Abstract
Glycoprotein (GP) Ib is the functionally dominant subunit of the platelet GPIb-IX-V receptor complex, with the von Willebrand factor (vWF) binding site residing on the amino-terminus. A threonine for methionine-145 replacement of GPIb is associated with the human platelet antigen (HPA)-2 system. To study the structural and functional consequences of this mutation, both forms of GPIb were expressed as calmodulin fusion proteins in insect cells. Both recombinant proteins were recognized by their respective alloantibodies, independent of glycosylation or intactness of disulfide bonds, and gave similar results to platelet-derived GPIb in antibody detection assays. Resonant mirror studies showed that vWF binding was not affected by the HPA-2 mutation; however, vWF binding was partially inhibited by IgG HPA-2 antibodies. Our data are compatible with an involvement of the leucine-rich repeat domain of GPIb in vWF binding and indicate that recombinant GPIb may be used to detect HPA-2 antibodies. (Blood. 2000;95:205-211)
The platelet glycoprotein (GP)Ib-IX-V complex plays an essential role in the maintenance of normal hemostasis by mediating initial adhesion of platelets to the vessel wall via von Willebrand factor (vWF). Initial studies mapped the binding sites for vWF and α-thrombin to the amino-terminal part of GPIbα, and more recent data indicate the involvement of the leucine-rich repeat region (LRR) in vWF binding.1,2 In vivo vWF binding of the GPIb-IX-V complex requires both immobilization of vWF in the subendothelium and the presence of high shear forces.3 In vitro binding can be initiated by both ristocetin, a peptide antibiotic, and botrocetin, a snake venom protein.1
In earlier studies, we and others obtained evidence that the di-allelic human platelet antigen (HPA)-2 system is localized on the amino terminal 45 kDa fragment of GPIbα and that the alloantigens co-segregate with a cytosine to thymine mutation at position 434 (the codon for residue 145 in the fifth LRR) of the GPIbα gene.4-6 This strongly suggests that the alloantigens are based on the Thr/Met-145 exchange arising from this mutation. The two alleles, HPA-2a and HPA-2b, have respective frequencies in the Caucasian population of approximately 0.926 and 0.074.7-9Exposure to nonself forms of the HPA-2 antigens via either pregnancy or transfusion may lead to alloantibody formation that can cause neonatal alloimmune thrombocytopenia, refractoriness to platelet transfusions, and posttransfusion purpura.10
To date there is only limited proof that the HPA-2 B cell epitopes are based solely on this single amino acid substitution, as studies have been based on use of a single example of anti-HPA-2b.6Furthermore, the roles of glycosylation and disulfide bonds in the formation of the epitopes remain to be determined. Similarly, many of the studies on vWF binding to GPIbα have been performed with GPIbα in its native stoichiometry on the platelet membrane in the context of GPIβ, GPIX, and GPV.
Interestingly, HPA-2 antibodies abolish ristocetin-mediated agglutination of platelets.4 Why HPA-2 antibodies inhibit is not well understood, because residue 145 is located in the fifth LRR of GPIbα and is distant from the proposed vWF binding site (residues 252-287).11 Obvious explanations might be the presence of a second vWF binding site, or alternatively the antibody-mediated inhibition is based on allosteric hindrance.
In the current study, both forms of a truncated GPIbα (residues His1-Val289) with either a Thr or Met at position 145 were expressed in a baculovirus/insect cell system as calmodulin (CaM) fusion proteins. The use of CaM as a tag for proteins has been reported previously.12 The fusion proteins were used in studies designed to answer the above questions on the molecular nature of the HPA-2 alloantigens, the effect of the mutation on vWF binding, and the effect on the inhibition of vWF binding by HPA-2 antibodies.
Materials and methods
Platelets, human HPA-2 antibodies, and monoclonal antibodies
HPA-2a2a and -2b2b platelets from donors genotyped by PCR-SSP13 were supplied by the Platelet Immunology Laboratory of the National Blood Service (NBS), East Anglia Centre, Cambridge, UK. Negative control sera were obtained from a nontransfused group AB male blood donors. Sera containing anti-HPA-2a and -2b were kind gifts from Professor Shibata (Tokyo University, Tokyo, Japan), Mr. C. Hurd, and Mrs. C. Quader (NBS, East Anglia, and NBS, Sheffield, UK), and Dr. L. Porcelijn (CLB, Amsterdam, The Netherlands). All sera were stored at −20°C until required for use.
The GPIbα (CD42b)-specific monoclonal antibody (mAb) MB45 was a gift from Professor A. E. G. Kr von dem Borne (CLB).
Construction of plasmids
To facilitate protein purification and subsequent immobilization on a resonant mirror sample cuvette, both forms of GPIbα were expressed as CaM fusion proteins in a baculovirus/insect cell system. A complementary DNA fragment coding for the signal peptide and mature protein residues His1-Val289 of GPIbα was obtained by polymerase chain reaction from the plasmid pDXα,14which contained the full-length GPIbα gene. The sense and anti-sense oligonucleotide primers used for cloning were (1) 5′-ACCTCGAGATGCCCTCCTCCTCTTGCTG-3′(contains aXhoI site); and (2) 5′-GCGGCCGCTGTATGGCTTTGGTGGGGAA-3′12(contains a NotIsite). The complementary DNA fragment amplified using these primers was digested with the restriction enzymes XhoI and NotI and ligated into the vector pND162 that contained the CaM gene (kindly provided by Dr. D. Neri, Medical Research Council Laboratory of Molecular Biology, Cambridge, UK).12 The resulting plasmid was then digested with XhoI and EcoRI to release the fusion gene encoding a GPIbα fragment in frame with the CaM gene. The fusion construct was ligated into the baculovirus expression vector pAcSG2 and its sequence confirmed. A construct with a 145-Met codon was generated by mutagenesis using the QuickChange method15 (Stratagene, La Jolla, CA) and the mutation was verified by sequencing.
Expression, purification, and characterization of recombinant GPIb/CaM
Cell culture.
Sf9 and High Five insect cells were routinely propagated in Sf-900 II SFM and Express Five SFM media, respectively, as adherent cell monolayers in 25 cm2 tissue-culture flasks or as suspension cultures in spinner flasks at 27°C.
The Sf9 and High Five cells were either seeded at a density of 1 × 106 cells/mL in spinner flasks and grown to 2 × 106 cells/mL with constant stirring at 100 rpm, or seeded for adherence in flasks at a density of 1.2 × 105 cells/cm2 for Sf9 cells and 5 × 104 cells/cm2 for High Five cells and grown to 80% confluency.
Expression.
Recombinant baculovirus transfer vectors containing either fusion construct were transfected into Sf9 cells according to the manufacturer's protocol (PharMingen, San Diego, CA). Briefly, 0.5 μg of linearized BaculoGold virus DNA and 2.5 μg recombinant baculovirus transfer vector were mixed for 5 minutes in a microcentrifuge tube at room temperature and then 1 mL of transfection buffer was added. A 60-mm2 Falcon tissue culture dish was seeded with 2 × 106 Sf9 cells; after 20 minutes cells were ready for transfection, and medium was replaced with 1 mL Grace's medium and the above DNA mixture. The cells were incubated for 4 hours at 27°C, and the medium was then replaced with fresh medium. After 5 days of culturing at 27°C, the medium was collected for plaque purification or further virus amplification. High Five cells were cultured in 100 mL spinner flasks with 50 mL cell culture medium at constant stirring (100 rpm) and infected with high titre recombinant virus stock at 10−20 multiplicities of infection. The infected High Five cells were grown for 72 hours, and, following centrifugation, the medium was collected for protein purification. To produce nonglycosylated GPIbα/CaM protein, cells were cultured in the presence of tunicamycin at 10 μg/mL.
Purification of CaM tagged GPIbα.
Fifty mL of culture medium was applied to a 5 mL column of W-7 agarose 4B resin (Sigma, Poole, UK) that was equilibrated in TBSC buffer (50 mM Tris, 150 mM NaCl, and 1 mM CaCl2, pH 7.4) with a flow rate of 25 mL/h. Nonspecifically bound material was removed by washing with high salt buffer (50 mM Tris, 600 mM NaCl, and 1 mM CaCl2, pH 7.4). Bound GPIbα/CaM fusion protein was eluted with 20 mM EGTA in TBS buffer (50 mM Tris, 150 mM NaCl, pH 7.4). Eluted protein solution was concentrated by ultrafiltration (Amicon, Beverly, MA), and protein concentration was determined by the method of Lowry.16
Gel electrophoresis and immunoblotting.
Purified recombinant proteins (containing Thr-145 or Met-145) were separated on 10% minigels for 45 minutes according to the method of Laemmli,17 but substituting 2-amino-2-methyl-1,3-propanediol for Tris base. To determine the importance of the disulfide bridges for the HPA-2 epitopes, recombinant proteins were treated with reducing reagents before electrophoresis. The gels were either stained with Coomassie Blue or transferred to nitrocellulose filters for Western blots. For immunoblotting, the membranes were blocked with 0.01% Tween-20 in phosphate buffered saline (PBS) for 30 minutes and then reacted overnight at 4°C with a 1 in 10 dilution of anti-HPA-2a and -2b sera, with continuous shaking. After extensive washing with blocking buffer, the membranes were incubated with a 1 in 1000 dilution of peroxidase-labeled anti-human IgM (for anti-HPA-2a) or IgG (for anti-HPA-2b) for 2 hours followed by extensive washing. Binding of antibody to the recombinant proteins was detected by the ECL method (Amersham, Arlington Height, IL).
Detection of HPA-2-specific antibodies
Enzyme-linked immunoabsorbent assay (ELISA).
A buffer containing 10 mM Tris, 145 mM NaCl, pH 7.4, 0.5% Nonidet P40, 0.05% Tween 20, 0.5 mM CaCl2, and 0.2% bovine serum albumin (BSA) was used for all wash steps and for diluting reagents, except where stated otherwise. Human sera were diluted 1 in 20 with the use of an ELISA specimen diluent (Abbott Laboratories, Abbott Park, IL). Microplate wells were coated with goat anti-mouse IgG (Jackson ImmunoResearch Laboratories Inc, West Grove, PA) diluted in sodium carbonate buffer, pH 9.6 by incubating overnight at 4°C. The plates were then washed twice and subsequently blocked by incubating with 3% BSA for 60 minutes at 37°C, followed by five washings. The CD42b mAb MB45 (1 in 1000 dilution of ascites) was added to the microplate wells and incubated for 60 minutes at 22°C, followed by five washes. The Thr- and Met-145 forms of recombinant GPIbα (2.5 μg/mL) were added to the wells and incubated for 30 minutes. After five more washes, the diluted human sera were added and incubated for 30 minutes. Following five more washes, bound human antibody was detected with an alkaline phosphatase conjugated goat anti-human IgG or IgM reagent (Jackson) with the use of p-Nitrophenyl phosphate substrate in Tris buffer, and optical density was measured in an ELISA reader (MR5000) at 405 nm.
Monoclonal antibody-specific immobilization of platelet antigens (MAIPA) assay.
The MAIPA was performed as previously described.18 19Briefly, platelets from either HPA-2a2a or -2b2b donors were incubated with sera, washed, and then incubated with mAb MB45. The platelets were washed again, solubilized, and centrifuged, and the clear lysates added to microplate wells that had been previously coated with goat anti-mouse IgG to capture the mAb-GPIb/IX complex. Human IgG bound to the complex was detected with alkaline phosphatase conjugated goat anti-human IgG (Jackson). The optical density was measured in an ELISA reader (MR5000) at 405 nm.
Resonant mirror technology.
An IAsys Auto+ resonant mirror biosensor (Labsytems Affinity Sensors, Cambridge, UK) was also used to study the reactivity of HPA-2 antibodies and vWF binding to both forms of the recombinant GPIbα/CaM fusion protein in the presence and absence of ristocetin (see below).
vWF binding studies
The 23-mer CaM-binding oligopeptide (CAAARWKKAFIAVSAANRFKKIS) cross-linked to BSA20 21 was coupled to the carboxymethylated dextran surface of the sample cuvette in 10 mM sodium acetate, pH 5, according to the manufacturer's instructions. This coupling allowed capture of the CaM-tagged GPIbα in the presence of calcium, with an insignificant off rate. For subsequent assays, the recombinant GPIbα/CaM fusion protein was removed from the cuvette surface by the addition of 10 mM EDTA. All experiments were performed in the presence of Tris buffered saline (50 mM Tris-HCl, 150 mM NaCl) containing 20 mM CaCl2 at 25°C. The factor VIII concentrate Alphanate (Alpha Therapeutic Corporation, Los Angeles, CA) was used as a source of vWF and diluted according to the manufacturer's instructions to give a final vWF concentration of 5 μg/mL.
Binding of vWF to GPIbα.
Following capture of Thr-145 GPIbα vWF (5 μg/mL) and ristocetin (1.5 mg/mL; Sigma Aldrich Company Ltd, UK) were added to the cuvette, and binding of vWF was observed. Thr-145 GPIbα and vWF were then removed by washing of the cuvette with a buffer containing 10 mM EDTA, and the experiment repeated with Met-145 GPIbα.
Inhibition of vWF binding to GPIbα by affinity purified anti-HPA-2b.
Microtitre plate wells were coated with 2.0 μg/mL of BSA-CaM binding peptide in 100 μL of binding buffer 100 mM NaCl,50 mM Tris-HCl, pH 7.4 and incubated overnight at 4°C. The plates were then blocked with 3% BSA for 2 hours at room temperature. After blocking, 1.0 μg/mL of Met-145 GPIbα/CaM fusion protein was added to the wells in a 100-μL volume of binding buffer and incubated overnight at 4°C. After washing with buffer, vWF at various concentrations and ristocetin (1.5 mg/mL) were added to the wells and incubated overnight at 4°C. vWF binding to GPIbα without ristocetin was included as a negative control. Incubation in the presence of purified IgG (1.0 mg/mL) prepared from a serum sample containing anti-HPA-2b was used to measure the inhibitory capacity of anti-HPA-2b on vWF binding, and purified IgG from an inert serum was included as a negative control. After extensive washing, bound vWF was detected with a rabbit anti-vWF, 1:1000 that was incubated for 2 hours at room temperature. After washing with buffer, the bound rabbit antibodies were detected by horseradish peroxidase-conjugated pig anti-rabbit antiserum 1:1000 at room temperature for 1 hour. After five washings with buffer, 100 μL of peroxidase substrate o-Phenylenediamine buffer was added to the wells for color development, which was measured at 450 nm in a multichannel photometer (ThermoMax, Molecular Devices Corporation, Menlo Park, CA).
Results
Binding of antibodies to GPIb/CaM fusion protein
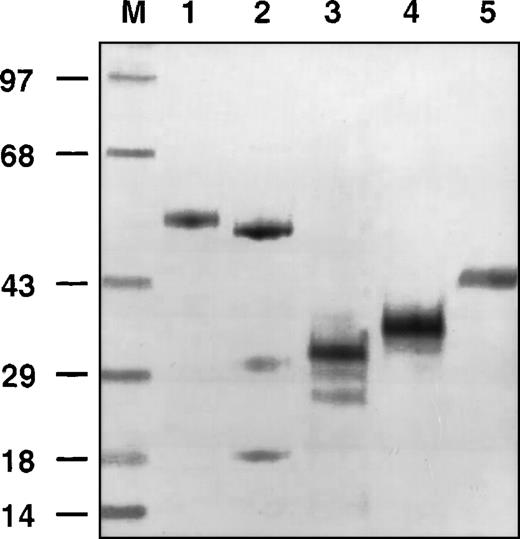
Insect cells infected with recombinant baculovirus expressed high levels of protein (8.0 mg/L after single-step purification) with the expected molecular mass of 60 and 56 kDa for glycosylated and nonglycosylated, amino-terminal 289 residues of GPIbα fused with CaM (Figure 1, lanes 1 and 2, respectively). The relative molecular masses of the recombinant amino-terminal GPIbα without CaM differed in the presence and absence of tunicamycin, (Figure 1, lanes 3 and 4, respectively) with a reduction occurring in the presence of tunicamycin. The recombinant GPIbα migrated with a molecular mass 3-4 kDa lower than corresponding platelet-derived fragment (Figure 1, lanes 4 and 5, respectively) because of differential glycosylation of two N-linked sites in the recombinant GPIbα.
Gel electrophoresis of different forms of glycoprotein (GP)Ib.
Molecular weight markers (M). Recombinant GPIbα/calmodulin (CaM) (lane 1). Recombinant GPIbα/CaM in the presence of tunicamycin (lane 2). Recombinant GPIbα in the presence of tunicamycin (lane 3). Recombinant GPIbα (lane 4). Platelet-derived, elastase digested GPIbα (lane 5).
Gel electrophoresis of different forms of glycoprotein (GP)Ib.
Molecular weight markers (M). Recombinant GPIbα/calmodulin (CaM) (lane 1). Recombinant GPIbα/CaM in the presence of tunicamycin (lane 2). Recombinant GPIbα in the presence of tunicamycin (lane 3). Recombinant GPIbα (lane 4). Platelet-derived, elastase digested GPIbα (lane 5).
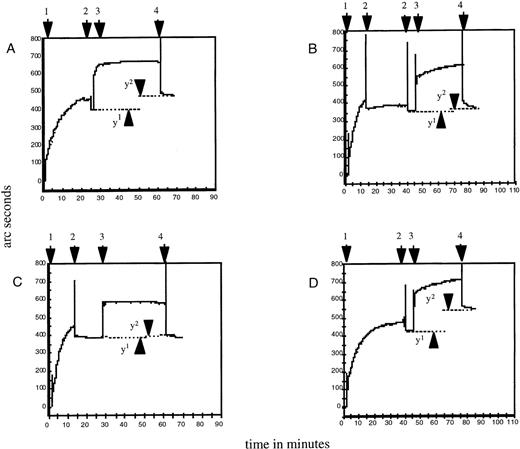
Binding of HPA-2–specific antibodies to the Thr-145 and Met-145 forms of the recombinant GPIbα/CaM fusion protein at coating concentrations of 0.625-10 μg/mL was determined by ELISA assay with the use of single samples of polyclonal IgM anti-HPA-2a and IgG anti-HPA-2b. Binding was shown to be specific for the expected protein or antibody combinations, giving dose-response curves reaching a plateau at about 7.5 μg/mL (Figure 2A). As a satisfactory signal-to-noise ratio was obtained at 2.5 μg/mL, this concentration was used for all further assays. Further specific binding of HPA-2 antibodies to their respective forms of recombinant GPIbα in ELISA was demonstrated with a panel of anti-HPA-2b (see below). This reactivity was confirmed in resonant mirror studies in which specific antibody binding to the relevant form of GPIbα was indicated by an increased response in arc seconds after washing, when compared with the response with the irrelevant GPIbα. (Figure3, A to D).
Enzyme-linked immunoabsorbent assay studies of antibody binding to recombinant glycoprotein (GP)Ib.
(A) Binding of anti-human platelet antigen (HPA)-2a and -2b to Thr-145 and Met-145 forms. (B) Binding of three examples of anti-HPA-2b and a negative control serum to glycosylated and nonglycosylated (NG) Met-145.
Enzyme-linked immunoabsorbent assay studies of antibody binding to recombinant glycoprotein (GP)Ib.
(A) Binding of anti-human platelet antigen (HPA)-2a and -2b to Thr-145 and Met-145 forms. (B) Binding of three examples of anti-HPA-2b and a negative control serum to glycosylated and nonglycosylated (NG) Met-145.
Binding of human platelet antigen (HPA)-2 antibodies to both forms of recombinant glycoprotein (GP)Ib measured by resonant mirror studies.
Addition of recombinant GPIbα to biosensor cuvette (1). Washing with TBS/Ca++ (2). Addition of serum diluted 1 in 10 (3). Washing with TBS/Ca++(4). Distance between points y1 and y2 represent specifically bound antibody remaining after TBS/Ca++ wash, expressed as arc seconds. (A) Incubation of anti-HPA-2a with Thr-145, y2 − y1 = 83 arc seconds. (B) Incubation of anti-HPA-2a with Met-145, y2 − y1 = 21 arc seconds. (C) Incubation of anti-HPA-2b with Thr-145, y2 − y1 = 7 arc seconds. (D) Incubation of anti-HPA-2b with Met-145, y2 − y1 = 128 arc seconds.
Binding of human platelet antigen (HPA)-2 antibodies to both forms of recombinant glycoprotein (GP)Ib measured by resonant mirror studies.
Addition of recombinant GPIbα to biosensor cuvette (1). Washing with TBS/Ca++ (2). Addition of serum diluted 1 in 10 (3). Washing with TBS/Ca++(4). Distance between points y1 and y2 represent specifically bound antibody remaining after TBS/Ca++ wash, expressed as arc seconds. (A) Incubation of anti-HPA-2a with Thr-145, y2 − y1 = 83 arc seconds. (B) Incubation of anti-HPA-2a with Met-145, y2 − y1 = 21 arc seconds. (C) Incubation of anti-HPA-2b with Thr-145, y2 − y1 = 7 arc seconds. (D) Incubation of anti-HPA-2b with Met-145, y2 − y1 = 128 arc seconds.
Glycosylation and disulfide bonds are not critical for the epitopes recognized by HPA-2 antibodies
Three HPA-2b antisera all gave similar results with glycosylated and nonglycosylated recombinant GPIbα/CaM fusion proteins by ELISA, suggesting that glycosylation is not relevant for the HPA-2 B cell epitopes (Figure 2B).
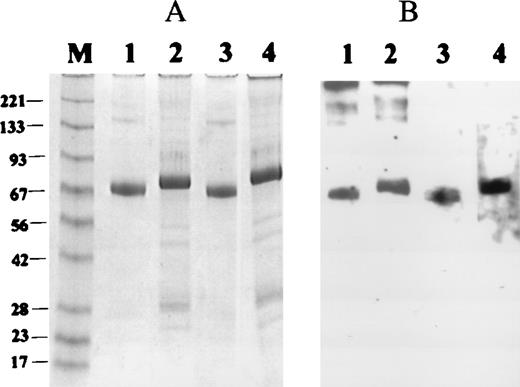
The 42 kDa amino-terminal segment of GPIbα contains three disulfide loops, one amino-terminal to the LRR (Cys-4 to Cys-17) and two located carboxy-terminal to this region (Cys-209 to Cys-248; Cys-211 to Cys-264).1 To determine the possible importance of the disulfide bonds in the expression of the HPA-2 epitopes, the two forms of recombinant GPIbα were analyzed by Western blotting. Both anti-HPA-2a and anti-HPA-2b were found to only react with 145-Thr and 145-Met, respectively, under the denaturing conditions of the SDS-PAGE (data not shown). The antibodies were further tested under reducing and nonreducing conditions and transferred to nitrocellulose in which binding was then analyzed by Western blotting (Figure4). The data demonstrated that HPA-2a and HPA-2b antibodies were able to distinguish their respective alloantigens on Western blots under both nonreducing and reducing conditions.
Western blots of Thr-145 and Met-145 forms of recombinant glycoprotein (GP)Ib under reducing and nonreducing conditions.
(A) Coomassie blue stain of recombinant GPIbα under nonreducing conditions (Thr-145, lane 1, and Met-145, lane 3) or reducing conditions (Thr-145, lane 2, and Met-145, lane 4). (B) Western blot of duplicate of panel A with anti-human platelet antigen (HPA)-2a (lanes 1 and 2), and anti-HPA-2b (lanes 3 and 4).
Western blots of Thr-145 and Met-145 forms of recombinant glycoprotein (GP)Ib under reducing and nonreducing conditions.
(A) Coomassie blue stain of recombinant GPIbα under nonreducing conditions (Thr-145, lane 1, and Met-145, lane 3) or reducing conditions (Thr-145, lane 2, and Met-145, lane 4). (B) Western blot of duplicate of panel A with anti-human platelet antigen (HPA)-2a (lanes 1 and 2), and anti-HPA-2b (lanes 3 and 4).
Comparison between platelet-derived and recombinant GPIb for HPA-2 antibody detection
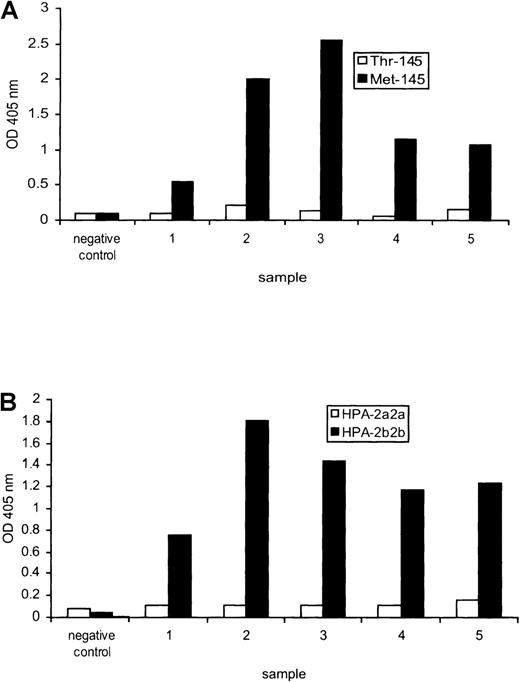
The standard technique for the detection and identification of HPA-2 antibodies is the MAIPA assay with the use of HPA-2 typed platelets as a source of GPIbα complexed with GPIbβ and GPIX. We compared the MAIPA with our recombinant protein-based antibody detection ELISA with the use of five HPA-2b antisera. All five reacted strongly with the Met-145 form of the protein, but not the Thr-145 form, when compared with the negative control (Figure 5A). Similar results were obtained by MAIPA, with all five sera reacting strongly with HPA-2b2b platelets (Figure 5B). Three of the anti-HPA-2b sera (samples 2, 3, and 4) were titrated to compare sensitivity of the two assays: sample 2 had a titre of 64 by ELISA and 128 by MAIPA, whereas samples 3 and 4 had identical titers of 256 and 64, respectively, by both techniques.
Comparison between recombinant and platelet-derived glycoprotein (GP)Ib for human platelet antigen (HPA)-2 antibody detection.
(A) An inert negative control serum and five different sera containing anti-HPA-2b tested against the Thr-145 and Met-145 forms of recombinant GPIbα by enzyme-linked immunoabsorbent assay. (B) The same sera tested against platelet-derived GPIbα by monoclonal antibody-specific immobilization of platelet antigens.
Comparison between recombinant and platelet-derived glycoprotein (GP)Ib for human platelet antigen (HPA)-2 antibody detection.
(A) An inert negative control serum and five different sera containing anti-HPA-2b tested against the Thr-145 and Met-145 forms of recombinant GPIbα by enzyme-linked immunoabsorbent assay. (B) The same sera tested against platelet-derived GPIbα by monoclonal antibody-specific immobilization of platelet antigens.
Effect of Thr/Met-145 mutation on vWF binding and inhibition by HPA-2 antibodies
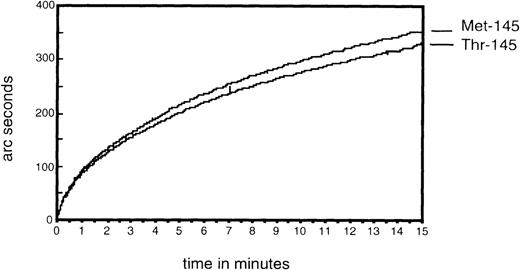
The two forms of recombinant GPIbα/CaM fusion protein were used to investigate possible effects of the Thr/Met-145 mutation on vWF binding. In the presence of ristocetin, and with equal amounts of recombinant GPIbα immobilized on the biosensor cuvette, vWF was observed to bind equally to both the Thr-145 and Met-145 forms (Figure6). No significant uptake of vWF was seen in the absence of ristocetin.
Demonstration of von Willebrand factor (vWF) binding to the Thr-145 and Met-145 forms of glycoprotein (GP)Ib with the use of resonant mirror technology.
Recombinant GPIbα-calmodulin was coupled to bovine serum albumin-peptide immobilized on the carboxymethylated dextran surface and binding of vWF to both forms of GPIbα measured.
Demonstration of von Willebrand factor (vWF) binding to the Thr-145 and Met-145 forms of glycoprotein (GP)Ib with the use of resonant mirror technology.
Recombinant GPIbα-calmodulin was coupled to bovine serum albumin-peptide immobilized on the carboxymethylated dextran surface and binding of vWF to both forms of GPIbα measured.
HPA-2 alloantibodies inhibit ristocetin-induced platelet agglutination but not collagen-induced platelet aggregation.4 The mechanism of this inhibition is not known. We investigated this finding further with the use of anti-HPA-2b in an inhibition ELISA. vWF binding to GPIbα-145-Met was measured at a single IgG concentration of 1 mg/mL and a range of vWF concentrations from 6.25 to 500 ng/well. Inhibition, which reached a plateau at about 40%, was observed (Figure 7).
Inhibition of von Willebrand factor (vWF) binding to recombinant glycoprotein (GP)Ib by IgG anti-human platelet antigen (HPA)-2b determined by enzyme-linked immunoabsorbent assay.
vWF was allowed to bind to GPIbα-Met-145 immobilized on microtitre plate wells via bovine serum albumin-peptide in the presence of ristocetin (⧫) and IgG anti-HPA-2b (▴). Bound vWF was detected with a polyclonal antibody. IgG obtained from an inert group AB serum was used as a negative control (▪). vWF did not bind in the absence of ristocetin (x).
Inhibition of von Willebrand factor (vWF) binding to recombinant glycoprotein (GP)Ib by IgG anti-human platelet antigen (HPA)-2b determined by enzyme-linked immunoabsorbent assay.
vWF was allowed to bind to GPIbα-Met-145 immobilized on microtitre plate wells via bovine serum albumin-peptide in the presence of ristocetin (⧫) and IgG anti-HPA-2b (▴). Bound vWF was detected with a polyclonal antibody. IgG obtained from an inert group AB serum was used as a negative control (▪). vWF did not bind in the absence of ristocetin (x).
Discussion
High-level expression (8 mg/L after purification) of both the Thr- and Met-145 forms of truncated GPIbα (amino acids His1-Val289) fused to CaM was achieved in insect cells. The CaM tag allowed single-step purification and reversible calcium dependent binding to the carboxymethylated dextran surface for resonant mirror studies. Characterization by SDS gel electrophoresis showed proteins of the expected size (Figure 1), whose migration was decreased on reduction (Figure 4). Synthesis of the Met-145 form in the presence of tunicamycin caused a reduction in molecular mass, most likely caused by the absence of the two N-linked glycan moieties at residues 21 and 159 (Figure 1). The baculoviral protein also had a reduced molecular mass when compared with the platelet-derived fragment of GPIbα obtained by elastase digestion, indicating different glycosylation in the recombinant protein. As already mentioned, differences in glycosylation relate to the two N-linked glycan moieties that have molecular masses of around 2-3 kDa each, giving a maximum difference of 6 kDa between glycosylated and nonglycosylated fragments.
The absent or alternative glycosylation does not seem to have a detrimental effect on the binding of HPA-2 alloantibodies. Indeed, the observation that these antibodies bind to both the glycosylated and nonglycosylated recombinant forms of GPIbα (Figure 2B) indicates that glycosylation is not required for the expression of their respective epitopes and corroborates proper folding of the recombinant protein. That the glycan moieties are of no significance to the binding of HPA-2 antibodies is comparable with HPA-1a antibodies, which react with the E. coli produced amino-terminal 66 amino acids of the leucine-33 form of GPIIIa (β3 integrin subunit).22 In contrast, binding of HPA-3a23and HPA-5b24 alloantibodies to the isoleucine-843 form of the αIIb and the lysine-505 form of the β1 integrin, respectively, is dependent on glycosylation.
Antibody detection by MAIPA and ELISA indicate that a sensitivity can be achieved with the recombinant antigens comparable to that obtained with the native platelet derived GPIbα. If the nonspecific binding of IgG to the solid phase can be reduced by modification of the antibody detection ELISA, the requirement to dilute serum samples 1 in 20 might be unnecessary, thus avoiding possible loss of sensitivity because of antibody dilution. Although the recombinant GPIbα/CaM fusion proteins were bound to the solid phase via the CD42b mAb MB45 to enable comparison with MAIPA, this situation has the potential problem that human sera with anti-mouse activity25 26 will give false-positive results. As an alternative, the CaM tag could be used to bind the recombinant protein to a solid phase. The resonant mirror results clearly show that rapid, but reversible binding, can be achieved with specific uptake of HPA-2a and -2b antibodies from 1 : 10 diluted serum samples (Figure 3, panels A to D).
The specific reactivity of the HPA-2 antibodies with their corresponding Thr-145 or Met-145 form of the protein make it highly unlikely that the reactions were with the CaM fragment, which was common to both forms of the recombinant GPIbα. The apparent minor cross-reactivity exhibited by some HPA-2b antibodies showing minor reactivity with the Thr-145 GPIbα in ELISA, and similarly anti-HPA-2a with the Met-145 GPIbα in the biosensor studies, is also unlikely to be because of reactivity with the CaM portion, as it also occurred with platelet-derived GPIbα in the MAIPA assays. It most likely reflects similar reactivity already described within the HPA-2 system,27 or minor variations in binding that are not significantly different from variation in negative controls.
By use of a panel of anti-HPA-2b and an anti-HPA-2a in multiple assay systems, the current study ruled out the possibility that the other subunits of the GPIb-IX-V complex are associated with or required for the formation of both the HPA-2a and HPA-2b B cell epitopes and showed that the epitopes are not dependent on disulfide bond formation (Figure4). The latter is in contrast with the results of experiments with the Thr-145 and Met-145 forms of a 302-residue GPIbα fragment expressed in mammalian cells, which showed that a single example of the antibody only reacted with its respective antigen by dot blot using nonreduced recombinant protein.6 This discrepancy is most likely a reflection of the different immunoassays used for antibody detection, but it may also be a feature of the single-antibody sample used in the study.
Previous work with HPA-2a and -2b platelets did not produce evidence that the Thr/Met-145 polymorphism has an effect on ristocetin-mediated platelet agglutination.7 However, a small effect of the Thr/Mer mutation on the binding kinetics of vWF to GPIbα might have been masked in the aggregation assay. The recombinant forms of GPIbα were, therefore, used to investigate the possible effects of the mutation in finer detail. The resonant mirror studies of vWF binding to CaM immobilized GPIbα confirmed that the binding kinetics to both forms of GPIbα are identical (Figure 6). However, studies of coronary heart disease and cerebral vascular disease have found an increased frequency of the HPA-2b allele, suggesting that the mutation does have a functional effect.28,29 These conflicting observations may reflect differences between in vitro ristocetin-induced vWF binding and binding in vivo. Alternatively, the HPA-2 polymorphism could be genetically linked to two other polymorphisms that influence GPIbα structure (variable number of tandem repeats polymorphism)30,31 and expression level (Kozak polymorphism)32 and have potential effect on thrombotic disease.
In conclusion, our studies demonstrate that the two forms of recombinant GPIbα/CaM fusion protein are useful for the detection of HPA-2 antibodies and that disulfide bond formation and glycosylation are not important for formation of the B cell epitopes. At the molecular level, there is no evidence that the Thr-/Met-145 mutation alone affects vWF binding. That HPA-2 antibodies inhibit vWF binding is suggestive of an involvement of the LRR in vWF binding, as it is unlikely that the inhibition is explained by allosteric hindrance.
Acknowledgments
We gratefully acknowledge the continuous support of the Apheresis Clinic and Platelet Immunology Reference Laboratory at the National Blood Service East Anglia Centre during this study and thank Dr N. A. Watkins for helpful discussions during preparation of the manuscript. The generous donation of monoclonal antibodies and HPA-2 antibodies by Prof von dem Borne, Mr C. Hurd, Dr L. Porcelijn, Mrs C. Quader, and Prof Shibata is greatly acknowledged.
J. Davies was supported by grant G9410995 from the Medical Research Council, UK; C. Q. Li and S. F. Garner were supported by a grant from the English National Blood Service.
Reprints:Willem H. Ouwehand, Division of Transfusion Medicine, Department of Hematology, University of Cambridge, National Blood Service East Anglia, Long Road, Cambridge, CB2 2PT, UK; e-mail:who1000@cam.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.