) activity, cGMP levels, and tumor angiogenesis has been recently described in head and neck28 and gynecological cancers.29 30 We have investigated the mechanism of NO-mediated regulation of the human VEGF gene in human glioblastoma and hepatoma cells. Our results show that the VEGF gene transcription is activated by NO as well as by hypoxia via the HIF-1 binding site and an adjacent “ancillary” sequence within the HRE of this gene. This response to NO is mediated, at least in part, by activation of the HIF-1 complex independent of the GC/cGMP pathway.
Materials and methods
Transient expression assays
The sequence phVEGF1 (provided by Dr A. Minchenko31) contains the promoter and 5′-flanking sequence of the human VEGF gene between positions -2279 and +54, cloned into the pGL2-basic vector (Promega). A series of deletion mutants was prepared by restriction endonuclease digestion and religation. The sequence pT81luc0 (L Cicatiello and A Weisz, unpublished data), modified from pT81luc (provided by Dr S.K. Nordeen32) contains the Herpes Simplex virus thymidine kinase (HSV-TK) gene promoter, upstream of the luciferase coding sequence. pHREL, pHRE, and related mutants of pHRE (pHREm1 to 3) were prepared by amplifying a specific segment of the 5′-flanking region of the human VEGF gene with polymerase chain reaction (PCR) and cloning it in a single copy, upstream of the HSV-TK promoter of pT81luc0. The pHRA and pHRB sequences were prepared by ligating commercially synthesized oligonucleotides to pT81luc0. The pSV-nlslacZ sequence (SV40-driven promoter) was used as a control for monitoring transfection efficiency; it contains lacZ coding sequences. Constructs (5 μg of the reporter plasmid and 1 μg of pSV-nlslacZ) were transfected into human glioblastoma A-172 or hepatoma Hep3B cells (Japanese Collection of Research Bioresources, Tokyo, Japan) at 20%-30% confluence in a 10-cm tissue culture plate, with 20 μL of lipofectin (Life Technologies, Rockville, MD).
After incubation at 37°C for 15 hours, the DNA-containing medium was replaced with normal culture medium. The cells were then incubated at 37°C before harvesting under normoxic conditions (21% O2) or following exposure to either hypoxic conditions (1% O2); the NO donors, S-nitroso-N-acetyl-D, L-penicillamine (SNAP), 3-(hydroxy-1-(1-methylethyl)-2-nitrosohydrazino)-1-propanamine (NOC5); or sodium nitroprusside (SNP). SNAP was dissolved in dimethyl sulfoxide (DMSO), and NOC5 and SNP were dissolved in phosphate-buffered saline (PBS) immediately before use. An aliquot of SNAP was added 12 hours before harvest. However, because the half-life of NOC5 (25 minutes) is much shorter than that of SNAP (8 hours), the first half-dose of NOC5 was added at 12 hours, and the remaining dose was added 6 hours before cell harvest in A-172 cells. Harvested cells were dissolved in 200 μL of 0.25 mol/L Tris-Cl, pH 7.5. Cell lysis was performed by 4 freeze-thaw cycles.
Luciferase activity was determined by mixing 100 μL of cell extract with 225 μL of luciferin reagent. The luciferin reagent was prepared by mixing 75 μL of luciferin stock solution (0.5 mmol/L D-luciferin [SIGMA, St Louis, MO], 25 mmol/L glycylglycine [ph 7.8]) and 150 μL of luciferase assay buffer (25 mmol/L glycylglycine [ph 7.8], 15 mmol/L KPO4, 15 mmol/L MgSO4, 4 mmol/L EGTA, 2 mmol/L ATP, and 1 mmol/L dithio treitol [DTT]). Luminescence was measured for 20 seconds in a luminometer (Luminescencer-JNR; ATTO, Tokyo, Japan), and results were expressed as relative light units. We measured β-galactosidase activity by using 50 μL of cell extract and a 690-μL mixture of 25.4 mmol/L Tris- Cl (pH 7.5); 1.4 mmol/L MgCl2; 58 mmol/L NaPO4 (pH 7.5); 1.4% β-mercaptoethanol; and 1.1 mg/mLo-nitrophenyl-β-D-galactopyranoside (SIGMA). Incubation was completed at 37°C for 0.5-1 hour. To determine theA420, we stopped the reaction by adding 50μL of 1 mol/L Na2CO3. The relative luc activity (mean ± standard error of the mean) was defined as luciferase activity standardized by β-galactosidase activity. Fold induction was defined as the ratio of the relative luc activity of stimulated cells to that of unstimulated controls.
Preparation of nuclear and whole-cell extracts
Cells at 60%-70% confluence were incubated at 37°C before harvest, given the following: under normoxic (21% O2) or hypoxic (1% O2) conditions (8 hours for A-172 cells and 12 hours for Hep3B cells), or with DMSO (0.1%) or SNAP (0.5 mmol/L in 0.1% DMSO) under normoxic conditions (3 hours for A-172 cells and 8 hours for Hep3B cells). The cells were scraped free and centrifuged at 270g for 10 minutes at 4°C. Nuclear extracts were prepared with buffers A and C, as previously described,33except that dialysis procedures were omitted. The pellet was resuspended in buffer A and incubated on ice for 10 minutes before being homogenized by pipetting 5-8 times with a syringe. The nuclei were pelleted by centrifugation at 12 000g for 2 minutes at 4°C. They were then resuspended in ice-cold buffer C and mixed by rotation for 30 minutes at 4°C. After centrifugation at 16 000g for 10 minutes at 4°C, the supernatant was stored at –70°C, pending electrophoretic mobility shift assay (EMSA) and Western blot analysis. Whole-cell extracts were prepared as previously described.34 In brief, the cells were harvested in 10 buffer (40 mmol/L Tris-Cl [pH 7.9], 10 mmol/L EDTA [pH 8.0], and 150 mmol/L NaCl). The cell pellet was resuspended in whole-cell extract buffer (10 mmol/L Hepes [pH 7.9]; 400 mmol/L NaCl; 0.1 mmol/L EDTA; 5% [vol/vol] glycerol; 1 mmol/L DTT; and 1 mmol/L phenylmethylsulfonyl fluoride). It was then centrifuged at 16 000g for 30 minutes at 4°C. The supernatant was stored at –70°C until required. Protein concentration was determined by assay (Bio-Rad Protein assay; Bio-Rad Laboratories, Hercules, CA).
Electrophoretic mobility shift assay
Nuclear extracts (5 μg) from the control or stimulated cells were incubated with 3 × 104 cpm of a32P-labeled double-stranded oligonucleotide probe and 0.1 μg of denatured calf thymus DNA, in modified buffer Z+ (58.5 mmol/L KCl) for 30 minutes at room temperature, as previously.12Electrophoresis was performed on 5% nondenaturing polyacrylamide gels at 25 mA in 1 × TAE at 4°C. Autoradiography of gels was performed (Bioimage Analyzer BAS 2000; Fuji Photo Film Co., Tokyo, Japan). Competition experiments were performed with 10-fold to 250-fold molar excess of unlabeled oligonucleotides, relative to the labeled probe. For SS assays, 1 μL each of antiserum specific for HIF-1α (provided by Dr DM Livingston35), HIF-1β (Affinity Bioreagents, Golden, CO), or c-Myc (Calbiochem, La Jolla, CA) were added to the binding reaction mixture without the labeled probe. These mixtures were incubated for 30 minutes at 4°C. The labeled probe was then added, and incubation continued for 30 minutes at room temperature.
Western blot analysis
For Western blots, anti-HIF-1α monoclonal antibody (mAb) (Novus Biologicals, Littleton, CO) was used according to the manufacturer's protocol. In brief, 30 μg of nuclear or whole-cell extracts per lane were resolved using SDS/6% polyacrylamide gels. The proteins were then transferred onto nitrocellulose membranes in the blotting buffer (5% [vol/vol] methanol, 25 mmol/L Tris, 120 mmol/L glycine). Membranes were blocked with 5% nonfat dried milk, 2% bovine serum albumin, and TBS-T (50 mmol/L Tirs-Cl [pH 7.5], 150 mmol/L NaCl, and 0.1% Tween-20). Endogenous HIF-1α protein was probed with 1:1000 dilution of anti–HIF-1α mAb. Horseradish peroxidase-conjugated anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a secondary antibody at a dilution of 1 in 5000 in nonfat dried milk/TBS-T. The protein complexes were visualized by enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Statistical analysis
The results are expressed as mean ± standard error of the mean (SEM). Comparison of 2 means was performed by the use of unpaired Student's t tests. Statistical significance was assumed at a value of P < 0.05.
Results
Analysis of the human VEGF promoter response to nitric oxide
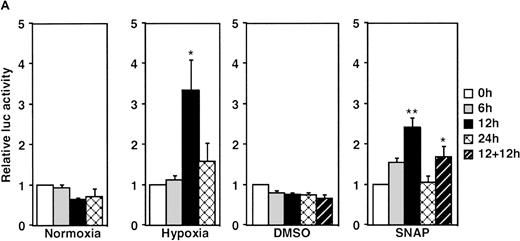
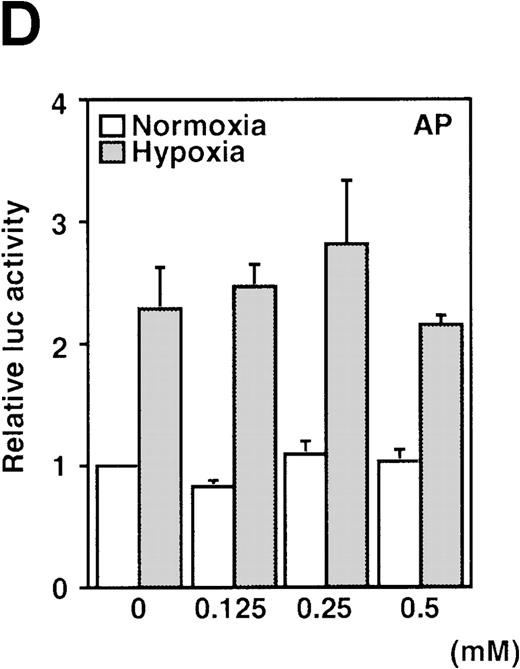
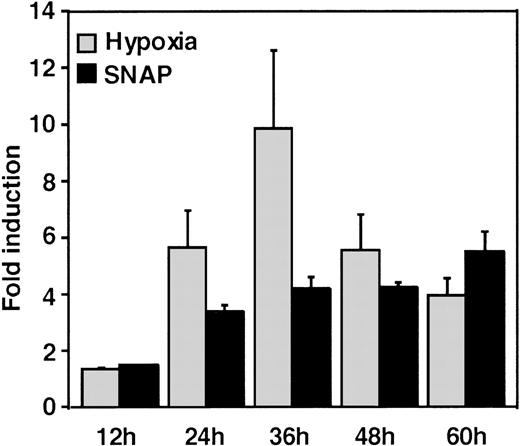
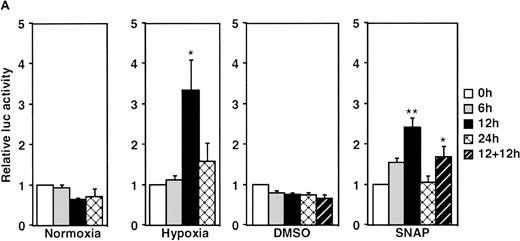
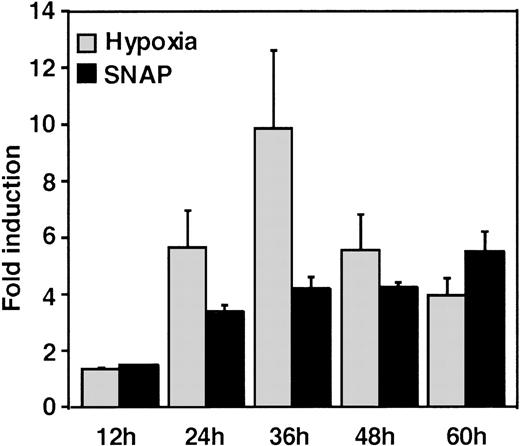
A luciferase reporter phVEGF1 was used to test the effect of NO donors on the activity of the human VEGF promoter in A-172 cells. SNAP enhanced the activity of the transfected promoter in a dose-dependent manner within 6 to 12 hours (Figure 1). The chemically distinct NO donor 3-(2-hydroxy-1-(1-methylethyl)-2-nitrosohydrazino)-1-propanamine (NOC5) was as effective as SNAP in inducing the reporter gene activation (Figure 1C), whereas acetylpenicillamine (AP), the non–NO-releasing analog of SNAP, did not elicit any promoter response at concentrations up to 0.5 mmol/L (Figure 1D). These results suggest that NO enhances the transcription of the VEGF gene. Similar dose-response correlations and induction kinetics were observed in the same cells for the endogenous VEGF gene activation by these NO donors.27 The response of the transfected VEGF promoter to SNAP was transient. It was maximal after 12 hours (P < 0.01 versus control) and decreased by 24 hours (Figure 1A). This was due to the relatively limited half-life of NO release by this compound in aqueous media.36 The late decrease in the promoter activity observed with a single dose of SNAP was prevented by a second application of this compound after the first 12 hours of stimulation (P < 0.05 versus control) (Figure 1A). For comparison, the effect of hypoxia (1% O2) on phVEGF1 expression was also determined in A-172 cells under the same experimental conditions. As shown in Figure 1A, transcription of the transfected reporter gene was enhanced by hypoxia (P < 0.01 versus control). This is consistent with the fact that this reporter contains the HRE of the VEGF gene.9,10 12 The response of the transfected VEGF promoter to SNAP and hypoxia in human hepatoma Hep3B cells was also tested. Maximum induction was obtained in 36 hours after exposure to hypoxia and 24 hours or later after exposure to SNAP (Figure2).
Effect of NO and hypoxia on the expression of the VEGF reporter gene in A-172 cells.
(A) Time-course of human VEGF promoter activity in A-172 cells. The cells were exposed to normoxia (21% O2), hypoxia (1% O2), DMSO (0.1%), or SNAP (0.5 mmol/L in 0.1% DMSO) for 0, 6, 12, or 24 hours. In the result labeled 12 + 12h, a half dose of SNAP was added at 24 hours, and the remaining dose was added 12 hours before harvesting the cells. *P < 0.05, **P < 0.01 versus corresponding controls; n = 6 independent experiments. (B-D) Effect of SNAP (B), NOC5 (C), and AP (D) on human VEGF promoter activity in A-172 cells. The cells were stimulated for 12 hours under normoxic or hypoxic conditions. The final concentration of DMSO was 0.1% in (B) and (D). (NOC5 was dissolved in PBS.) (B): n = 8; *P < 0.05, **P < 0.01 versus control in normoxia, #P < 0.01 versus control in hypoxia. (C): n = 8; *P < 0.01 versus control in normoxia, #P < 0.01 versus control in hypoxia. (D): n = 6; no significant difference. Relative luc activity represents the mean ± SEM of the ratio of luciferase/β-galactosidase activity.
Effect of NO and hypoxia on the expression of the VEGF reporter gene in A-172 cells.
(A) Time-course of human VEGF promoter activity in A-172 cells. The cells were exposed to normoxia (21% O2), hypoxia (1% O2), DMSO (0.1%), or SNAP (0.5 mmol/L in 0.1% DMSO) for 0, 6, 12, or 24 hours. In the result labeled 12 + 12h, a half dose of SNAP was added at 24 hours, and the remaining dose was added 12 hours before harvesting the cells. *P < 0.05, **P < 0.01 versus corresponding controls; n = 6 independent experiments. (B-D) Effect of SNAP (B), NOC5 (C), and AP (D) on human VEGF promoter activity in A-172 cells. The cells were stimulated for 12 hours under normoxic or hypoxic conditions. The final concentration of DMSO was 0.1% in (B) and (D). (NOC5 was dissolved in PBS.) (B): n = 8; *P < 0.05, **P < 0.01 versus control in normoxia, #P < 0.01 versus control in hypoxia. (C): n = 8; *P < 0.01 versus control in normoxia, #P < 0.01 versus control in hypoxia. (D): n = 6; no significant difference. Relative luc activity represents the mean ± SEM of the ratio of luciferase/β-galactosidase activity.
Time-course of human VEGF promoter activity in Hep3B cells.
The cells were exposed to the same conditions as Figure 1 for 12, 24, 36, 48, or 60 hours. All data within 24-60 hours are significantly greater than for controls (P < 0.01). n = 6 independent experiments. Fold induction by hypoxia or by SNAP represents the ratio of relative luc activity in cells at 1% O2, or 0.5 mmol/L SNAP in 0.1% DMSO, to those in 21% O2 or 0.1% DMSO, respectively.
Time-course of human VEGF promoter activity in Hep3B cells.
The cells were exposed to the same conditions as Figure 1 for 12, 24, 36, 48, or 60 hours. All data within 24-60 hours are significantly greater than for controls (P < 0.01). n = 6 independent experiments. Fold induction by hypoxia or by SNAP represents the ratio of relative luc activity in cells at 1% O2, or 0.5 mmol/L SNAP in 0.1% DMSO, to those in 21% O2 or 0.1% DMSO, respectively.
The response seen in Hep3B cells was more intense than the response to either stimulus in A-172 cells. This was also seen in this cell line under hypoxia.12 However, it should be noted that the relatively low quantitative responses of phVEGF1 to NO and hypoxia that we observed appear to be a general feature of the response of both chromosomal10 and transfected VEGF genes to inducers.10,37 The overall response of this gene to a variety of stimuli, including NO, results from a combination of transcriptional activation and mRNA stabilization.10,27,38 39
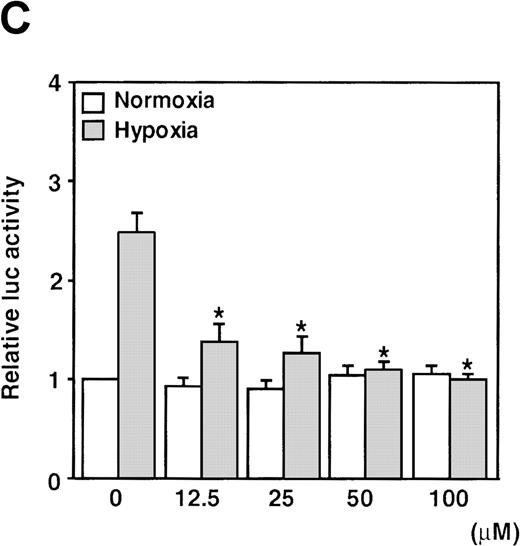
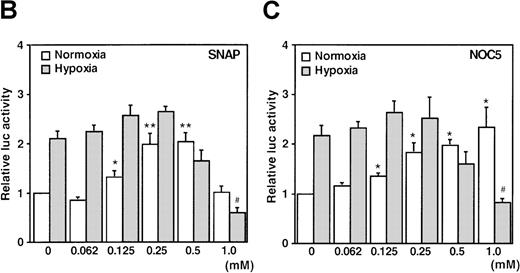
When cells were stimulated with either SNAP or NOC5 under hypoxic conditions, maximal promoter activation was achieved by lower concentrations of NO donors, as compared with normoxic conditions (Figure 1B and C). However, 0.5 mmol/L of either donor, the optimal concentration for reporter activation under normoxia, inhibited hypoxic induction of the reporter gene (Figure 1B and C).
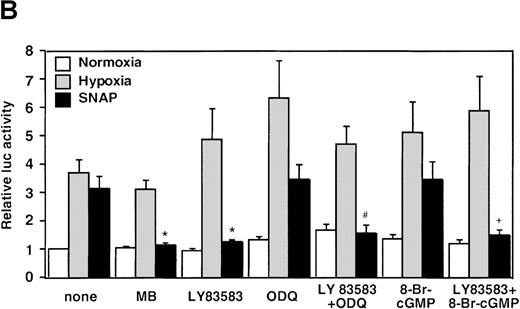
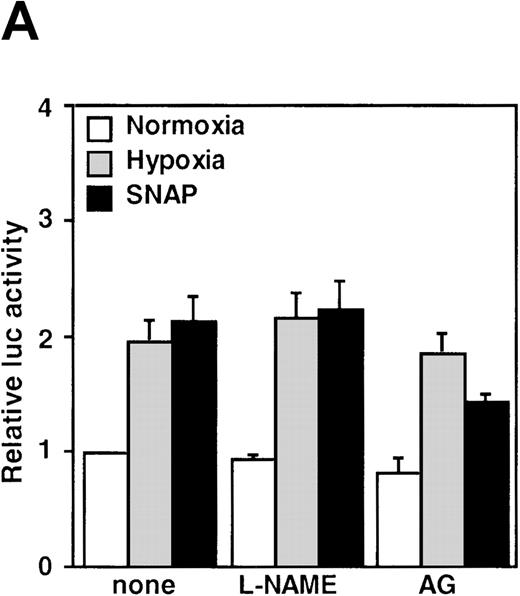
The NOS inhibitors NG-nitro-L-arginine methyl ester (L-NAME, 2 mmol/L) or aminoguanidine (AG, 5 mmol/L) did not interfere with hypoxia-induced VEGF promoter activation (Figure3A). This indicates that NO production by endogenous NOS is not essential for transcriptional regulation of the VEGF gene in hypoxic cells. Studies with GC inhibitors have suggested that VEGF mRNA accumulation in response to NO donors is mediated by an increase in intracellular cGMP levels.27 The response of the reporter to NO in the presence of GC inhibitors was therefore analyzed. When cells were incubated with SNAP for 12 hours in the presence of either methylene blue (MB, 25 μmol/L) or 6-anilino-5,8-quinolinequinone (LY83 583, 1.25 μmol/L), NO-induced promoter activation was completely inhibited (P < 0.01 versus control in SNAP). In contrast, acting alone, 1H-[1,2,4]oxidiazolo[4,3-a]quinoxalin-1-1 (ODQ, 25 μmol/L), another specific GC inhibitor, did not inhibit the NO-induced transcriptional activation even at the higher concentration. However, LY83 583 (1.25 μmol/L) could attenuate the activation to the levels of the untreated cells in the presence of ODQ (25 μmol/L) (P < 0.05 versus ODQ+SNAP) (Figure 3B). In addition, to test if an increase of cGMP levels could enhance VEGF promoter activity, 8-Br-cGMP (a protein kinase G activator) was added to the culture medium at a concentration of 800 μmol/L, in the presence or absence of LY83 583 (1.25 μmol/L). In both cases, 8-Br-cGMP did not show any effect on NO-induced promoter activity. Hypoxic induction of the same reporter gene was unaffected by either GC inhibitor or by 8-Br-cGMP when tested under the same experimental conditions (Figure 3B). Despite the suppression of NO-induced VEGF promoter activation by MB and LY83 583, these results suggest that the NO-induced activation is not mediated by the GC/cGMP pathway.
Effects of inhibitors on human VEGF promoter activity in A-172 cells.
The cells were exposed to the same conditions as Figure 1for 12 hours. (A) Effect of the NOS inhibitor (2 mmol/L L-NAME or 5 mmol/L AG). n = 8; no significant difference. (B) Effect of the GC inhibitor and/or protein kinase G activator (25 μmol/L MB, 1.25 μmol/L LY83 583, or 25 μmol/L ODQ and/or 800 μmol/L 8-Br-cGMP). n = 6; *P < 0.01 versus control in SNAP, #P < 0.05 and +P < 0.01 versus ODQ in SNAP, and 8-Br-cGMP in SNAP, respectively. (C): n = 6; *P < 0.01 versus control in hypoxic conditions. (C) Effect of SNP. *P < 0.01 versus control in hypoxic conditions.
Effects of inhibitors on human VEGF promoter activity in A-172 cells.
The cells were exposed to the same conditions as Figure 1for 12 hours. (A) Effect of the NOS inhibitor (2 mmol/L L-NAME or 5 mmol/L AG). n = 8; no significant difference. (B) Effect of the GC inhibitor and/or protein kinase G activator (25 μmol/L MB, 1.25 μmol/L LY83 583, or 25 μmol/L ODQ and/or 800 μmol/L 8-Br-cGMP). n = 6; *P < 0.01 versus control in SNAP, #P < 0.05 and +P < 0.01 versus ODQ in SNAP, and 8-Br-cGMP in SNAP, respectively. (C): n = 6; *P < 0.01 versus control in hypoxic conditions. (C) Effect of SNP. *P < 0.01 versus control in hypoxic conditions.
It has been recently reported that NO suppresses hypoxic induction of VEGF gene by using SNP as an NO donor in human hepatoma cell lines.23 25 Our studies with SNP demonstrated that it attenuated the VEGF promoter activation by hypoxia in a dose-dependent manner. Moreover, it had no significant effect under normoxia at concentrations up to 100 μmol/L in A-172 cells (Figure 3C) and Hep3B cells (data not shown), in contrast to our results with SNAP and NOC5.
Identification of the NO-response elements of the human VEGF gene
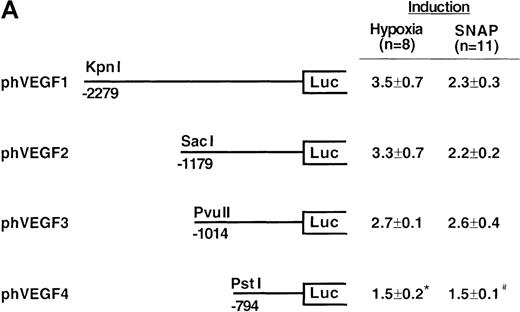
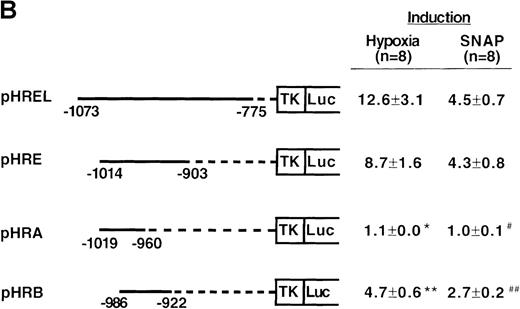
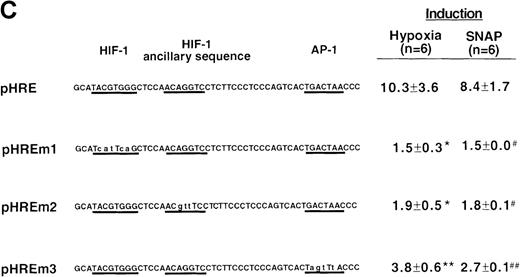
To determine the NO-response element of the human VEGF promoter, we constructed a series of deletion mutants of phVEGF1 and tested their response to SNAP (0.5 mmol/L for 12 hours) following transient transfection in A-172 cells. Removal of DNA sequences between positions -2279 and -1014 did not cause any significant change in the hypoxia- or NO-induced activation of the VEGF reporter gene (Figure4A). A further deletion to -794 (phVEGF4) significantly reduced the promoter response to either stimulus (P < 0.01 and < 0.05 versus phVEGF3 in hypoxia and in SNAP, respectively). It should be noted that a similar stepwise decrease in the response of the VEGF reporter genes to hypoxia has been reported previously.9,11,12 To confirm that NO-response element is indeed located in the -1014 and -794 region, we tested the ability of this sequence to confer NO-inducibility to the HSV-TK promoter. As shown in Figure 4B, this is the case. Deletion of the VEGF promoter sequence between -960 and -903 (pHRA) completely lost the response to SNAP (P < 0.01 versus pHRE in SNAP). Analysis of an additional recombinant (pHRB) further locates the NO-response element between positions -986 and -922 (P < 0.01 versus pHRA in SNAP). It overlaps with the HRE of the VEGF gene. Sequence comparisons between human, mouse, and rat VEGF genes in this DNA region reveal a high degree of evolutionary conservation. In particular, there is conformity in 4 sequences. These correspond to the HIF-1 binding site 5′-TACGTGGG (-975 to -968); the AP-1 site 5′-TGACTAA (-937 to -931); the “NF-κB–like' sequence 5′-GGGTTTTGCC (-1,000 to -991); and the sequence 5′-ACAGGTC (-962 to -956), which we call the HIF-1 ancillary sequence. The last has been previously suggested to be essential for hypoxic induction of the VEGF promoter.9
Localization of VEGF 5′-flanking sequences that mediate transcriptional response to hypoxia or SNAP.
VEGF sequences were cloned to the promoterless pGL2 basic vector (A) or 5′ to an HSV-TK promoter-luciferase transcription unit of pT81luc0 (B, C). The locations of restriction sites are shown relative to the transcription start site. Relative luc activity is defined as the mean ratio of luciferase/β-galactosidase activity ± SEM. Fold induction, by hypoxia or SNAP, represents the ratio of relative luc activity in cells at 1% O2 or 0.5 mmol/L SNAP in 0.1% DMSO to those at 21% O2 or 0.1% DMSO, respectively. (A) n = 8 (hypoxia) or n = 11 (SNAP); *P < 0.05 versus phVEGF1 and <0.01 versus phVEGF3 in hypoxia, #P < 0.01 versus phVEGF1 and < 0.05 versus phVEGF3 in SNAP. (B) n = 8; *P < 0.01 versus pHRB in hypoxia, **P < 0.05 versus pHRE in hypoxia, #P < 0.01 versus pHRB in SNAP, and ##P < 0.05 versus pHREL in SNAP. (C) Nucleotides of transcription factor binding sites (HIF-1 binding site, HIF-1 ancillary sequence, AP-1) are underlined (-975 to -968, -962 to -956, and -937 to -931, respectively), and substituted bases are shown in lowercase letters. n = 6; *P < 0.01 versus pHRE and < 0.05 versus pHREm3 in hypoxia, **P < 0.05 versus pHRE in hypoxia, #P < 0.01 versus pHRE and < 0.05 versus pHREm3 in SNAP, and ##P < 0.05 versus pHRE in SNAP.
Localization of VEGF 5′-flanking sequences that mediate transcriptional response to hypoxia or SNAP.
VEGF sequences were cloned to the promoterless pGL2 basic vector (A) or 5′ to an HSV-TK promoter-luciferase transcription unit of pT81luc0 (B, C). The locations of restriction sites are shown relative to the transcription start site. Relative luc activity is defined as the mean ratio of luciferase/β-galactosidase activity ± SEM. Fold induction, by hypoxia or SNAP, represents the ratio of relative luc activity in cells at 1% O2 or 0.5 mmol/L SNAP in 0.1% DMSO to those at 21% O2 or 0.1% DMSO, respectively. (A) n = 8 (hypoxia) or n = 11 (SNAP); *P < 0.05 versus phVEGF1 and <0.01 versus phVEGF3 in hypoxia, #P < 0.01 versus phVEGF1 and < 0.05 versus phVEGF3 in SNAP. (B) n = 8; *P < 0.01 versus pHRB in hypoxia, **P < 0.05 versus pHRE in hypoxia, #P < 0.01 versus pHRB in SNAP, and ##P < 0.05 versus pHREL in SNAP. (C) Nucleotides of transcription factor binding sites (HIF-1 binding site, HIF-1 ancillary sequence, AP-1) are underlined (-975 to -968, -962 to -956, and -937 to -931, respectively), and substituted bases are shown in lowercase letters. n = 6; *P < 0.01 versus pHRE and < 0.05 versus pHREm3 in hypoxia, **P < 0.05 versus pHRE in hypoxia, #P < 0.01 versus pHRE and < 0.05 versus pHREm3 in SNAP, and ##P < 0.05 versus pHRE in SNAP.
To determine the role of each of these sequence elements in the NO-mediated responses, we tested responses of pHRE and its related mutants to SNAP. The response of pHRE was quantitatively and qualitatively comparable to that of phVEGF3 (Figure 4A). Mutation in the HIF-1 binding site (pHREm1) or in the HIF-1 ancillary sequence (pHREm2) completely abolished the NO-induced activation of the promoter (P < 0.01 versus pHRE), and mutation in the AP-1 site (pHREm3) inhibited, partially but significantly, the response to NO (P < 0.05 versus pHRE) (Figure 4C). As controls, the response of these reporters to hypoxia was also measured under the same conditions and was found to be superimposable to that of NO. The effects of NO and hypoxia on these reporters were also the same when tested in Hep3B cells (data not shown). Taken together, these results indicate that the NO-response and hypoxia-response sequences of the human VEGF gene co-localize with the HIF-1 binding site; the HIF-1 ancillary sequence; and in part, the AP-1 site.
Characterization of NO-responsive nuclear proteins that bind to the HIF-1 site of the human VEGF gene
To identify the transcription factor(s) present in A-172 cells that may interact with the NO-response element, and in particular with the HIF-1 site, we analyzed the in vitro binding of nuclear proteins to a labeled WT HIF (-985 to -960) double-stranded oligonucleotide (Figure5A). Several DNA-protein complexes were detected, as shown in Figure 5B. Four bands are present when using nuclear extracts from both control and SNAP-stimulated cells (NS, C1, C2, C3). The lowest band (NS) does not represent a specific complex since it can interact with a variety of wild-type and mutated oligonucleotides. The other 3 complexes (C1, C2, C3) represent proteins interacting specifically with the HIF-1 site or with its flanking sequences. These 3 complexes are detected using probes containing the HIF-1 site from either VEGF9,12 or erythropoietin (Epo)12,40,41 genes. It has been suggested that the complexes represent constitutive binding of ATF-1 and CREB-1 transcription factors within or near HIF-1 sites.42
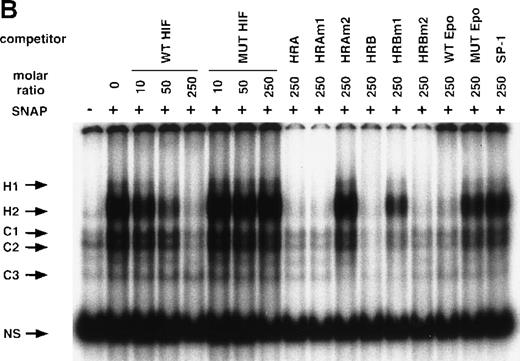
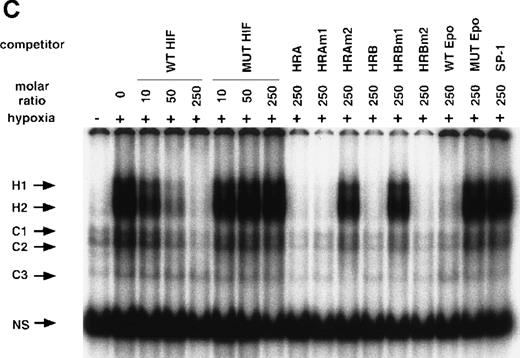
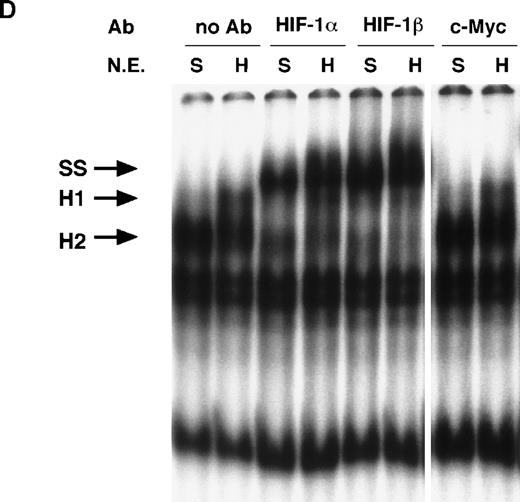
NO and hypoxia-enhance HIF-1 binding activity.
(A) Oligonucleotide sequences for EMSA. Nucleotides of functional transcription factor binding sites are underlined, and substituted bases are shown in lowercase letters. WT HIF was used as a labeled probe. (B, C) EMSAs showing the binding specificity of nuclear factors from SNAP-treated cells (B) or hypoxia-treated cells (C). Nuclear extracts (5 μg) from A-172 cells, treated by 0.1% DMSO or 0.5 mmol/L SNAP (B), or under normoxic or hypoxic (1% O2) conditions (C), were incubated with WT HIF probe for 30 minutes in the presence of no competitor0 or 10-, 50-, or 250-fold molar excess of unlabeled competitor oligonucleotides. SNAP-induced or hypoxia-induced (H1 and H2), constitutive (C1, C2, C3), and nonspecific (NS) complexes are indicated. (D) SS of HRE binding complexes. Nuclear extracts (5 μg) from A-172 cells treated by 0.5 mmol/L SNAP or by hypoxia were incubated with labeled WT HIF probe, in the presence or absence of mAbs against HIF-1α, HIF-1β, or c-Myc as potential supershifting reagents. The shifted complexes (SS) are indicated. Ab = antibody, N.E. = nuclear extract, S = SNAP, and H = hypoxia.
NO and hypoxia-enhance HIF-1 binding activity.
(A) Oligonucleotide sequences for EMSA. Nucleotides of functional transcription factor binding sites are underlined, and substituted bases are shown in lowercase letters. WT HIF was used as a labeled probe. (B, C) EMSAs showing the binding specificity of nuclear factors from SNAP-treated cells (B) or hypoxia-treated cells (C). Nuclear extracts (5 μg) from A-172 cells, treated by 0.1% DMSO or 0.5 mmol/L SNAP (B), or under normoxic or hypoxic (1% O2) conditions (C), were incubated with WT HIF probe for 30 minutes in the presence of no competitor0 or 10-, 50-, or 250-fold molar excess of unlabeled competitor oligonucleotides. SNAP-induced or hypoxia-induced (H1 and H2), constitutive (C1, C2, C3), and nonspecific (NS) complexes are indicated. (D) SS of HRE binding complexes. Nuclear extracts (5 μg) from A-172 cells treated by 0.5 mmol/L SNAP or by hypoxia were incubated with labeled WT HIF probe, in the presence or absence of mAbs against HIF-1α, HIF-1β, or c-Myc as potential supershifting reagents. The shifted complexes (SS) are indicated. Ab = antibody, N.E. = nuclear extract, S = SNAP, and H = hypoxia.
The remaining 2 upper bands (H1 and H2) were faint in extracts from untreated cells, but quite intense following cell exposure to SNAP (Figure 5B). These labeled complexes were inhibited by an excess of unlabeled oligonucleotides containing the wild-type HIF-1 site from either the human VEGF or Epo genes (WT HIF, HRA, HRAm1, HRB, HRBm2, WT Epo). However, they were not inhibited by an excess of oligonucleotides containing mutation in the HIF-1 site (MUT HIF, HRAm2, HRBm1, MUT Epo) or by an SP-1-binding oligonucleotide of unrelated sequence (SP-1) (Figure 5B). Nuclear extracts from hypoxic A-172 cells showed a similar pattern of binding (Figure 5C). However, the intensity of H2 is generally stronger than that of H1 in the NO-treated cells, while both are similarly enhanced in the hypoxic cells.
To verify the presence of HIF-1 protein in H1 and H2 complexes, we then used antibodies against both HIF-1α and HIF-1β/ARNT in supershift (SS) assays. As shown in Figure 5D, both NO-induced and hypoxia-induced H complexes were indeed completely supershifted by either anti–HIF-1α or anti–HIF-1β antibodies but not by unrelated anti–c-Myc antiserum.
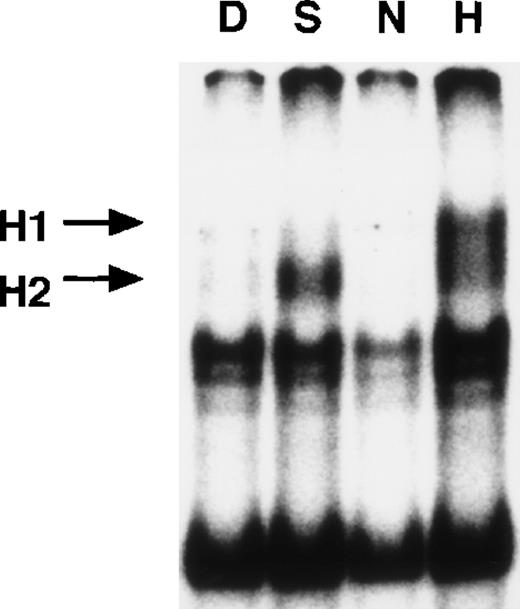
Gel shift assays of Hep3B nuclear extracts were also performed for HIF-1 binding to the WT HIF probe. H1 and H2 complexes were quite visible in NO-treated and hypoxia-treated cell extracts, and the patterns of relative amounts of these bands were quite similar to those seen in A-172 nuclear extracts (Figure 6). The SS assay demonstrated that these inducible bands also contained HIF-1α and β protein (data not shown). These results indicate that NO and hypoxia induce HIF-1 binding activity in A-172 and Hep3B cells.
EMSA with nuclear extracts from Hep3B cells.
Nuclear extracts (5 μg) from Hep3B cells, treated by 0.1% DMSO or 0.5 mmol/L SNAP (8 hours) or under normoxic or hypoxic conditions (12 hours), were incubated with WT HIF probe for 30 minutes at room temperature. SNAP-induced or hypoxia-induced complexes (H1 and H2) are indicated. D = DMSO, S = SNAP, N = normoxia, and H = hypoxia.
EMSA with nuclear extracts from Hep3B cells.
Nuclear extracts (5 μg) from Hep3B cells, treated by 0.1% DMSO or 0.5 mmol/L SNAP (8 hours) or under normoxic or hypoxic conditions (12 hours), were incubated with WT HIF probe for 30 minutes at room temperature. SNAP-induced or hypoxia-induced complexes (H1 and H2) are indicated. D = DMSO, S = SNAP, N = normoxia, and H = hypoxia.
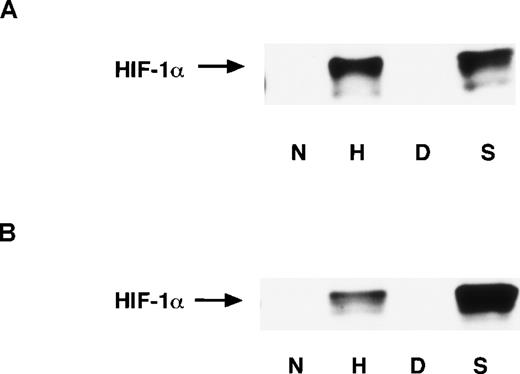
It has been reported that the amount of HIF-1α protein is significantly increased under hypoxic conditions. This response depends upon the stabilization of HIF-1α rather than increased HIF-1α mRNA levels, and the abundance of HIF-1α protein primarily determines the enhancement of HIF-1 binding activity.34 43 To examine whether NO affects the HIF-1α accumulation, we performed Western blot analysis with mAb anti–HIF-1α. The nuclear and whole-cell extracts were prepared from A-172 cells. NO as well as hypoxia significantly induced HIF-1α accumulation in both the nucleus and the whole cell (Figures 7A and 7B). This indicates that the abundance of HIF-1α also accounts for NO-induced HIF-1 activation.
Discussion
NO regulation of VEGF gene transcription and HIF-1 binding activity
In the present work, NO and hypoxia have been found to significantly induce the expression of a human VEGF reporter gene in glioblastoma and hepatoma cells. We have previously shown that VEGF mRNA rapidly accumulates following exposure to NO donors in these cells, and that this is prevented by pretreating the cells with the RNA polymerase inhibitor actinomycin D.27 These results suggest that NO activates the transcription of the endogenous VEGF gene as well as the transfected VEGF reporter gene.
The transcription factor HIF-1 plays a central role in hypoxic induction of the VEGF gene by binding to its target DNA sequence. NO-induced VEGF expression is also, at least in part, mediated by activation and subsequent binding of HIF-1. Therefore, NO and hypoxia may share common features in the pathways of VEGF induction.
SNAP stimulates the VEGF reporter expression for 60 hours in Hep3B cells (Figure 2), in contrast to its transient effect on the same reporter expression in A-172 cells (Figure 1A). We cannot explain clearly the reason for this difference. It may be that the pathway from NO to VEGF promoter activation is different between these cell lines or that SNAP has different kinetics of NO release in these 2 cell lines due to their different redox status. To confirm that NO is responsible for these effects, we performed a further experiment using a specific NO scavenger, 2-(4-carboxyphenyl)-4,4,5,5-tetramethyl-imidazoline-1-oxyl-3-oxide (carboxy-PTIO) (Dojindo Laboratories, Osaka, Japan). When stable transformants of both A-172 and Hep3B cells with phVEGF1 were pretreated with this, the reporter activation by SNAP was completely blocked, but this effect was not produced by hypoxia (our unpublished data). This result indicates that NO was an effector in both cases.
Optimal concentrations of NO donors for VEGF reporter gene expression in normoxia cause an inhibitory effect on the gene activation by hypoxia (Figure 1B and C). This may be attributed to higher concentrations of NO released from NO donors under hypoxia than under normoxia, as exposure to excessive amounts of NO could be toxic.44 This indicates that the final effect of NO on VEGF expression (activation or suppression) could also depend on the redox status of the cellular environment.
HIF-1α protein levels were massively upregulated in various cell lines under hypoxia, while HIF-1α mRNA levels were unchanged under the same conditions. Similarly, in the present work a dramatic increase in HIF-1α protein levels was seen in NO-treated A-172 cells (Figure7A and B), although we were unable to detect a significant change in HIF-1α mRNA. Thus, an increase in HIF-1α mRNA may not be the main mechanism for HIF-1α protein accumulation, but rather post-transcriptional or post-translational mechanisms34 43 may be involved. HIF-1α is rapidly degraded under normoxia, while it is stabilized and immediately translocated to the nucleus under hypoxia. Our results demonstrate that HIF-1α protein levels were elevated not only in the nucleus, but also in the whole cell (Figure 7A and B). This suggests that accumulation of HIF-1α, as well as its translocation to the nucleus, may play a central role in NO-induced and hypoxia-induced activation of this transcription factor.
Expression of HIF-1 proteins in A-172 cells.
Nuclear extracts (A) and whole-cell extracts (B) were prepared from A-172 cells under the following conditions: untreated or hypoxia-treated (12 hours), DMSO-treated (8 hours), or SNAP-treated (8 hours). The cells were subjected to Western blot analysis using mAb anti-HIF-1α.
Expression of HIF-1 proteins in A-172 cells.
Nuclear extracts (A) and whole-cell extracts (B) were prepared from A-172 cells under the following conditions: untreated or hypoxia-treated (12 hours), DMSO-treated (8 hours), or SNAP-treated (8 hours). The cells were subjected to Western blot analysis using mAb anti-HIF-1α.
Although HIF-1 binding activity and HIF-1α accumulation are similarly induced by NO and hypoxic stimulation, EMSA showed that relative amounts of doublet bands are different in extracts from NO-treated and hypoxia-treated cells. There are some possible mechanisms that could account for this difference. It may be attributable to the different status of phosphorylation. DNA binding of HIF-1 is regulated by protein phosphorylation,45 and the status of phosphorylation can affect the mobility of the target protein in polyacrylamide gels. It is also possible that different coactivators may be involved in HIF-1 activation by hypoxia when compared with that activated by NO.
A number of NO effects appear to be mediated by soluble GC, heme-containing proteins that react directly with NO and thereby induce an increase in intracellular cGMP levels.18,19 An involvement of cGMP in NO-induced activation of the endogenous VEGF promoter was suggested by results obtained with the GC inhibitors MB and LY8 358 3.27 However, as described here, another GC inhibitor (ODQ) did not attenuate NO-induced activation of the transfected VEGF promoter (Figure 3B). Moreover, 8-Br-cGMP did not mimic VEGF reporter gene induction by NO, even when used in conjunction with a NO donor and LY83 583. Hypoxic induction of this reporter gene and HIF-1 binding activity were unaffected by either MB or LY83 583 (Figure 3B). These results indicate that NO-induced activation of the gene promoter and the HIF-1 factor does not occur via cGMP-mediated signal transduction. They also show that NO and hypoxia act through distinct pathways, or via different molecular components of a single pathway, because of the different responses to MB and LY83 583.
Controversial effect of NO on VEGF expression and angiogenesis
The role of NO in angiogenesis is controversial. Nitric oxide donors inhibit angiogenesis in the chick chorioallantoic membrane, the tube formation in the matrigel tube formation assay,46 and the growth and metastatic properties of the Lewis lung tumor in mice.47 Nitric oxide donors inhibited VEGF expression in the arterial wall in response to balloon angioplasty21 and in rat lungs during acute and chronic hypoxia.22 In contrast, there are some observations that NO enhances the expression of angiogenic activity. Nitric oxide synthase activity correlates positively with tumor growth and vascular density.28-30Human colon tumor cell lines transfected with a NOS-encoding gene grew faster and were more vascularized than the parent cell lines in vivo.48 Nitric oxide produced in vascular endothelium has also been suggested as a downstream mediator for VEGF receptors in angiogenesis.49 Exogenous NO and endogenous NO, elicited by substance P, enhanced angiogenesis in vivo. Nitric oxide also enhanced the proliferation and migration of endothelial cells in vitro.50 Moreover, promoting endothelial NOS activity accelerated in vivo angiogenesis.51
Some recent reports show an inhibitory effect of NO on VEGF expression.23-25 Sogawa et al23 and Huang et al25 demonstrated that SNP suppresses hypoxia-induced VEGF gene activation and HIF-1 binding activity. As shown in this work, SNP inhibits the hypoxic induction of the VEGF gene in a dose-dependent manner in glioblastoma and hepatoma cell lines, in contrast to the effects of SNAP and NOC5. This contradiction is clearly due to the specific nature of SNP. SNAP and NOC5 are chemically distinct compounds that generate NO radicals spontaneously. In contrast, after donating NO, SNP disintegrates into ferrocyanide, ferricyanide, iron ions, and cyanide, each of which has a variety of biological effects.52 There is no definite explanation for the cause of the inhibitory effect of SNP, as ferrocyanide and ferricyanide at concentrations up to 100 μmol/L made no change on VEGF promoter activity in A-172 cells (our unpublished data). Therefore, SNP is far from an ideal NO donor. Sogawa et al23 also used S-nitrosoglutathione (GSNO) and 3-morpholinosydnonimine (SIN-1) only under hypoxic conditions.
We also examined the effects of these compounds and found that they showed remarkable induction of the VEGF reporter gene under normoxic conditions in both A-172 and Hep3B cells (our unpublished data). These results suggest that SNP has a distinct effect on the promoter activity, when compared with other NO donors, and that its inhibitory effect may not simply be attributable to NO itself. In another recent report,24 the cell lines used were not tumor cells but were vascular endothelial and smooth muscle cells. SNAP downregulated VEGF expression by inhibiting PKC-induced AP-1 binding activity in smooth muscle cells.21 Nitric oxide inhibits proliferation and migration of endothelial cells.53,54 These contradictory data indicate that NO has both inhibitory and activating effects on angiogenesis, depending upon the cellular environment and the types of cells in which assays are performed. Nitric oxide chemistry is highly redox-sensitive. This may also explain the contradictory effects of NO on HIF-1 activation in different cell systems and why MB and LY83 583 (but not ODQ) are inhibitory, as the former 2 compounds are known to generate superoxide anions.55 56
Conclusions and implications
Our results imply a direct involvement of NO in the control of angiogenesis through its regulation of VEGF expression, where HIF-1α activity appears to be essential.57 Moreover, the identification of HIF-1 as an additional molecular target of NO opens a new path for the molecular characterization of the effects of this intercellular mediator on gene transcription. Furthermore, these findings also suggest a role of NO and its redox derivatives in tissue reactions to hypoxia. Indeed, given the importance of HIF-1 in the genomic responses of hypoxic cells, these results establish a direct link between NO and the adaptation of normal and neoplastic cells and tissues to low oxygen tension. This helps explain why NO donors can exert such diverse beneficial therapeutic actions, for example, in cardiovascular diseases,58 in ischemic brain injury,59 and following surgically related ischemic-reperfusion injuries.60 On the other hand, there is a strong positive correlation between NO production and tumor angiogenesis.28-30 61 These last observations suggest that there may be possible risks of long-term treatments with pharmacological agents that potentiate NO in patients suffering from (or at risk of) cancer, where enhanced angiogenesis would be hazardous.
Acknowledgments
We thank Alexander Minchenko for providing the human VEGF promoter DNA, Steven K. Nordeen and Luigi Cicatiello for providing the pT81luc0 vector, and David M. Livingston for providing anti–HIF-1α antibodies.
Supported by a Research Resident Fellowship from the Foundation for the Promotion of Cancer Research and by a Grant from the Ministry of Health and Welfare for the Second-term Comprehensive 10 Year Strategy for Cancer Control.
A.W., F. D'A., and R.A. were Foreign Research Fellows of the Foundation for the Promotion of Cancer Research, Tokyo, Japan.
Reprints:Hiroyasu Esumi, Investigative Treatment Division, National Cancer Center Research Institute East, 6-5-1 Kashiwanoha, Kashiwa, Chiba, Japan; e-mail: hesumi@east.ncc.go.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.