Stat6 transcription factor is a critical mediator of IL-4-specific gene responses. Tyrosine phosphorylation is required for nuclear localization and DNA binding of Stat6. The authors investigated whether Stat6-dependent transcriptional responses are regulated through IL-4-induced serine/threonine phosphorylation. In Ramos B cells, the serine/threonine kinase inhibitor H7 inhibited IL-4-induced expression of CD23. Treatment with H7 did not affect IL-4R-mediated immediate signaling events such as tyrosine phosphorylation of Jak1, Jak3, insulin receptor substrate (IRS)-1 and IRS-2, or tyrosine phosphorylation and DNA binding of Stat6. To analyze whether the H7-sensitive pathway was regulating Stat6-activated transcription, we used reporter constructs containing different IL-4 responsive elements. H7 abrogated Stat6-, as well as Stat5-, mediated reporter gene activation and partially reduced C/EBP-dependent reporter activity. By contrast, IL-4-induced transcription was not affected by wortmannin, an inhibitor of the phosphatidyl-inositol 3′-kinase pathway. Phospho-amino acid analysis and tryptic phosphopeptide maps revealed that IL-4 induced phosphorylation of Stat6 on serine and tyrosine residues in Ramos cells and in 32D cells lacking endogenous IRS proteins. However, H7 treatment did not inhibit the phosphorylation of Stat6. Instead, H7 inhibited the IL-4-induced phosphorylation of RNA polymerase II. These results indicate that Stat6-induced transcription is dependent on phosphorylation events mediated by H7-sensitive kinase(s) but that it also involves serine phosphorylation of Stat6 by an H7-insensitive kinase independent of the IRS pathway.
Extracellular signals, including small polypeptide cytokines, play a central role in the regulation of cell proliferation and differentiation. Cytokine receptors activate various cellular signal transduction pathways, leading to specific gene responses. Interleukin-4 (IL-4) is a pleiotropic cytokine produced mainly by Th2 cells, mast cells, and basophils.1 The biologic responses to IL-4 stimulation are dependent on the cell type and on the differentiation stage of the cell. Initially, IL-4 was identified as a growth and differentiation factor for B cells, yet it exerts distinctive biologic effects on various cell types, among them T cells, myeloid cells, hepatocytes, keratinocytes, and epithelial cells.1 In B cells, IL-4 acts as a co-mitogen. IL-4 also induces the expression of the Fc receptor for IgE (CD23) and major histocompatibility complex (MHC) class II molecules, and it stimulates the transcription of immunoglobulin heavy-chain germline Igε and Igγ1 genes, leading to class switching and to IgE and IgG1 synthesis.2-4
The biologic responses to IL-4 are initiated on IL-4 binding to its plasma membrane receptor. The IL-4 receptor (IL-4R) is composed of 2 subunits, the widely expressed IL-4Rα chain and the common γc chain.5-7 The latter is also used by the IL-2 family cytokines IL-2, IL-7, IL-9, and IL-15. In addition, in endothelial cells, IL-4Rα has been shown to form a functional receptor complex with IL-13R.8 IL-4Rα is primarily responsible for ligand binding. Dimerization of IL-4Rα alone is sufficient for transducing IL-4-specific signal transduction, though the presence of γc increases receptor-binding affinity and facilitates signal transduction.9,10 All receptor chains implicated in IL-4 signaling belong to the cytokine receptor superfamily, and they do not possess intrinsic catalytic activity.5-7 Instead, IL-4 stimulation activates the receptor-associated Jak1 and Jak3 tyrosine kinases, resulting in phosphorylation of the receptor at specific tyrosine residues, recruitment of signaling proteins to the receptor complex, and activation of signal transduction.11 The best-characterized downstream signaling pathways from the IL-4R complex involve activation of phosphatidyl-inositol 3′-kinase (PI 3-K) and Stat6 (signal transducer and activator of transcription) signaling pathways.
The adapter proteins insulin receptor substrate-1 (IRS-1) and IRS-2 are the major tyrosine-phosphorylated substrates in IL-4R signaling. They are required for IL-4-induced cell proliferation in 32D myeloid progenitor cells.12,13 Upon IL-4 stimulation IRS-1 and IRS-2 are recruited to the IL-4R complex by the phosphorylated tyrosine residue Y497 in the IL-4Rα, and they undergo tyrosine phosphorylation by Jak1.14,15 The tyrosine-phosphorylated IRS proteins provide docking sites for SH2 domain-containing cellular signaling proteins, including the regulatory subunit p85 of PI 3-K and Grb-2.16,17 An important biologic role for PI 3-K in IL-4 signaling appears to be in the regulation of cell survival.18 The downstream effectors of activated PI 3-K include serine/threonine kinases Akt/protein kinase B, protein kinase C, and p70 ribosomal protein S6 kinase, of which Akt has been shown to mediate the effect of PI 3-K on cell survival by preventing apoptosis.19-21
IL-4 stimulation results in tyrosine phosphorylation and activation of Stat6.22,23 Stat6 functions as a crucial mediator of IL-4-specific gene responses, as attested by the similar phenotypes of the Stat6 and IL-4 knockout mice with deficient Th2 differentiation and absent IgE responses.24-27 Stat6 activation has been shown to involve 3 tyrosine residues in the IL-4Rα, and possibly also the IRS docking site Y497, and Stat6 is considered to bind to these phosphorylated residues through its SH2 domains.22,28-30Once recruited to the receptor complex, Stat6 becomes tyrosine phosphorylated and forms SH2-mediated homodimers that are translocated to the nucleus and bind to specific DNA elements on IL-4-responsive genes.31,32 Recognition of specific promoter elements is an important determinant of IL-4/Stat6 signaling specificity, and Stat6 displays strong binding preference for sequences in which the dyad symmetric Stat recognition elements (TTC-GAA) are separated by 4 nucleotides, as found in the promoter regions of Igε, CD23, and IL-4Rα genes.33-35
The family of Stat transcription factors consists of 7 mammalian members activated by various cytokines.36,37 The Stat proteins share a conserved overall structure, including a central DNA binding domain followed by the linker domain and the SH2 domain. The carboxy-terminal portions of the proteins contain the single phosphorylated tyrosine residue absolutely required for their DNA binding activity. The carboxy-terminal domain is also required for the transcriptional activity of the Stats, as initially demonstrated in studies involving interferon (IFN) and Stat1.38 Stat1 mRNA is expressed as 2 alternatively spliced isoforms, Stat1α and Stat1β, that differ by the 38 carboxy-terminal residues lacking in Stat1β.39 In IFN-γ signaling, both isoforms become tyrosine phosphorylated and bind identical DNA target sequences, but only Stat1α is transcriptionally active. Subsequent studies with naturally occurring variants and deletion mutants of different Stats have demonstrated that the carboxy-termini of Stat2, Stat3, Stat5, and Stat6 are similarly required for transactivation of their respective cytokine-responsive target promoters.40-46 The mechanisms coupling Stats to transcriptional activation are still poorly understood, but posttranslational modifications through serine phosphorylation have been shown to be involved in this regulation. Stat1, Stat3, and Stat5a/b become serine phosphorylated in response to cytokine or growth factor stimulation.47-53 In Stat1 and Stat3, this phosphorylation has been shown to regulate their transcriptional activation. Thus far, the regulatory mechanisms involved in Stat6-mediated transcriptional activation and the occurrence of serine/threonine phosphorylation of Stat6 are unknown. In this study, we investigated the mechanisms regulating Stat6-mediated transcriptional responses in IL-4 signaling. We found that IL-4-induced transcriptional activation required the convergence of tyrosine and multiple serine/threonine kinase pathways.
Materials and methods
Cell culture and transfections
COS-7, HepG2, and 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco BRL, Life Technologies, Gaithersburg, MD) and antibiotics. Ramos 2G6 cells were grown in RPMI medium (Gibco BRL) containing 10% FBS and antibiotics. Transfections of 293T and HepG2 cells were made by the calcium-phosphate coprecipitation method, and COS-7 cells were electroporated with a Bio-Rad gene pulser at 260 V/960 μF.
DNA constructs
Igε-reporter (RE) construct was generated by cloning 4 copies of the annealed oligonucleotides (5′-P-TCGAGCTGTTGCTCAATCGACTTCCCACCAAGAACAG-3′ and 5′-P-TCGACTGTTCTTGGGAAGTCGATTGAGCAACAG-3′) as direct repeats into SalI site of pfLUC-plasmid, which carries c-fosminimal promoter in front of the Photinus pyralis luciferase gene. Stat6-RE and C/EBP reporter constructs were created by cloning annealed oligonucleotides (Stat6-RE: 5′-P-TCGACATCGACTTCCCAAGAACAGAATCGACTTCCCAAGAACAGAATCGACTTCCCAAGAACAGT-3′ and 5′-P-TCGAACTGTTCTTGGGAAGTCGATTCTGTTCTTGGGAAGTCGATTCTGTTCTT-GGGAAGTCGATG-3′) and (C/EBP-RE to give 5′-P-TCGAGCTGTTGCTCAATCGACGCTGTTGCTCAATCGACGCTGTTGCTCAATCGAC-3′ and 5′-P-TCGAGTCGATTGAGCAACAGCGTCGATTGAGCAACAGCGTCGATTGAGCAACAGC-3′) into the SalI site of pLUC-plasmid. Drs M. Heim, B. Groner, and T. Wood kindly donated the Stat6 and Stat5 expression vectors, the Stat5-Jak2-VP16 construct,54 and the Stat5-responsive SPI-luciferase construct.55
Antibodies
Antiphosphotyrosine antibody (clone 4G10) was from Upstate Biotechnology (Lake Placid, NY). Rabbit polyclonal anti-Jak1 and anti-Jak3 antibodies, previously characterized,11 were a kind gift of Dr. J. Ihle. Monoclonal anti-Jak1 antibody (Transduction Laboratories, Lexington, KY) was used for Western blot analysis. Anti-Stat6 antibody (M-20) and anti-RNA polymerase II antibody (C-21) were purchased from Santa Cruz Technology (Santa Cruz, CA), and antibodies against IRS-1 and IRS-2 were purchased from Upstate Biotechnology.
Immunoprecipitation and Western blot analysis
Cells were lysed in Triton lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 10% glycerol, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, 50 mmol/L NaF, 1 mmol/L Na3VO4) or in RIPA lysis buffer (50 mmol/L Tris-HCl, pH 8, 150 mmol/L NaCl, 1% NP-40, 0.5% DOC, 0.1% sodium dodecyl sulphate [SDS]), supplemented with phenylmethylsulfonyl fluoride and aprotinin. Immunoprecipitations from equal protein amounts were carried out as previously described.56 Protein concentrations of the lysates were measured using the Bio-Rad Protein Assay system (Bio-Rad Laboratories, Hercules, CA). Immunoprecipitates were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane (Micron Separation, Westborough, MA). Immunodetection was performed using specific primary antibodies, biotinylated antimouse or antirabbit secondary antibodies (Dako A/S, Glustrup, Denmark), and streptavidin-biotin horseradish peroxidase-conjugate and electrochemiluminescence detection (Amersham Pharmacia Biotech, Buckinghamshire, UK).
Electrophoretic gel mobility shift assay
For electrophoretic gel mobility shift assay, nuclear extracts were prepared as described earlier.56 COS-7 cells were transfected with 1 μg Stat6 plasmid and 1 μg β-galactosidase plasmid to monitor the transfection efficiency. Annealed Igε oligonucleotide235′-CGATCAAGACCTTTCCCAAGAAATCT-3′ was end-labeled by T4 polynucleotide kinase using [γ-32P]-adenosine triphosphate. Nuclear extracts (6μg), poly-dI-dC (240 ng/μL), bovine serum albumin (1.5 mg/mL), and 32P-labeled Igε oligonucleotide (0.5 ng) were incubated for 30 minutes at room temperature, and the reactions were resolved in 4.5% TBE (0.25×) PAGE, followed by autoradiography.
Luciferase assay
For luciferase assays, 3 × 106 COS-7 cells were transfected with 5 μg Igε-RE or Stat6-RE luciferase reporter constructs and 2 μg β-galactosidase plasmid. After electroporation, cells were divided into 6-well plates, 50 000 cells per plate. 293T and HepG2 cells were transfected by the calcium phosphate coprecipitation method. 293T and COS-7 cells were cotransfected with Stat6 expression plasmid. Cells were grown for 24 hours, and O/N serum-starved cells were stimulated with 10 ng/mL recombinant human IL-4 (PeproTech EC, London, UK) or 4 IU/mL erythropoietin (Eprex, Janssen-Cilag) for 6 hours. H7 (1-(5-isoquinolinylsulfonyl)2-methylpiperazine) (Sigma Chemical, St. Louis, MO) and wortmannin (Sigma) were added to the cultures, when indicated, 30 minutes before cytokine stimulus. Forty-eight hours after transfection, cells were lysed in Reporter Lysis Buffer (Promega, Madison, WI), and luciferase activity was determined using the Luciferase Assay System (Promega) according to manufacturer's instructions. The luciferase values were normalized against measured β-galactosidase activities.
Phospho-amino acid analysis and phosphopeptide map
Phospho-amino acid analysis and phosphopeptide maps were done as described earlier.57 58 Briefly, Ramos 2G6 cells were starved overnight and labeled with32P-orthophosphate (Amersham) in phosphate-free medium (Sigma) for 3 hours. Cells were stimulated as indicated and lysed in Triton lysis buffer, and Stat6 was immunoprecipitated. Immunocomplexes were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)-membrane (Amersham). Proteins were visualized by autoradiography, and bands corresponding to Stat6 were excised for further analysis. For the phospho-amino acid analysis, proteins were hydrolyzed in 6 N HCl for 2 hours at 110°C. Lysates were lyophilized and resolved in pH 1.9 buffer containing standard phospho-amino acids (o-Phospho-DL-serine, o-Phospho-DL-threonine, o-Phospho-DL-tyrosine; Sigma). Phospho-amino acids were separated by 2-dimensional electrophoresis. Standard amino acids were visualized by ninhydrin staining, and autoradiography was used to detect phosphorylated amino acids. For phosphopeptide mapping, proteins were digested with sequencing-grade Trypsin (Promega) at 37°C O/N. Washed fragments were applied to thin-layer cellulose plates, separated in the first dimension by electrophoresis at pH 8.9 and in the second dimension by ascending chromatography using Phospho-buffer (37.5% n-butanol, 25% pyridine, 7.5% acetic acid). Phosphopeptides were visualized by autoradiography.
RA polymerase II phosphorylation in vivo
Ramos cells were labeled in phosphate-free medium containing 10 μCi/mL 32P-orthophosphate as described above and were treated with IL-4 and H7 as indicated, and then nuclear extracts were prepared.59 Ten micrograms nuclear extract was separated on 5% SDS-PAGE and transferred to PVDF-membrane. Phosphorylated proteins were detected by autoradiography. RNA polymerase II was detected by immunoblotting with anti-RNA polymerase II antibody using unlabeled lysates from the same experiment. For immunoprecipitation,32P-orthophosphate-labeled Ramos cells were lysed in RIPA buffer, and 1000 μg lysate was immunoprecipitated with 2 μg anti-RNA polymerase II antibody. Immunoprecipitates were separated on 6% SDS-PAGE, and phosphorylated proteins were detected by autoradiography.
FACS analysis
Cells were suspended in RPMI + 10% FBS and first stained with 2 μg mouse antihuman CD23 antibody (Pharmingen, San Diego, CA) or with 2 μg control antibody, isotype-matched mouse antihuman-Nef mAb (a kind gift from Dr V. Ovod) for 30 minutes at 4°C and then washed twice with phosphate-buffered saline. Secondary antibody, 5 μg fluorescein isothiocyanate-conjugated goat antimouse antibody (Dako), was added to the cells for 30 minutes. Cells were washed again twice with phosphate-buffered saline and analyzed with FACScan (Becton Dickinson, San Jose, CA).
Results
H7 inhibits IL-4-induced CD23 expression but not the immediate signaling events in Ramos B cells
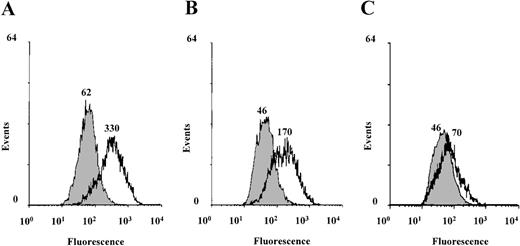
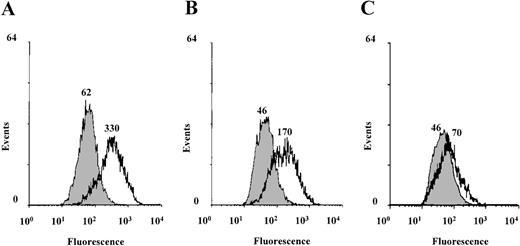
To investigate the role of serine/threonine phosphorylation in Stat6-mediated IL-4 transcriptional activation, we used the serine/threonine kinase inhibitor (1-(5-isoquinolinesulfonyl)-2-methylpiperazine (H7). H7 binds to the adenosine triphosphate binding site in the kinase domain and inhibits many, but not all, serine/threonine kinases.60 For example, the cytokine-induced ERK and p70 S6 kinases are not affected by H7.60,61 IL-4-induced transcriptional activation was studied in the human Burkitt lymphoma B-cell line Ramos 2G6, which expresses abundant IL-4R.62 Stimulation of B cells with IL-4 has been shown to induce the expression of the low-affinity IgE Fc receptor (CD23) in a Stat6-dependent manner.4,25-27 62Ramos cells were stimulated with IL-4 for 20 hours in the presence or absence of H7, and the surface expression of CD23 was analyzed by using FACS. As shown in Figure 1, IL-4-induced CD23 expression was inhibited by H7 in a concentration-dependent manner, indicating that the IL-4- and Stat6-regulated expression of CD23 requires serine/threonine kinase activity.
IL-4-induced CD23 expression is inhibited by H7 in Ramos B cells.
Ramos 2G6 cells were untreated (filled curves) or treated with IL-4 (10 ng/mL) (open curves) for 20 hours in the absence or presence of H7. (A) No H7. (B) 10 μmol/L H7. (C) 25 μmol/L H7. CD23 expression was analyzed by staining with anti-CD23 mAb and fluorescein isothiocyanate-conjugated goat antimouse mAb. The fluorescence intensity was determined using a FACScan, and the mean fluorescence intensities are indicated in the figure. Nonspecific binding was determined by staining the cells with an isotype-matched irrelevant mAb, and the median fluorescence channel for negative control antibody was 17. One representative of 3 experiments is shown.
IL-4-induced CD23 expression is inhibited by H7 in Ramos B cells.
Ramos 2G6 cells were untreated (filled curves) or treated with IL-4 (10 ng/mL) (open curves) for 20 hours in the absence or presence of H7. (A) No H7. (B) 10 μmol/L H7. (C) 25 μmol/L H7. CD23 expression was analyzed by staining with anti-CD23 mAb and fluorescein isothiocyanate-conjugated goat antimouse mAb. The fluorescence intensity was determined using a FACScan, and the mean fluorescence intensities are indicated in the figure. Nonspecific binding was determined by staining the cells with an isotype-matched irrelevant mAb, and the median fluorescence channel for negative control antibody was 17. One representative of 3 experiments is shown.
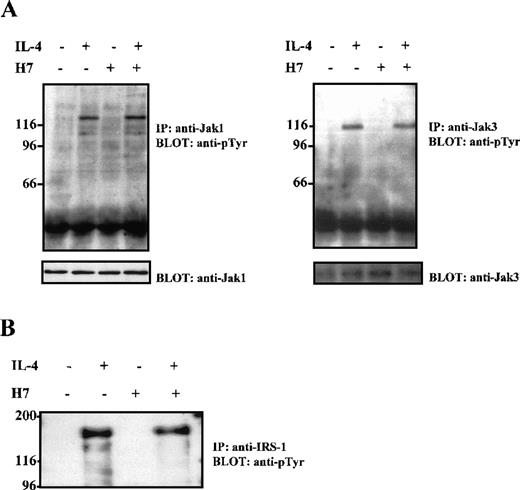
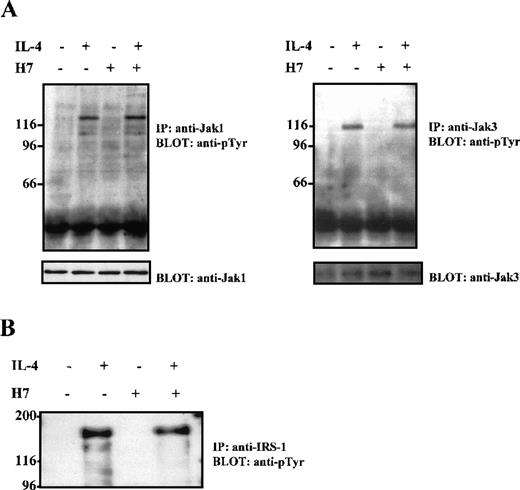
On IL-4 stimulation, Stat6 becomes tyrosine phosphorylated in the IL-4R complex, presumably by the receptor-associated Jak1 and Jak3 tyrosine kinases. To examine whether the effect of H7 on CD23 expression was caused by the inhibition of the IL-4R-induced immediate signaling events, the consequence of H7 treatment on IL-4-induced Jak1 and Jak3 activation was investigated in Ramos cells. Extracts from these cells were immunoprecipitated with anti-Jak1 and anti-Jak3 antisera, separated by SDS-PAGE, and subjected to anti-phosphotyrosine immunoblot analysis. As shown in Figure 2A, pretreatment of cells with H7 for 30 minutes before IL-4 stimulation did not inhibit either Jak1 or Jak3 tyrosine phosphorylation. For control, the filter was blotted again with anti-Jak1 and anti-Jak3 antisera; the treatments did not affect the protein levels. In IL-4R signaling, the receptor-associated IRS-1/IRS-2 proteins play a critical role for several downstream signaling pathways. The effect of H7 on IRS-1 and IRS-2 tyrosine phosphorylation was analyzed in Ramos cells from immunoprecipitated proteins; H7 treatment did not affect IL-4-induced tyrosine phosphorylation of IRS-1 or IRS-2 (Figure 2B and data not shown).
H7 does not inhibit IL-4-induced Jak1, Jak3, or IRS-1 tyrosine phosphorylation.
Ramos cells were starved O/N and were untreated or treated with H7 (100 μmol/L) for 30 minutes and then were stimulated with IL-4 (100 ng/mL) for 10 minutes as indicated. (A) Jak1 and Jak3 were immunoprecipitated, separated by 7.5% SDS-PAGE, and immunoblotted with anti-phosphotyrosine antibody. Shown in the lower panel are protein levels from the same filters blotted again with anti-Jak1 and anti-Jak3. (B) Ramos cells treated as in (A), but immunoprecipitation was carried out with anti-IRS-1 antibody before anti-phosphotyrosine immunoblotting.
H7 does not inhibit IL-4-induced Jak1, Jak3, or IRS-1 tyrosine phosphorylation.
Ramos cells were starved O/N and were untreated or treated with H7 (100 μmol/L) for 30 minutes and then were stimulated with IL-4 (100 ng/mL) for 10 minutes as indicated. (A) Jak1 and Jak3 were immunoprecipitated, separated by 7.5% SDS-PAGE, and immunoblotted with anti-phosphotyrosine antibody. Shown in the lower panel are protein levels from the same filters blotted again with anti-Jak1 and anti-Jak3. (B) Ramos cells treated as in (A), but immunoprecipitation was carried out with anti-IRS-1 antibody before anti-phosphotyrosine immunoblotting.
H7-sensitive pathway does not regulate IL-4-induced tyrosine phosphorylation and DNA-binding activity of Stat6
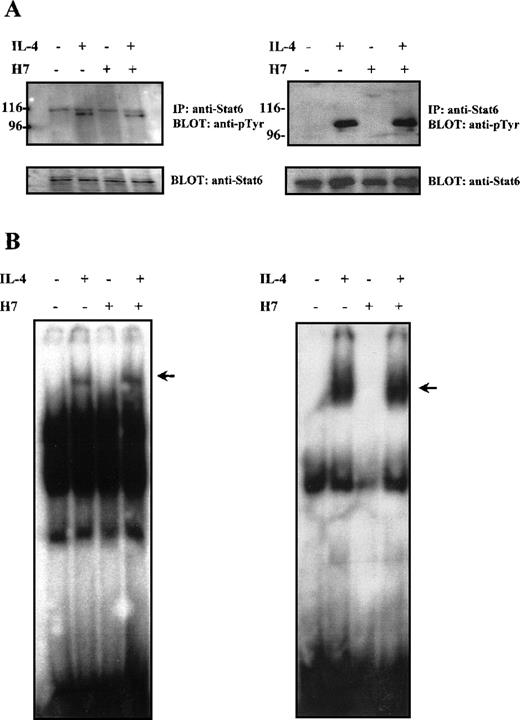
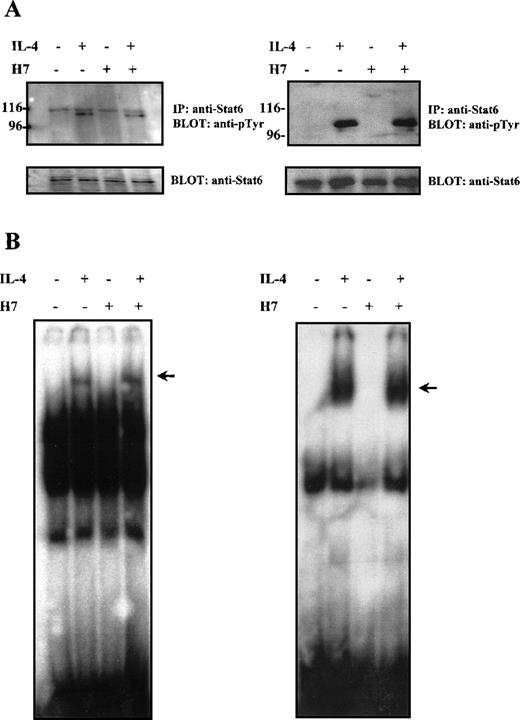
Most of the prevailing evidence regarding the effect of Stat serine phosphorylation suggests that serine phosphorylation primarily affects transcriptional activity of the proteins. However, in the case of Stat3, serine phosphorylation has been reported to regulate negatively the degree of tyrosine phosphorylation and of DNA binding capacity.48,63 64 We analyzed the effect of H7 on tyrosine phosphorylation of Stat6 in Ramos cells and in HepG2 cells. In both cell types, IL-4 stimulation resulted in rapid tyrosine phosphorylation of Stat6, which could not be prevented by H7 treatment (Figure3A).
Effect of H7 on IL-4-induced Stat6 tyrosine phosphorylation and DNA binding.
(A) HepG2 (left) and Ramos cells (right) were untreated or treated with H7 (100 μmol/L) for 30 minutes and then stimulated with IL-4 (100 ng/mL) for 10 minutes as indicated. Stat6 was immunoprecipitated and separated by 7.5% SDS-PAGE and was subjected to anti-phosphotyrosine immunoblotting. Shown in the lower panel are protein levels after anti-Stat6 reblotting. (B) Lysates from Ramos cells (left) and COS-7 cells transfected with Stat6 (right), treatments as in (A), and nuclear lysates were analyzed in the mobility shift assay using32P-labeled Igε oligonucleotide. The Stat6 binding complex is indicated with an arrow.
Effect of H7 on IL-4-induced Stat6 tyrosine phosphorylation and DNA binding.
(A) HepG2 (left) and Ramos cells (right) were untreated or treated with H7 (100 μmol/L) for 30 minutes and then stimulated with IL-4 (100 ng/mL) for 10 minutes as indicated. Stat6 was immunoprecipitated and separated by 7.5% SDS-PAGE and was subjected to anti-phosphotyrosine immunoblotting. Shown in the lower panel are protein levels after anti-Stat6 reblotting. (B) Lysates from Ramos cells (left) and COS-7 cells transfected with Stat6 (right), treatments as in (A), and nuclear lysates were analyzed in the mobility shift assay using32P-labeled Igε oligonucleotide. The Stat6 binding complex is indicated with an arrow.
The effect of H7 on Stat6 DNA binding activity was analyzed in Ramos B cells and in COS7 cells. COS7 cells express a functional IL-4R but do not endogenously express Stat6.44 Therefore, they were transiently transfected with Stat6 expression plasmid. The Stat6 DNA-binding activity was analyzed in electrophoretic mobility shift assay from nuclear cell lysates using the Stat6-specific Igε oligonucleotide as a probe.23 In both cell lines, IL-4 induced rapid DNA binding activity of Stat6, which was unaffected by H7 treatment (Figure 3B). Taken together, these results indicated that the H7-sensitive kinase(s) did not affect IL-4-induced activation events promoting tyrosine phosphorylation, nuclear localization, or DNA binding of Stat6; therefore, they suggested that the effect of H7 on Stat6-dependent gene expression resulted from transcriptional inhibition.
IL-4-induced activation of Igɛ reporter gene is dependent on the H7-sensitive pathway
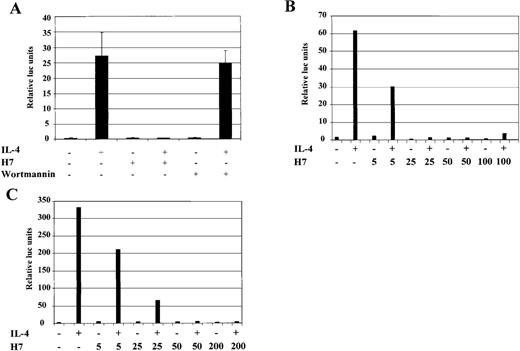
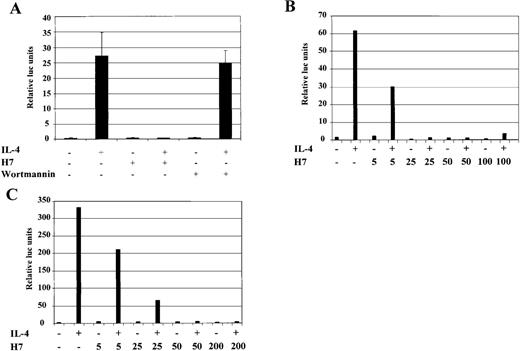
To study in more detail the observed serine/threonine kinase dependency of IL-4-induced gene expression, we used the previously characterized IL-4 responsive cell lines HepG2, COS-7, and 293T, which are more feasible to transfect with defined reporter constructs than B cells.44,46 Analysis of the promoter region of the human Igε heavy-chain constant region gene has defined the minimal IL-4-responsive element, composed of C/EBP and Stat6 binding elements, to nucleotides −168 to −138 relative to the first RNA initiation site.46 A luciferase reporter construct (Igε-RE) was prepared that consisted of 4 copies of this IL-4-responsive element placed upstream of a minimal heterologous promoter. In agreement with previous reports, 46 IL-4 induced pronounced transcriptional activation of the Igε-RE construct in HepG2 cells (Figure 4A), whereas the control vector carrying the minimal promoter without the Igε element was not activated (data not shown). When HepG2 cells were treated simultaneously with H7 and IL-4, Igε-RE reporter activation was found to be completely abrogated (Figure 4A). We also tested whether the prolonged H7 treatment would affect IL-4R signaling proteins, but no effects were found on Stat6 DNA binding or on Jak1 and Stat6 protein levels (data not shown).
IL-4-induced activation of Igɛ-RE reporter is inhibited by H7 but not by wortmannin.
(A) HepG2 cells were transfected with Igε-RE reporter construct (0.5 μg) together with a β-galactosidase vector (0.2 μg). Forty-eight hours after transfection, the cells were treated with IL-4 (10 ng/mL), H7 (200 μmol/L), or wortmannin (1 μmol/L) as indicated for 6 hours, and β-galactosidase and luciferase activities were measured. The luciferase values were normalized against β-galactosidase. Mean values of 3 independent experiments with SD are shown. (B) HepG2 cells transfected with Igε-RE reporter and treated with IL-4 and different concentrations of H7 for 6 hours, as indicated, and luciferase and β-galactosidase activities were measured. (C) Human 293T cells were transfected with Stat6 expression vector (0.15 μg), together with Igε-RE and β-galactosidase vectors. H7 and IL-4 treatments were performed as described above, and luciferase and β-galactosidase values were measured.
IL-4-induced activation of Igɛ-RE reporter is inhibited by H7 but not by wortmannin.
(A) HepG2 cells were transfected with Igε-RE reporter construct (0.5 μg) together with a β-galactosidase vector (0.2 μg). Forty-eight hours after transfection, the cells were treated with IL-4 (10 ng/mL), H7 (200 μmol/L), or wortmannin (1 μmol/L) as indicated for 6 hours, and β-galactosidase and luciferase activities were measured. The luciferase values were normalized against β-galactosidase. Mean values of 3 independent experiments with SD are shown. (B) HepG2 cells transfected with Igε-RE reporter and treated with IL-4 and different concentrations of H7 for 6 hours, as indicated, and luciferase and β-galactosidase activities were measured. (C) Human 293T cells were transfected with Stat6 expression vector (0.15 μg), together with Igε-RE and β-galactosidase vectors. H7 and IL-4 treatments were performed as described above, and luciferase and β-galactosidase values were measured.
PI 3-K activates several serine/threonine kinases, including Akt, various isoforms of PKC, and p70 S6 kinase.19-21 We wanted to determine whether the PI 3-K-dependent pathways regulated the activation of the Igε-RE by using the irreversible PI 3-K inhibitor wortmannin.65 However, wortmannin did not have any effect on IL-4-induced reporter activity at concentrations that completely block PI 3-K catalytic activity (Figure 4A).66 In contrast, when the concentration dependency of the H7 effect was analyzed, it was found to inhibit the IL-4-induced transcriptional activation already at 25 μmol/L concentration, which is similar to the concentration required for inhibition of CD23 induction in Ramos cells (Figures 1,4B).
To investigate whether IL-4-induced transcriptional activation was similarly dependent on H7 in other IL-4 responsive cell types, the Igε-RE reporter was transfected to 293T cells, which express the necessary IL-4R signaling components but lack Stat6 protein.46 In agreement with previous reports,46 IL-4 activated the Igε-RE reporter only when cotransfected with Stat6 (data not shown). The Igε-RE reporter activation was inhibited by H7 at the same concentration dependency level observed in HepG2 cells (Figure 4C). Taken together, these results indicate that IL-4-induced transcriptional activation was dependent on H7-sensitive kinase(s) and that the PI 3-K-dependent pathway did not regulate this kinase.
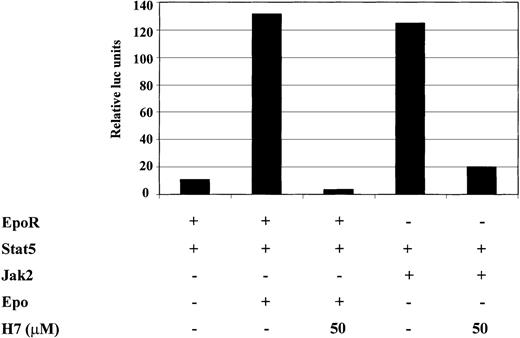
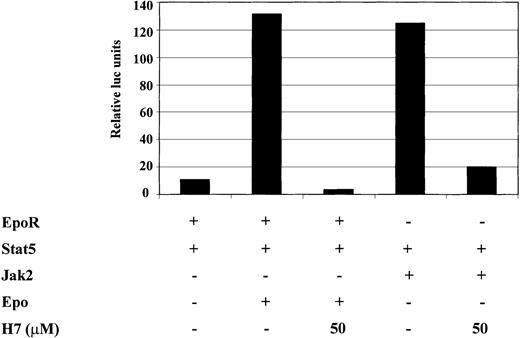
Next we wanted to investigate whether the H7-sensitive kinase was regulated directly by the cytokine receptor or by the downstream Jak kinases. Cotransfection of Jak1, Jak3, or their combination failed to induce significant Stat6-dependent transcription (data not shown). Therefore, we studied Stat5, which is activated by multiple cytokines such as erythropoietin (Epo) in a Jak2-dependent manner. For this purpose, 293T cells were transfected with Stat5, together with either Jak2 or EpoR. H7 completely inhibited Epo-induced and Jak2-induced activation of the Stat5-dependent reporter (Figure5). These results showed that H7 inhibited Stat5-driven transcription and suggested that the H7-sensitive kinase functioned downstream of Jak kinases in the signaling cascade.
H7 inhibits Jak2- and erythropoietin-induced activation of the Stat5-dependent reporter.
293T cells were transfected with 1 μg Stat5-responsive SPI luciferase construct and 0.2 μg β-galactosidase, together with 0.25 μg erythropoietin (Epo)R expression vector, 0.5 μg Stat5 expression vector, and 1 μg Jak2 expression vector, as indicated. Where indicated, O/N-starved 293T cells were treated with IL-4 (10 ng/mL) or Epo (4 IU/mL) for 6 hours. H7 (50 μmol/L) was added either 30 minutes before cytokine stimulation or 20 hours before harvesting in Jak2-transfected cells. β-Galactosidase and luciferase values were measured, and luciferase values were normalized against β-galactosidase values. The experiment was repeated twice and yielded identical results.
H7 inhibits Jak2- and erythropoietin-induced activation of the Stat5-dependent reporter.
293T cells were transfected with 1 μg Stat5-responsive SPI luciferase construct and 0.2 μg β-galactosidase, together with 0.25 μg erythropoietin (Epo)R expression vector, 0.5 μg Stat5 expression vector, and 1 μg Jak2 expression vector, as indicated. Where indicated, O/N-starved 293T cells were treated with IL-4 (10 ng/mL) or Epo (4 IU/mL) for 6 hours. H7 (50 μmol/L) was added either 30 minutes before cytokine stimulation or 20 hours before harvesting in Jak2-transfected cells. β-Galactosidase and luciferase values were measured, and luciferase values were normalized against β-galactosidase values. The experiment was repeated twice and yielded identical results.
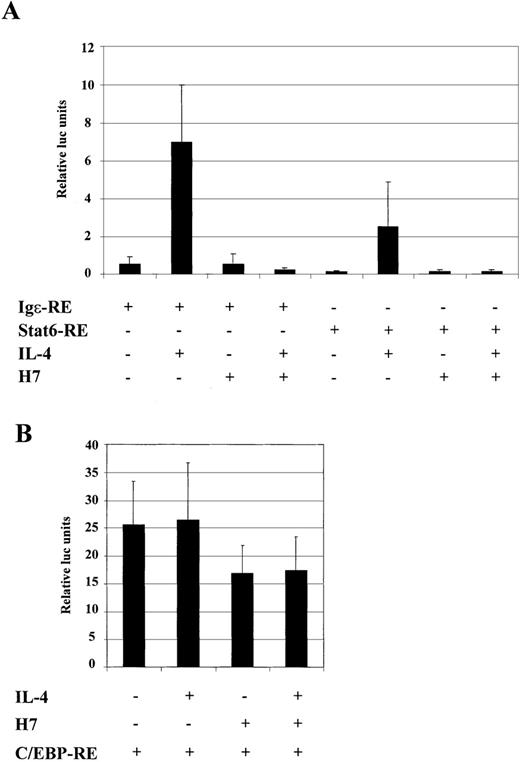
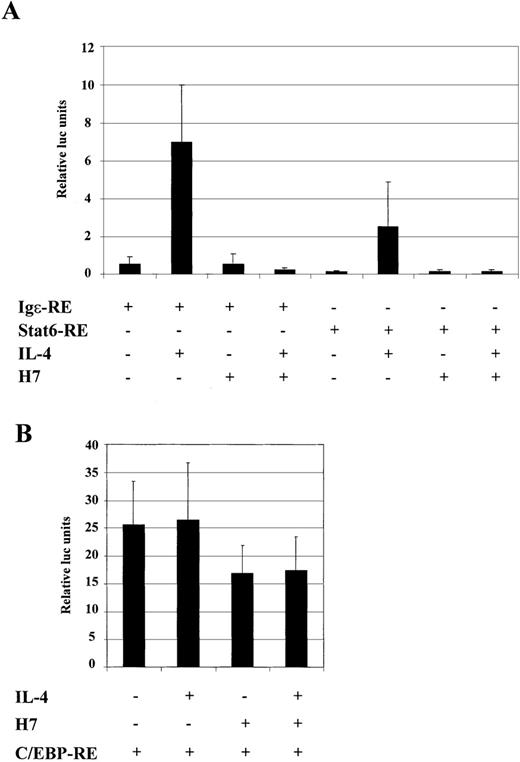
The Igε-RE reporter construct contains binding sites for Stat6 and C/EBP, and H7-sensitive kinase could potentially affect either of these factors. To confirm that the dramatic effect of H7 on the expression of the Igε-RE reporter was directed to Stat6-dependent transcription rather than inhibition of the synergistic action of the C/EBP protein, we prepared luciferase reporter constructs that contained either Stat6 binding sites (Stat6-RE) or C/EBP binding sites (C/EBP-RE). As shown in Figure 6A, IL-4 activated Igε-RE and Stat6-RE, though Igε-RE was more efficiently induced. Nevertheless, IL-4 induction of both reporter constructs was completely inhibited by H7. In lymphoid cells and in HepG2 cells, activation of the Igε element has been shown to be dependent on both Stat6 and C/EBP sites. In our experiments Stat6-RE reporter was not significantly induced in HepG2 cells (2- to 3-fold; data not shown).67 68 The difference between HepG2 and COS-7 cells in activation of the Stat6-RE construct might have been caused by higher Stat6 protein levels in transfected COS-7 cells or, alternatively, by differences in expression of transcriptional coactivators. The C/EBP-RE reporter was constitutively active in HepG2 cells, and its activity was not affected by IL-4 stimulation (Figure 6B). Interestingly, the C/EBP-RE reporter activity was only partially inhibited by H7 (Figure 6B). Taken together, these results indicated that the H7-sensitive kinase regulated the Stat6-dependent transcription.
H7 inhibits IL-4-stimulated transcription of Stat6-RE reporter construct.
Luciferase construct from human Igε promoter containing 3 repeats of Stat6 binding sites (3 × AATCGACTTCCCAAGAACAG) (Stat6-RE) and 4 repeats of C/EBP binding sites (4 × GCTGTTGCTCAATCGAC) (C/EBP-RE) were analyzed. (A) Stat6-RE and Igε reporter constructs were transfected into COS-7 cells, together with β-galactosidase and Stat6 expression vectors. (B) C/EBP-RE reporter construct was transfected into HepG2 cells, together with β-galactosidase expression vector. O/N-starved cells were treated with IL-4 (10 ng/mL) and H7 (100 μmol/L), as indicated. β-Galactosidase and luciferase values were measured. Luciferase values were normalized against β-galactosidase, and mean values of 3 (A) and 7 (B) independent experiments with SD are shown.
H7 inhibits IL-4-stimulated transcription of Stat6-RE reporter construct.
Luciferase construct from human Igε promoter containing 3 repeats of Stat6 binding sites (3 × AATCGACTTCCCAAGAACAG) (Stat6-RE) and 4 repeats of C/EBP binding sites (4 × GCTGTTGCTCAATCGAC) (C/EBP-RE) were analyzed. (A) Stat6-RE and Igε reporter constructs were transfected into COS-7 cells, together with β-galactosidase and Stat6 expression vectors. (B) C/EBP-RE reporter construct was transfected into HepG2 cells, together with β-galactosidase expression vector. O/N-starved cells were treated with IL-4 (10 ng/mL) and H7 (100 μmol/L), as indicated. β-Galactosidase and luciferase values were measured. Luciferase values were normalized against β-galactosidase, and mean values of 3 (A) and 7 (B) independent experiments with SD are shown.
IL-4 induces serine phosphorylation of Stat6
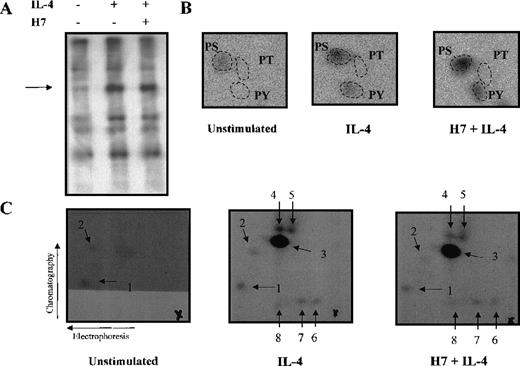
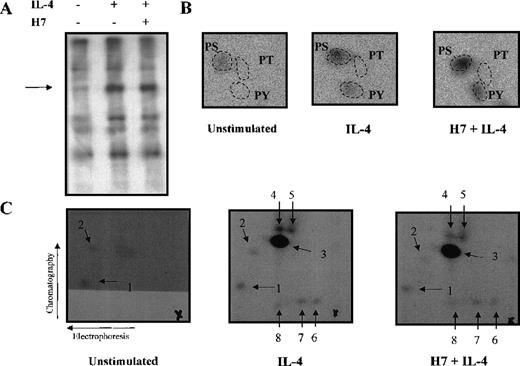
The results obtained using H7 kinase inhibitor suggested that Stat6-dependent transcriptional activation is subject to secondary modification through serine/threonine phosphorylation. To assess directly whether IL-4 induced serine/threonine phosphorylation of Stat6, Ramos cells were metabolically labeled with32P-orthophosphate and were left untreated or were stimulated with IL-4 for 15 minutes. Stat6 was immunoprecipitated, and the phosphorylated bands corresponding to Stat6 were subjected to phospho-amino acid analysis (Figure 7A, B). In nonstimulated cells, Stat6 was phosphorylated at low levels only on serine residues. IL-4 stimulation resulted in increased phosphorylation on serine and tyrosine residues, whereas no phosphorylation on threonine was detected. The magnitude of IL-4-induced serine phosphorylation of Stat6 was relatively low, approximately 2-fold, but it was similar to the level of induction of serine phosphorylation observed in other Stats.51-53 64 Treatment of Ramos cells with H7 for 30 minutes before IL-4 stimulation did not have any significant effect on the level of IL-4-induced serine or tyrosine phosphorylation (Figure 7B).
Phospho-amino acid analysis and phosphopeptide map of Stat6.
Ramos cells were starved O/N and metabolically labeled with32P-orthophosphate in phosphate-free medium for 3 hours. Cells were stimulated with IL-4 (100 ng/mL) for 15 minutes and were treated with H7 (200 μmol/L) for 30 minutes before stimulation, if indicated. Cells were lysed in Triton lysis buffer, and Stat6 was immunoprecipitated. Immunocomplexes were separated in 7.5% SDS-PAGE and transferred to PVDF-membrane. (A) Autoradiography of the precipitates. (B) Phospho-amino acid analysis of the excised and hydrolyzed Stat6 bands. (C) For the phosphopeptide map, the Stat6 proteins excised from PVDF-membrane were digested with trypsin. Fragments were separated in the first dimension by electrophoresis at pH 8.9 and in the second dimension by chromatography. Phosphopeptides visualized by autoradiography are shown (numbered 1-8), and origins are marked with a cross.
Phospho-amino acid analysis and phosphopeptide map of Stat6.
Ramos cells were starved O/N and metabolically labeled with32P-orthophosphate in phosphate-free medium for 3 hours. Cells were stimulated with IL-4 (100 ng/mL) for 15 minutes and were treated with H7 (200 μmol/L) for 30 minutes before stimulation, if indicated. Cells were lysed in Triton lysis buffer, and Stat6 was immunoprecipitated. Immunocomplexes were separated in 7.5% SDS-PAGE and transferred to PVDF-membrane. (A) Autoradiography of the precipitates. (B) Phospho-amino acid analysis of the excised and hydrolyzed Stat6 bands. (C) For the phosphopeptide map, the Stat6 proteins excised from PVDF-membrane were digested with trypsin. Fragments were separated in the first dimension by electrophoresis at pH 8.9 and in the second dimension by chromatography. Phosphopeptides visualized by autoradiography are shown (numbered 1-8), and origins are marked with a cross.
To study further the mechanism of Stat6 phosphorylation, additional experiments were performed in 32D cells that do not express endogenous IRS proteins.12 In untreated cells, low levels of basal serine phosphorylation of Stat6 were detected. IL-4 induced serine and tyrosine phosphorylation of Stat6, and this induction was insensitive to H7 treatment (data not shown). Taken together, the results with 32D cells confirmed the observations made in Ramos cells and further indicated that the serine kinase responsible for phosphorylation of Stat6 was not dependent on the IRS signaling pathway.
To confirm that phosphorylation of Stat6 was not affected by H7, we analyzed tryptic phosphopeptide maps of Stat6 from in vivo labeled Ramos cells. In nonstimulated cells, 2 weakly phosphorylated peptides were detected, and IL-4 stimulation resulted in the occurrence of 1 major and 5 minor phosphopeptides (Figure 7C). However, pretreatment of the cells with H7 did not affect the phosphopeptide pattern of Stat6, indicating that phosphorylation of Stat6 was mediated by H7-insensitive kinase.
IL-4-induced RNA polymerase II phosphorylation is inhibited by H7
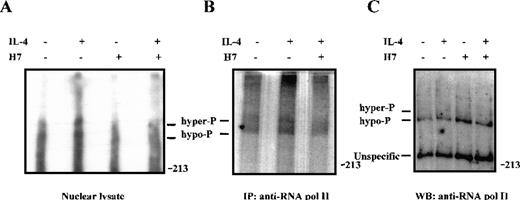
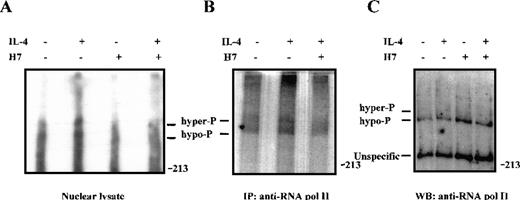
Because phosphorylation of Stat6 was not regulated by H7, we considered the possibility that H7 inhibited other phosphorylation events in IL-4-induced transcriptional regulation. The general transcription factor TFIIH-associated cyclin-dependent kinase (cdk) activating kinase (CAK), as well as cdk7, cdk8, and cdk9, are sensitive to H7 treatment in vitro.69,70 These kinases have been shown to phosphorylate the C-terminal domain (CTD) of RNA polymerase II, resulting in enhanced transcription elongation.70-72 To investigate the regulation of RNA polymerase II phosphorylation in IL-4 signaling, 32P-orthophosphate-labeled Ramos cells were treated with H7 and IL-4, and phosphorylation of RNA polymerase II was analyzed from nuclear cell lysates and from immunoprecipitated proteins. The hyperphosphorylated form of RNA polymerase II has slightly retarded mobility in SDS-PAGE.69 70 As shown in Figure 8, IL-4 stimulation induced phosphorylation of RNA polymerase II, and this phosphorylation was inhibited by pretreatment of the cells with H7 for 30 minutes.
IL-4-induced phosphorylation of RNA polymerase II in vivo is inhibited by H7.
32P-orthophosphate-labeled Ramos cells were left untreated or were pretreated with H7 (100 μmol/L) for 30 minutes and stimulated with IL-4 (100 ng/mL) for 15 minutes, as indicated. (A) Nuclear extracts (10 μg) were separated on 5% SDS-PAGE and transferred to PVDF membrane. Autoradiography of the membrane: hypophosphorylated (hypo-P) and hyperphosphorylated (hyper-P) forms of RNA polymerase II are indicated. (B) RNA polymerase II was immunoprecipitated from32P-orthophosphate-labeled Ramos cells and separated on SDS-PAGE before autoradiography. (C) 50 μg nuclear lysates from unlabeled Ramos cells were electrophoresed on SDS-PAGE, transferred to PVDF membrane, and subjected to immunoblotting with anti-RNA polymerase II antiserum.
IL-4-induced phosphorylation of RNA polymerase II in vivo is inhibited by H7.
32P-orthophosphate-labeled Ramos cells were left untreated or were pretreated with H7 (100 μmol/L) for 30 minutes and stimulated with IL-4 (100 ng/mL) for 15 minutes, as indicated. (A) Nuclear extracts (10 μg) were separated on 5% SDS-PAGE and transferred to PVDF membrane. Autoradiography of the membrane: hypophosphorylated (hypo-P) and hyperphosphorylated (hyper-P) forms of RNA polymerase II are indicated. (B) RNA polymerase II was immunoprecipitated from32P-orthophosphate-labeled Ramos cells and separated on SDS-PAGE before autoradiography. (C) 50 μg nuclear lysates from unlabeled Ramos cells were electrophoresed on SDS-PAGE, transferred to PVDF membrane, and subjected to immunoblotting with anti-RNA polymerase II antiserum.
Discussion
The central role of IL-4 in the regulation of immune responses, including IgE production and Th2 differentiation, has centered wide interest toward the molecular mechanisms of IL-4 actions. In IL-4R signaling, the principal activation event is the catalytic activation of Jak tyrosine kinases, which leads to receptor phosphorylation, recruitment of SH2 and phosphotyrosine-binding domain-containing signaling proteins, and activation of various signaling pathways. IL-4 stimulation also results in the activation of serine/threonine kinases,73 but the role of these pathways in the regulation of IL-4-dependent gene responses is still poorly understood. In this study we provide evidence that IL-4-induced Stat6-dependent transcriptional activation was regulated by the convergence of tyrosine and serine/threonine kinase pathways. However, this IL-4-induced transcriptional regulation appeared to be complex and to involve the regulation of Stat6-dependent gene responses through H7-sensitive and H7-insensitive kinases.
The serine/threonine kinase inhibitor H7 efficiently inhibited the Stat6-dependent gene expression in Ramos cells without affecting the immediate activation events, including Jak1, Jak3, and IRS protein tyrosine phosphorylation. In addition, tyrosine phosphorylation and DNA binding activity of Stat6 were not affected by H7 treatment, suggesting that the effect of H7 may be directed to the inhibition of transcription. The natural promoters of IL-4-regulated genes contain, in addition to Stat6 binding sites, response elements for several other transcription factors, such as C/EBP, NF-κB, and BSAP, which are also potential targets for the observed H7 inhibition. To gain more insight into the target molecules for the H7-sensitive kinase, we used specified promoter elements in the context of reporter genes and studied their activation requirements in IL-4 signaling. The minimal IL-4 response element of the human Igε promoter, consisting of Stat6- and C/EBP binding sites, was efficiently inhibited by H7 in different IL-4-responsive cell types. Additional experiments with Igε-RE, Stat6-RE, and C/EBP-RE reporters confirmed the previously reported synergistic role of C/EBP on Stat6-mediated gene induction67,68 and demonstrated that the Stat6-dependent transcription was the target of H7 inhibition. The C/EBP-RE reporter was only partially inhibited by H7. Similar results were obtained with another constitutively active Stat5-Jak2-VP16 fusion construct54 (data not shown). Thus, these constitutively active transactivation domains are less sensitive to H7 treatment than IL-4- and Stat6-activated transcription. In addition, H7 inhibited Epo- and Jak2-induced activation of a Stat5-dependent reporter, suggesting that activation of the H7-sensitive kinase occurred downstream of Jak kinases. These results also indicate that H7 affected a process required for Stat5 and Stat6 to activate transcription.
The transactivation domain of Stat6 has been shown to be regulated independently of its DNA-binding activity. By using GAL4-Stat6 fusion constructs, Groner et al44 demonstrate that in B cells, IL-4 stimulation enhanced the activity of the transactivation domain, suggesting that IL-4-induced signals provide regulation at transcriptional level. The transactivation domain contains 8 positionally conserved serine residues between mouse and human Stat6; their phosphorylation is a potential mechanism of transcriptional regulation. In line with this hypothesis, the phosphorylation of Ser727 in Stat1 and Stat3 is required for their optimal transcriptional response. In Stat1, Ser727 is critical for association with nuclear proteins that participate in transcriptional activation.47,74 In Ramos and 32D cells, IL-4 stimulated phosphorylation of Stat6 on tyrosine and on serine residues, whereas no phosphorylation was observed on threonine residues. This phosphorylation pattern is similar to what has been observed for the cytokine-induced phosphorylation of Stat1, Stat3, and Stat5.47,50-53 Phosphorylation of Stat3 and Stat5a occurs at least on 2 serine residues, whereas Stat1 is primarily phosphorylated on a single Ser727 site.47 50-53 Tryptic phosphopeptide maps demonstrated the presence of 2 constitutively phosphorylated peptides in Stat6, and IL-4 stimulated prominent phosphorylation of 1 peptide and low levels of phosphorylation of 5 additional peptides.
The identity of the serine kinases responsible for Stat phosphorylation has remained enigmatic, but it now appears that several serine kinase pathways participate in the regulation of Stat-mediated transcriptional responses. Analysis of the kinetics of Stat1 and Stat5 serine phosphorylation have led to the conclusion that serine phosphorylation is a downstream event from Jak activation and tyrosine phosphorylation of Stat and that it occurs in the cytoplasm.50,75 However, lipopolysaccharide stimulation of macrophages also induces Stat1 Ser727 phosphorylation, apparently without Jak involvement, emphasizing the complexity of the signaling pathways involved in Stat regulation.76 As an example of this heterogeneity, IL-2-induced Stat3 Ser727 phosphorylation is insensitive to H7 treatment, but IL-2 also induces H7-sensitive phosphorylation, which affects the DNA binding of Stat3.48In T cells, IL-2 stimulates serine phosphorylation of Stat5, which is inhibited by H7 as measured by a change in its electrophoretic mobility.49 In contrast, prolactin-induced serine phosphorylation of Stat5a (Ser725) and Stat5b (Ser730) was recently found to be insensitive to H7 treatment.53 Thus, the regulation of serine phosphorylation of Stats may involve receptor- or cell type-specific components. Much attention has been centered on the role of ERK kinases in Stat phosphorylation. ERK kinases are responsible for Stat3 Ser727 phosphorylation in growth factor receptor signaling and for the basal phosphorylation of Stat5a,53,64but distinct serine kinases mediate the phosphorylation of Stats in other cases.50,75 76 This also appears to be true for the phosphorylation of Stat6 because, in most cell types, IL-4 is unable to activate the Ras/Raf/MAPK/ERK pathway. In addition, the carboxy-terminus of Stat6 lacks the consensus ERK kinase phosphorylation site (PXS/TP). Our results show that the kinase responsible for serine phosphorylation of Stat6 is H7 insensitive and that this phosphorylation is independent of the IRS pathway. IRS proteins mediate the activation of PI 3-K. Therefore, the PI 3-K-regulated kinases, such as Akt, PKC, and p70 S6, are not likely candidates for mediating Stat6 serine phosphorylation. Thus, the IL-4-stimulated serine phosphorylation of Stat6 may involve currently undiscovered signaling pathways.
The functional role of Stat6 serine phosphorylation cannot be evaluated based on current information. Such evaluation must await the characterization of the serine phosphorylation sites in Stat6. However, H7 treatment did not affect this phosphorylation, and thereby Stat6 itself does not appear to be the target for the observed transcriptional effect of H7. Instead, we found that IL-4 induced the phosphorylation of RNA polymerase II in Ramos cells, which was inhibited by H7. This is in accordance with a recent study59 showing that nerve growth factor-stimulated immediate early gene induction and RNA polymerase II phosphorylation were similarly blocked by H7. The CTD of RNA polymerase II is highly phosphorylated during transcriptional elongation, and phosphorylation of the CTD of RNA polymerase II is considered a regulatory event in the initiation of transcription.72 The CTD is phosphorylated by TFIIH-associated CAK and cdk kinases. Based on previous in vitro studies,69-71 inhibition of these kinases by H7 is a likely mechanism for the observed abrogation of RNA polymerase II phosphorylation in Ramos cells. Also, another kinase inhibitor, H8, which inhibits cdks and RNA polymerase II phosphorylation in vitro,70 was found to abrogate IL-4-induced gene transcription (data not shown). Although phosphorylation of RNA polymerase II is a target for the inhibitory effect of H7, we cannot exclude the possibility that H7-sensitive kinases could regulate other phosphorylation events in Stat6-dependent transcriptional activation. Identification of such phosphorylation events could provide further insight into the regulation of transcription in cytokine signaling.
Acknowledgments
The authors thank Paula Kosonen for technical assistance, Drs B. Groner, M. Heim, J. Ihle, and T. Wood for kindly providing reagents, and Dr Leena Valmu for helpful advice with phospho-amino acid analysis.
Supported by the Academy of Finland, the Sigrid Juselius Foundation, the Medical Research Fund of Tampere University Hospital, and the Emil Aaltonen Foundation.
Reprints:Olli Silvennoinen, Laboratory of Molecular Immunology, Institute of Medical Technology, University of Tampere, P.O. Box 607, FIN-33101 Tampere, Finland; e-mail:olli.silvennoinen@uta.fi.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.