The 2-phenylaminopyrimidine derivative STI571 has been shown to selectively inhibit the tyrosine kinase domain of the oncogenicbcr/abl fusion protein. The activity of this inhibitor has been demonstrated so far both in vitro with bcr/abl expressing cells derived from leukemic patients, and in vivo on nude mice inoculated with bcr/abl positive cells. Yet, no information is available on whether leukemic cells can develop resistance to bcr/ablinhibition. The human bcr/abl expressing cell line LAMA84 was cultured with increasing concentrations of STI571. After approximately 6 months of culture, a new cell line was obtained and named LAMA84R. This newly selected cell line showed an IC50 for the STI571 (1.0 μM) 10-fold higher than the IC50 (0.1 μM) of the parental sensitive cell line. Treatment with STI571 was shown to increase both the early and late apoptotic fraction in LAMA84 but not in LAMA84R. The induction of apoptosis in LAMA84 was associated with the activation of caspase 3–like activity, which did not develop in the resistant LAMA84R cell line. LAMA84R cells showed increased levels of bcr/abl protein and mRNA when compared to LAMA84 cells. FISH analysis with BCR- and ABL-specific probes in LAMA84R cells revealed the presence of a marker chromosome containing approximately 13 to 14 copies of the BCR/ABL gene. Thus, overexpression of the Bcr/Abl protein mediated through gene amplification is associated with and probably determines resistance of human leukemic cells to STI571 in vitro.
The blockade of Bcr/Abl kinase activity represents a rational strategy to treat leukemias caused by this oncogenic fusion protein. Expression of the bcr/abl oncogenic product has been found in virtually all patients with chronic myelogenous leukemia (CML) and in 30% to 50% of adult patients with acute lymphoblastic leukemia (ALL). bcr/abl bears a causal relationship with both CML and ALL1,2; in addition, increased bcr/abl levels have been associated with disease progression.3 The 2-phenylaminopyrimidine derivative STI571 is a recently designed tyrosine kinase inhibitor4 that inhibits the ATP binding in the kinase domain of both abl andbcr/abl in a competitive fashion.5 This compound is active both in vitro and in vivo against a variety of bcr/ablor v-abl transformed cells. In previous studies, we and others have demonstrated the selectivity of STI571 to inhibit the proliferative activity and the clonogenic potential of cell lines and fresh leukemic cells expressing the Bcr/Abl fusion protein.6,7 In addition we showed that the inhibition of Bcr/Abl kinase activity led to the development of apoptosis in leukemic cells. In particular, LAMA84, a human bcr/abl expressing cell line derived from a leukemic patient in blast crisis,8underwent apoptosis as early as 28 hours after treatment with STI571, while an incubation time of about 100 hours was needed in fresh samples obtained from CML patients in chronic phase.6
In vivo treatment of nude mice injected with human leukemic cells can achieve tumor eradication in 70% to 100% of treated mice, when schedules able to obtain continuous bcr/abl inhibition were used.9 These data constitute a significant promise for better treatments in patients with leukemias caused bybcr/abl,10 and initial clinical studies are already in progress.
While in vivo data indicate that the continuous presence of the STI571 drug in effective concentrations is needed to eradicate tumors in nude mice derived from bcr/abl expressing cells, some leukemia relapse can occur, depending on the initial tumor load.9 In this study, we selectively generated a bcr/abl positive cell line derived from LAMA84 and resistant to the effects of STI571, designated as LAMA84R. Furthermore, the molecular and cellular mechanisms that lead to such a resistance were investigated. We present evidence that the resistant, newly generated cell line contained a higher level ofbcr/abl expression and phosphorylation, when compared to the parental, sensitive cell line. Finally, the observed bcr/abloverexpression was found to be mediated through gene amplification.
Materials and methods
STI571
The 2-phenylaminopyrimidine derivative STI571 (molecular weight 590 d) was developed and kindly provided by Novartis (Basel, Switzerland). The stock solutions of this compound were prepared at 1 and at 10 mmol/L with distilled water, and filtered and stored at −20°C.
Cell lines
The erythromegakaryocytic cell line LAMA84, derived from the peripheral blood of a blast crisis CML patient,8 was maintained in RPMI 1640 (Bio Whittaker Europe, Verviers, Belgium) supplemented with 10% fetal calf serum (FCS). The LAMA84R cell line was maintained under the same culture conditions.
Induction of resistance in LAMA84
LAMA84 cells were initially cultured in the presence of 0.05 μM STI571. The concentration of the STI571 in the culture medium was subsequently increased by 0.1 μM every 3 to 4 weeks. After 4 months of culture, a LAMA84 subline growing at 0.6 μM STI571 was isolated and designated LAMA84R. Further culture showed that this cell line can grow in the presence of STI571 concentrations up to 1 μM (data not shown), whereas higher concentrations proved to be toxic for the cells.
Proliferation assay
The assay was performed using a standard 3H-thymidine (3HTdR) incorporation assay as described.9Briefly, 104 cells, from LAMA84 and LAMA84R, were seeded in 6 replicates in the presence of various concentrations of STI571 ranging from 0.1 to 10 μM, in round-bottomed 96-microtiter well plates (Costar, Cambridge, MA). After 60 hours at 37°C, 20 μL of RPMI/10% FCS containing 3HTdR (1 μCi/well) were added to each well. After an additional 18 hours, the cells were harvested and transferred to a filter (spot-on filtermat, Wallac, Turku, Finland).3HTdR uptake was determined by a 1205 betaplate liquid scintillation counter (Wallac).
Determination of apoptosis by FACS analysis
Quantitative determination and differentiation of viable, early, and late apoptotic cells were carried out using the Annexin V/Propidium iodide binding technique, as previously described.11 Briefly, 0.5 × 106 cells were incubated in the presence of 0.6 μM STI571. Controls were cultured in the absence of the drug. After 28 hours, samples were washed in phosphate-buffered saline (PBS) and kept for 15 minutes in an Annexin V/Propidium Iodide mixture prior to the analysis at the FACScan. Compensation for double fluorescence was performed by staining the samples either with Annexin V or with Propidium Iodide only.6
Caspase 3–like activity
Acetyl-Asp-Glu-Val-Asp-(amino-4-methylcoumarin) (DEVD-amc) hydrolytic activity was analyzed as previously described.12106 LAMA84 and LAMA84R cells were treated with 0.6 μM STI571 at various time points (7, 17, and 23 hours). Controls for each cell line were incubated in the absence of STI571. Cells were washed in PBS and lysed in 100 μL of lysis buffer, containing 10 mmol/L Hepes pH 7.4, 0.1% CHAPS, 2 mmol/L EDTA, and 2 mmol/L Dithiothreytol (DTT). Protein content was determined by the Bradford method and volumes equivalent to 3 μg of protein were incubated in 500 μL reaction buffer containing 20 mmol/L Hepes buffer pH 7.4, 10% glycerol, 2 mmol/L Dithiothreytol, and 20 μM DEVD-amc (Peptide Co, Kyoto, Japan) at 37°C for 2 hours. The reaction was stopped by the addition of 500 μL of reaction buffer without DEVD-amc. Samples were read immediately on a spectrophotofluorimeter, at an excitation wavelength of 380 nm and an emission wavelength of 460 nm.12
Western blot analysis
Immunoblotting was performed as described before.6 In general 106 LAMA84 and LAMA84R cells, the latter deprived overnight of STI571, were respectively incubated in a 24-well plate (Costar) at 37°C with various concentrations of STI571, ranging from 0.6 to 10 μM. Controls were incubated in the absence of the inhibitor. After 2 hours, cells were washed twice with cold PBS and subsequently lysed in 200 μL of 1× Laemmli's buffer (50 mmol/L Tris-HCl pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, 5% β-mercaptoethanol). Cell lysates, corresponding to 150,000 cells, were boiled at 95°C for 10 minutes, sonicated for 1 minute, and analyzed by SDS Electrophoresis on 7.5% polyacrylamide gels. Endogenous bcr-abl, tyrosine-phosphorylated bcr-abl,and the endogenous actin were detected with the corresponding mouse monoclonal antibody or rabbit antiserum and then visualized by enhanced chemioluminescence detection (ECL) (Amersham Corp Cleveland, OH), using horseradish peroxidase-linked goat anti-mouse or anti-rabbit immunoglobulin G as the secondary antibody (Amersham Corp.). The monoclonal anti-abl antibody (clone Ab-3) was purchased from Calbiochem. The monoclonal anti-phospho tyrosine antibody (clone 4G10) was purchased from Upstate Biotechnology. Rabbit polyclonal anti-actin was purchased from Sigma.
Densitometric analysis was performed with an Eagle Eye II Photodensitometer (Stratagene), and the intensities of bcr-abland tyrosine-phosphorylated bcr-abl bands were normalized against the actin expression level.
Bacterial expression of GST fusion proteins
The BL 21 Lys strain of Escherichia coli was transformed by electroporation with the vector PGEX-4T3 alone or encoding the CH1 domain of Shc.13 The transformed bacteria were cultured overnight at 37°C and then diluted 1:20 in 500 mL of Luria-Bertani medium. When the optical density reached 0.5, bacteria were incubated with 1 mmol/L isopropyl-[beta]-thiogalactopyranoside (IPTG) for 3 hours at 37°C. Cells were collected by centrifugation at 3000g for 30 minutes and resuspended in a lysis buffer containing 10 mL PBS, 1% Triton X-100, 1 mmol/L EDTA, 2 μg/mL aprotinin, 2 μg/mL leupeptin, and 1 mmol/L phenylmethylsulfonyl fluoride (PMSF). The cell suspension was sonicated and cellular debris removed by centrifugation at 10 000g for 10 minutes. The supernatant was mixed with 300 μL of glutathione-agarose beads (Pharmacia Biotech) and incubated for 1 hour at 4°C with constant rotation. The glutathione-bound protein was recovered by centrifugation at 3000g for 5 minutes and the pellet washed 3 times in a buffer containing 1× of PBS, 1% Triton X-100, 1 mmol/L EDTA, 2 μg/mL aprotinin, 2 μg/mL leupeptin, and 1 mmol/L PMSF; and 2 times in a solution containing 1× PBS, 2 μg/mL aprotinin, 2 μg/mL leupeptin, and 1 mmol/L PMSF. Finally, purified fusion proteins were eluted in a buffer containing 50 mmol/L Tris, 10 mmol/L glutathione, 2 μg/mL aprotinin, 2 μ/mL leupeptin, and 1 mmol/L PMSF.
Kinase assay
LAMA84 and LAMA84R cells were washed with cold PBS and lysed at 4°C in a buffer containing 25 mmol/L HEPES (pH 7.5), 0.3 mol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.1% SDS, 0.5 mmol/L DTT, 20 mmol/L β-glycerophosphate, 0.1 mmol/L Vanadate, 1% Triton X-100, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 0.5% sodium dehoxycholate, 2 μg/mL leupeptin, 2 μg/mL aprotinin. Cleared lysates were immunoprecipitated at 4°C for 2 hours with the polyclonal anti-abl, clone 24-11 (Santa Cruz Biotechnology). Immunocomplexes were recovered with the aid of Gamma-Bind Sepharose beads (Pharmacia) and washed 3 times with PBS containing 1% Nonidet P-40 and 1 mmol/L sodium orthovanadate, once with 100 mmol/L Tris (pH 7.5), 0.5 mol/L LiCl, and once with kinase reaction buffer (25 mmol/L Hepes [pH 7.5], 10 mmol/L MgCl2, 10 mmol/L MnCl2). bcr/abl activity was determined by resuspension in 30 μl of kinase reaction buffer containing 10 μCi (4500 Ci/mmol) of γ-32P] ATP per reaction, 20 μM of cold ATP, 2 mM of Dithiothreytol and 4 μg of GST (control) or GST-CH1-Shc as substrate, and 0 and 3 μM of STI571. After incubation at 30°C for 30 minutes, reactions were terminated by the addition of 15 μL of 5× Laemmli's buffer. Samples were heated at 95°C for 5 minutes, analyzed by SDS electrophoresis on 10% polyacrylamide gels, and visualized by autoradiography.
Sequencing of the bcr/abl ATP-binding domain amplified from LAMA84 and LAMA84R cell lines
The LAMA84 and LAMA84R total RNA was extracted using the Ultraspec-II RNA isolation system (Biotecx). Of the total RNA, 5 μg was subjected to RT-PCR using Oligo dT primers. Two primers, corresponding to the nucleotides 2638-2659 of the bcr coding sequence and to the nucleotides 1033-1053 of the Abl coding sequence, were synthesized for the polymerase chain reaction (PCR).
The amplified fragments were gel-purified and both strands were sequenced using an automated sequencer (ABI Prism 377, Perkin Elmer). The sequences were analyzed using the Sequencing Analysis 3.3 program and the alignments obtained with the ExPASy server (Swiss Institute of Bioinformatics Geneva, Switzerland).
Northern blot analysis
Northern blotting was carried out as described elsewhere.14 Total RNAs from LAMA84 and LAMA84R were extracted using the Ultraspec-II RNA isolation system (Biotecx). For each sample, 25 μg of RNA were separated on a 2% agarose gel. Transfer to a nylon filter (Biotrans, ICN, Costa Mesa, CA) was achieved by standard capillary transfer. A variety of breakpoints both in the ABL and in the BCR genes have been described.15 In LAMA84, the b3a2 junction is present involving breakpoints between BCR exons 3 and 4 and between ABL exons 1 and 2.15 A 40-mer oligonucleotide covering the BCR/ABL b3a2 junction was synthesized (5′TGG ATT TAA GCA GAG TTC AAA AGC CCT TCA CGC GCC AGT A 3′) with an 8-mer as primer (5′TACTGGCC3′). Synthesis of32P-labeled probes was achieved by extension of the 8-mer hybridized to the 40-mer in 10 μL of a primer extension mixture, containing 6 mmol/L Tris-HCl (pH 7.5); 6 mmol/L MgCl2; 6 mmol/L β-mercaptoethanol; 50 mmol/L NaCl; 30 μCi (α-32P) dATP; 100 mmol/L each of dGTP, dCTP, and dTTP; and 5 U of Klenow polymerase.
The murine β-actin cDNA was random primed (DECAprime II-Ambion) and used to rehybridize the same filter, as previously described.15 This probe was a kind gift from Dr Tommaso A. Dragani and is described elsewhere.16
Filters were extensively washed in 0.5% SSC (75 mmol/L NaCl, 7.5 mmol/L sodium citrate) at 60°C and exposed to HyperFilm (Amersham).
FISH analysis
Harvested cell suspensions were dropped onto slides according to standard laboratory techniques and slides were submitted to dual-color, dual target FISH. Cells were washed 1 minute with 70% acetic acid, dehydrated, and denatured in 70% formamide/2× SSC (300 mmol/L NaCl, 30 mmol/L sodium citrate) at 72°C for 2.5 minutes. The LSIbcr/abl translocation probe (Vysis Inc, Downers Grove, IL) was used. The probe was denatured and applied to selected areas of each slide. These areas were covered with glass coverslips and sealed with rubber cement. Hybridization was allowed to occur for 20 hours at 37°C. Posthybridization washes were performed at 46°C with 50% formamide/2× SSC (3 washes of 6 minutes), 2× SSC, and 2× SSC/0.1% NP40 (1 wash of 6 minutes). After rinsing in PBS, slides were stained with DAPI/DABCO. Analyses were performed on an Olympus B × 60 fluorescence microscope using single interference filter sets for red (Texas red), green (FITC), and blue (DAPI), and a dual-color (red/green) filter. The LSI bcr/abl translocation probe was built in that particular configuration so that the bcr DNA (DNA segment proximal to the 22q11.2 breakpoint) was labeled with SpectrumGreen and identified by a green fluorescent signal, and theabl DNA (DNA segment distal to the 9q34 breakpoint) was labeled with SpectrumOrange and identified as a red signal. In a cell without the t9;22 (q34;q11), 2 red and 2 green signals will be observed. In a cell carrying the t9;22 (q34;q11), 1 red signal (representing the abl gene), 1 green signal (representing the bcr gene), and 1 fused red/green signal (representing the chimeric BCR/ABL expected in the derivative chromosome 22, the Ph chromosome) will be observed. Samples were analyzed using QUIPS XL Genetics System (Vysis Inc) and IPLab (Signal Analytics Corporation, Vienna, VA) softwares, to obtain a quantitative analysis of fluorescence intensity.
MDR1 gene expression
Highly purified total RNA from approximately 107 cells was obtained with the Ultraspec-II RNA isolation system (Biotecx). Reverse transcription was carried out using 5 μM of Random Hexamers (Boehringer-Mannheim, Germany) in a 20 μL mixture containing 1 μg of RNA and 200 U of reverse transcriptase (GIBCO BRL). PCR was performed using 2 primers (sense: 5′-ATG TTG AGC CGG GCA GTG TGC-3′; antisense: 5′-CTG AAG AGC TGT CTG GGC TGT-3′) that amplified a 220-bp (base pair) fragment within the Multi Drug Resistant 1 (MDR1) gene and, as internal control, 2 specific primers for the β-actin gene (sense: 5′-ARG GAT GAT GAT ATC GCC GCG-3′; antisense: 5′-AAA GAA CAC GGC TAA GTG TGC-3′), which amplified a 500-bp fragment. The β-actin primers were added after the 14th cycle of the PCR.
Cell survival after exposure to doxorubicin
Proliferative cellular resistance to the doxorubicin was detected by cell count. LAMA84 and LAMA84R cells (107) and HL-60 cells (5 × 106) were seeded at various concentrations of doxorubicin ranging from 8 × 10−3 to 0.5 μg/mL. After 72 hours, the cells were collected and counted by an automatic cell counter (ZBI, Coulter Contron, Milan, Italy).
Results
Proliferative activity
The proliferative activity of LAMA84 and LAMA84R cells, incubated with increasing concentration of STI571 for 60 hours, was determined.
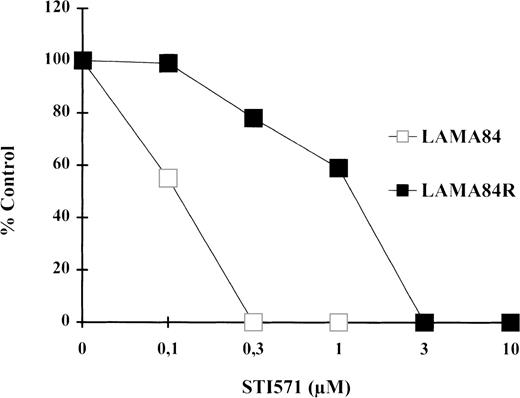
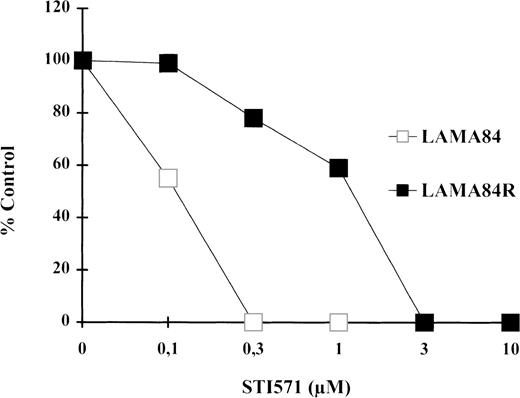
As depicted in Figure 1, STI571 treatment caused a dramatic inhibition of the proliferative rate of LAMA84 and of LAMA84R to a lesser extent. Our experiments showed that the IC50 of the inhibitory effect of STI571 on the proliferation of the LAMA84R was 10 times higher (1 μM) than the value calculated in the sensitive, parental cell line (0.1 μM). Moreover, further experiments showed that LAMA84R cells, cultured in the absence of STI571 for 4 weeks, did not present a reversion of the resistant phenotype (data not shown). Our data, taken together, let us conclude that in the LAMA84 model it is possible to select leukemic cells that present a resistance to the STI571 inhibitory effect of at least 10-fold in respect to the sensitive parental cells.
Proliferative activity as determined by 3HTdR uptake assay.
104 cells from LAMA84 and LAMA84R were incubated in 6 replicates in the presence of various concentrations of STI571 ranging from 0.1 to 10 μM. The proliferative assay was performed after 78 hours as described in “Materials and Methods.” The IC50 of the parental line LAMA84 (empty squares) and of the resistant line LAMA84R (black squares) was reached at 0.1 μM and 1 μM, respectively. The total counts per minute (cpm) of untreated samples of LAMA84 and LAMA84R were 161 275 ± 9276 (SD) cpm and 182 961 ± 11 552 (SD) cpm, respectively.
Proliferative activity as determined by 3HTdR uptake assay.
104 cells from LAMA84 and LAMA84R were incubated in 6 replicates in the presence of various concentrations of STI571 ranging from 0.1 to 10 μM. The proliferative assay was performed after 78 hours as described in “Materials and Methods.” The IC50 of the parental line LAMA84 (empty squares) and of the resistant line LAMA84R (black squares) was reached at 0.1 μM and 1 μM, respectively. The total counts per minute (cpm) of untreated samples of LAMA84 and LAMA84R were 161 275 ± 9276 (SD) cpm and 182 961 ± 11 552 (SD) cpm, respectively.
Induction of apoptosis
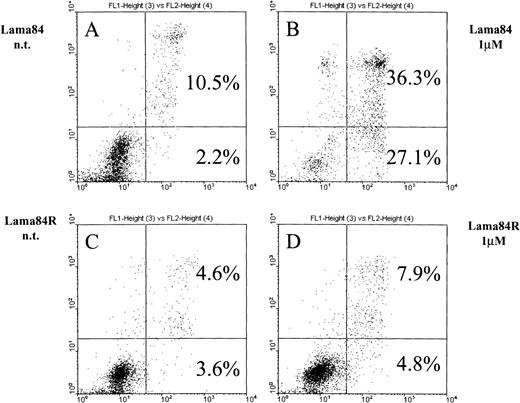
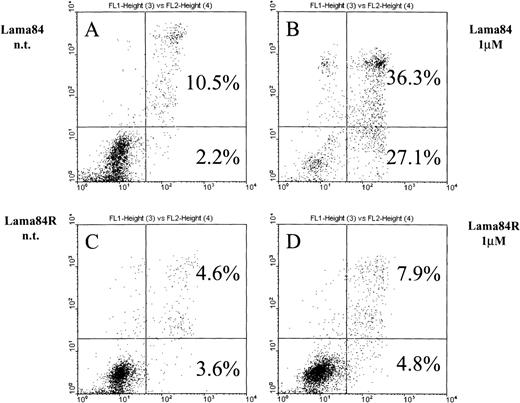
As previously shown,6 STI571 commits bcr/ablpositive cells to apoptosis. Figure 2 shows the induction of early apoptosis in LAMA84 but not in LAMA84R on incubation with the inhibitor for 28 hours. At this point, the early apoptotic fraction of the parental cell line LAMA84 detected by FACS analysis contained 27.1% of the cells compared to 4.8% detected in the LAMA84R cells. The combined proportion of early and late apoptotic LAMA84 cells (63.4%) was 5 times higher than the one calculated in the resistant cell line (12.7%). In fact, neither the early apoptotic fraction (4.8%) nor the late one (7.9%) of the treated LAMA84R cells showed a significant increase over the nontreated samples (3.6% and 4.6%, respectively). Taken together, these findings strongly suggest that LAMA84R cells showed resistance to apoptosis induced by concentrations of STI571 active on LAMA84 cells.
Induction of apoptosis in LAMA84 and LAMA84R cells.
Cells (0.5 × 106) were incubated with 1 μM STI571 and cultured over 28 hours. After the addition of annexin V and propidium iodide, samples were immediately submitted to double fluorescence analysis. In the sensitive LAMA84 line, STI571 committed 27.1% of cells to early apoptosis (2B) while in the nontreated sample (A), only 2.2% were found in the respective fraction. In addition, the late apoptotic fraction in the sensitive sample (36.3%) (B) also exceeded the nontreated control (10.5%) (A). In the resistant LAMA84R cells, both the early (4.8%) and the late (7.9%) apoptotic fraction of the treated sample did not significantly exceed the nontreated sample (3.6% and 4.6%, respectively) (C) and (D).
Induction of apoptosis in LAMA84 and LAMA84R cells.
Cells (0.5 × 106) were incubated with 1 μM STI571 and cultured over 28 hours. After the addition of annexin V and propidium iodide, samples were immediately submitted to double fluorescence analysis. In the sensitive LAMA84 line, STI571 committed 27.1% of cells to early apoptosis (2B) while in the nontreated sample (A), only 2.2% were found in the respective fraction. In addition, the late apoptotic fraction in the sensitive sample (36.3%) (B) also exceeded the nontreated control (10.5%) (A). In the resistant LAMA84R cells, both the early (4.8%) and the late (7.9%) apoptotic fraction of the treated sample did not significantly exceed the nontreated sample (3.6% and 4.6%, respectively) (C) and (D).
Caspase 3 like activity
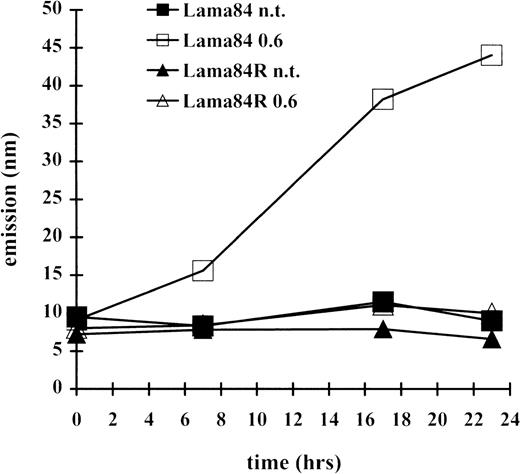
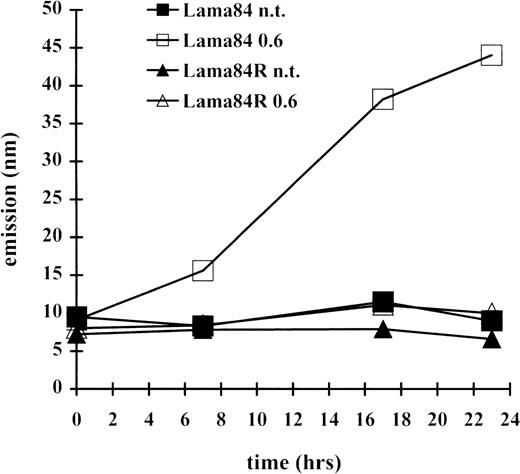
Since the induction of apoptosis frequently involves the activation of the caspase family of proteases, the induction of a caspase 3–like activity was measured at various times. As indicated in Figure3, incubation of LAMA84 cells with 0.6 μM induced caspase 3–like activity, already detectable after 7 hours of treatment, and with further increase at the 18- and 23-hour time points. In contrast, the caspase 3–like activity detected in the LAMA84R samples, at the various times examined, was comparable to that of the parental sensitive and the resistant untreated cell lines. Altogether, these data strongly indicate that STI571 treatment of sensitive bcr/abl-transformed cells leads to caspase 3–like activation and subsequently to the induction of apoptosis. However, this pathway appears to be inactive in the resistant cell line, under the same treatment conditions.
Induction of caspase 3–like activity in LAMA84 and LAMA84R.
The cleavage of the caspase 3 substrate DEVD-amc was evaluated in a fluorimetric assay, as described in “Materials and Methods.” Caspase 3–like activity was detected by reading samples spectrophotofluorimetrically at an excitation wavelength of 380 nm and an emission wavelength of 460 nm.
Induction of caspase 3–like activity in LAMA84 and LAMA84R.
The cleavage of the caspase 3 substrate DEVD-amc was evaluated in a fluorimetric assay, as described in “Materials and Methods.” Caspase 3–like activity was detected by reading samples spectrophotofluorimetrically at an excitation wavelength of 380 nm and an emission wavelength of 460 nm.
Expression and phosphorylation of the bcr/abl protein
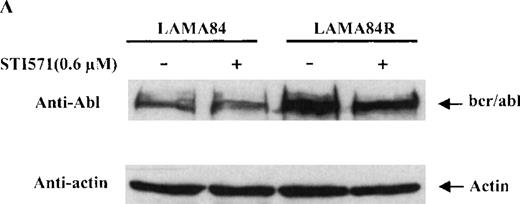
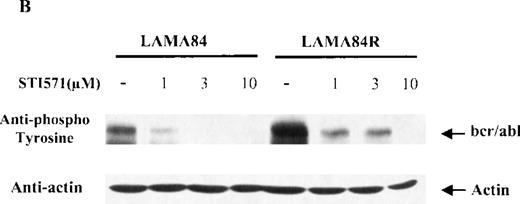
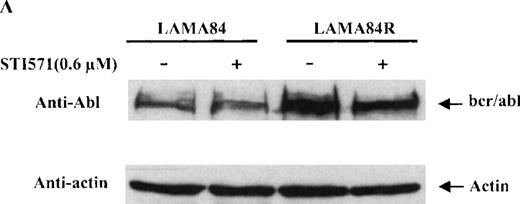
To determine the expression level of the bcr/abl fusion protein in the LAMA84 and LAMA84R cells, equal amounts of total cell lysates from the 2 leukemic cell lines were examined by immunoblot analysis using a specific anti-abl antiserum. The content of bcr/abl in the LAMA84R cell line was found to be approximately four-fold higher when compared to that of the parental cell line and was not influenced by the presence of STI571 (Figure 4A). The variation of the bcr/abl expression level in the 2 cell lines examined was not due to a difference in the amount of protein loaded, since similar levels of actin were detected in the same cell lysates (Figure 4a, lower panel).
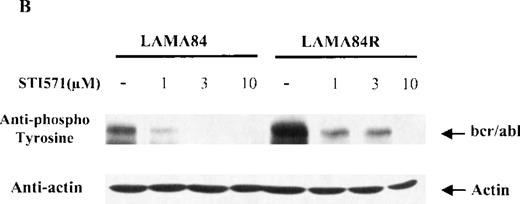
Expression, phosphorylation status, and enzymatic activity of bcr/abl in LAMA84 and LAMA84R cells.
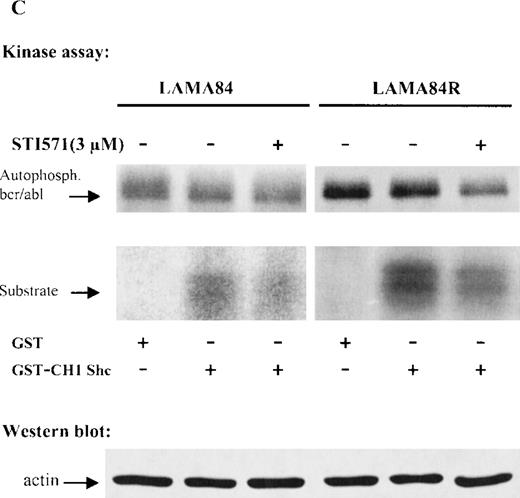
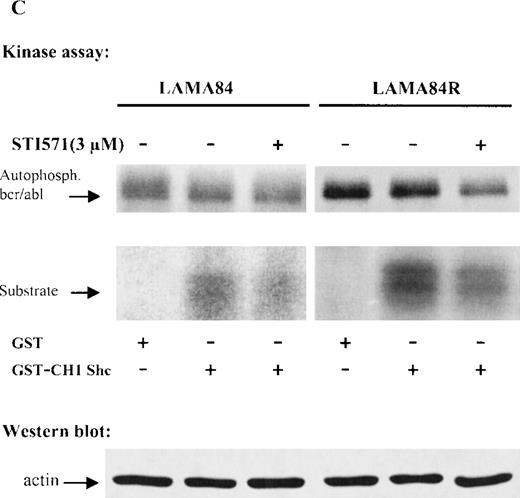
(A) The expression of endogenous bcr/abl and actin in extracts of LAMA84 and LAMA84R cells treated with 0.6 μM of STI571 or left untreated (control) was determined by Western blot analysis using the monoclonal anti-abl and the polyclonal anti-actin antibodies, as described in “Materials and Methods.” (B) The bcr/ablphosphorylation status in LAMA84 and LAMA84R cell lysates treated with 1, 3, and 10 μM of STI571 or left untreated (control) was examined by immunoblotting using the monoclonal anti-phospho tyrosine antibody, as mentioned in “Materials and Methods.” The level of actin expression in the same lysates was detected as described above. (C) Immunocomplex kinase assay of bcr/abl in LAMA84 and LAMA84R. Cell lysates from both cell lines were immunoprecipitated with an anti-Abl antibody. The immunoprecipitates were left untreated (control) or treated with 3 μM STI571 and then subjected to an in vitro kinase assay as described under “Materials and Methods.” The upper panel shows the bcr/abl autophosphorylation level. The bcr/abl kinase activity was determined as GST (negative control) and GST-CH1 Shc phosphorylation (middle panel). Total cell lysates were subjected to Western blot analysis using anti-actin antibody to demonstrate equal protein levels in the samples analyzed (lower panel).
Expression, phosphorylation status, and enzymatic activity of bcr/abl in LAMA84 and LAMA84R cells.
(A) The expression of endogenous bcr/abl and actin in extracts of LAMA84 and LAMA84R cells treated with 0.6 μM of STI571 or left untreated (control) was determined by Western blot analysis using the monoclonal anti-abl and the polyclonal anti-actin antibodies, as described in “Materials and Methods.” (B) The bcr/ablphosphorylation status in LAMA84 and LAMA84R cell lysates treated with 1, 3, and 10 μM of STI571 or left untreated (control) was examined by immunoblotting using the monoclonal anti-phospho tyrosine antibody, as mentioned in “Materials and Methods.” The level of actin expression in the same lysates was detected as described above. (C) Immunocomplex kinase assay of bcr/abl in LAMA84 and LAMA84R. Cell lysates from both cell lines were immunoprecipitated with an anti-Abl antibody. The immunoprecipitates were left untreated (control) or treated with 3 μM STI571 and then subjected to an in vitro kinase assay as described under “Materials and Methods.” The upper panel shows the bcr/abl autophosphorylation level. The bcr/abl kinase activity was determined as GST (negative control) and GST-CH1 Shc phosphorylation (middle panel). Total cell lysates were subjected to Western blot analysis using anti-actin antibody to demonstrate equal protein levels in the samples analyzed (lower panel).
Since the target of STI571 is represented by the kinase activity of thebcr/abl fusion protein, which is constitutively tyrosine phosphorylated in leukemic cells, we sought to determine the dose-response effect of STI571 on the bcr/abl phosphorylation levels in the LAMA84 and LAMA84R cells. Equal amounts of total cell lysates from the 2 leukemic cell lines, incubated with 1, 3, and 10 μM of STI571 for 2 hours, were examined by immunoblot analysis using an antiphosphotyrosine antiserum. As control, lysates from the 2 cell lines were left untreated. As shown in Figure 4B, the incubation of LAMA84 and LAMA84R cells with increasing concentrations of STI571 produced a reduction of bcr/abl tyrosine phosphorylation in both lines. Although the amount of phosphorylated bcr/abl was clearly higher in the untreated LAMA84R when compared to that of the parental cell line (due to the increased bcr/abl content), the incubation of the cells with 10 μM of STI571 produced an almost complete inhibition of bcr/abl tyrosine phosphorylation in both cell lines. Furthermore, to measure the protein expression levels in the same lysates analyzed, the membrane was probed with the anti-actin antibody. As shown in the lower panel of Figure 4B, similar levels of actin were observed in all the samples analyzed.
These data, taken together, suggest that LAMA84R cells contain higher levels of bcr/abl compared to the parental sensitive LAMA84 cells, while no intrinsic resistance to STI571 was evident, differing from other STI571-resistant cell lines.17
Determination of the enzymatic activity of bcr/abl isolated from LAMA84 and LAMA84R
To study whether the kinase activity of bcr/abl was sensitive to inhibition by STI571, immunoprecipitated bcr/ablfrom LAMA84 and LAMA84R was used as the source of activity. Thebcr/abl autophosphorylation and the phosphorylation of an exogenous substrate (CH1 domain of Shc) were evaluated. Figure 4C presents the result of a representative experiment. The level of activity was higher in LAMA84R lysates, according to the higherbcr/abl content of these cells. However, inhibition ofbcr/abl and Shc phosphorylation by STI571 (3 μM) could be observed in both lines. Differing from Western blot experiments (Figure4B), the inhibition of the kinase activity was never complete, even at 10 μM (not shown). However, it has to be considered, in the immunocomplex kinase assay, that very high, nonphysiological ATP concentrations (>20 μM) are present, which compete with STI571 for the ATP-binding pocket of bcr/abl. Thus, altogether, these results confirm the intrinsic sensitivity of the bcr/ablprotein derived from LAMA84R to STI571.
Sequencing of the LAMA84 and LAMA84R bcr/abl ATP binding domain
To determine whether a point mutation in the bcr/ablATP-binding domain was responsible for the resistance of the LAMA84R cells to the inhibitory effect of STI571 on the phosphorylation of the high endogenously expressed bcr/abl, sequencing of the cDNA portion corresponding to the ATP-binding region of the fusion protein was performed (Figure 5). Briefly, oligonucleotides, corresponding to the BCR2 exon (nucleotides 2638-2659 of the bcr coding sequence) and to an ABLregion situated carboxyl-terminal to the ATP-binding domain (nucleotides 1033-1053 of the c-abl coding sequence), were synthesized and used to amplify by RT-PCR, a 1,200-bp fragment within thebcr/abl cDNA from LAMA84 and LAMA84R. After purification, the fragment was subjected to automated sequencing in both strands, and the sequences encompassing the ATP-binding domain of the fusion product were analyzed. As shown in Figure 5, both sequences obtained from LAMA84 and LAMA84R were aligned and compared to the c-ablpredicted sequence (Gene Bank accession number: M14752). No mutations in the nucleotidic sequence of the bcr/abl ATP-interaction domain from LAMA84R were detected. These data led us to conclude that the LAMA84R-resistant phenotype might not be explained by a decrease of the STI571 binding affinity to the bcr/abl ATP-interaction domain.
Sequence alignment of the bcr/abl ATP-binding domain.
The predicted nucleotidic sequence of the ATP-binding domain ofbcr/abl (Gene Bank accession number: M14752) was aligned with the corresponding sequences obtained from the LAMA84 and LAMA84Rbcr/abl cDNAs. The ATP-binding domain is indicated in bold, and the numbers on the left refer to the position of the codon relative to the initiator ATG.
Sequence alignment of the bcr/abl ATP-binding domain.
The predicted nucleotidic sequence of the ATP-binding domain ofbcr/abl (Gene Bank accession number: M14752) was aligned with the corresponding sequences obtained from the LAMA84 and LAMA84Rbcr/abl cDNAs. The ATP-binding domain is indicated in bold, and the numbers on the left refer to the position of the codon relative to the initiator ATG.
Analysis of the BCR/ABL gene transcription in LAMA84 and LAMA84R
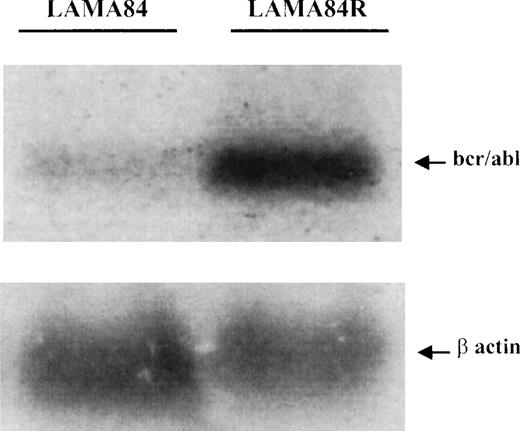
The increased level of the bcr/abl protein in the LAMA84R cells could be explained by an increased transcription of the fusion gene or a reduced degradation of the protein (posttranslational control). Therefore, to examine the expression of BCR/ABL at the mRNA level in the LAMA84 and LAMA84R cells, a specific oligonucleotide complementary to the b3-a2 sequence ofBCR/ABL14 was used as a probe to hybridize the total RNA extracted from the 2 cell lines. As shown in Figure6, the b3-a2 probe specifically detected a 8.5-kb transcript in both cell lines. LAMA84R cells clearly expressed higher amounts of bcr/abl mRNA. Densitometric analysis indicated that the resistant cell line contained a 4.6-fold higher level of transcript, compared to the parental, sensitive cell line. To further confirm that the same amount of RNAs was analyzed, the blot was rehybridized with a murine β-actin probe. Since similar bands were obtained, these results, taken together, supported the contention thatBCR/ABL was overtranscribed in the resistant LAMA84 cell line.
Determination of BCR/ABL mRNA in LAMA84 and LAMA84R cells.
Samples containing 25 μg of total RNA per lane, extracted from LAMA84 and LAMA84R, were fractionated and analyzed by Northern blotting. A highly specific 40mer, corresponding to a portion of the b3-a2 junction sequence of BCR/ABL,14 was used as a probe and detected an 85-Kb transcript (upper panel). The same membrane was rehybridized with a murine β-actin probe (lower panel).
Determination of BCR/ABL mRNA in LAMA84 and LAMA84R cells.
Samples containing 25 μg of total RNA per lane, extracted from LAMA84 and LAMA84R, were fractionated and analyzed by Northern blotting. A highly specific 40mer, corresponding to a portion of the b3-a2 junction sequence of BCR/ABL,14 was used as a probe and detected an 85-Kb transcript (upper panel). The same membrane was rehybridized with a murine β-actin probe (lower panel).
Genomic analysis of the BCR/ABL gene in LAMA84 and LAMA84R cells
To further study whether the overtranscription of theBCR/ABL gene in the LAMA84R cells was due to the presence of increased number of the fusion gene at the genomic level (gene amplification), the 2 cell lines were subjected to fluorescent in situ hybridization (FISH) analysis using 2 bcr- andabl-specific probes. The presence of more than 1 copy of the Philadelphia chromosome in LAMA84 was previously described8and was confirmed by our observations. Figure7 shows both interphase (7A, C) and metaphase (7B, D) images of LAMA84 and LAMA84R. The modal number in LAMA84 was 4 fused signals per cell (7B). The average number of fused signals in 216 interphase nuclei was 3.7 ± 1.3 and in 100 metaphase spreads was 4.1 ± 0.6. In LAMA84R, in addition to the above-mentioned single BCR/ABL signals, a cluster of fusion signals was observed in 98% of cells. This cluster appears in a single copy in 95% of nuclei, and in 2 copies in 3% of nuclei. When performing a comprehensive evaluation of the slides, we were able to detect a few metaphases in the specimen, and the amplified fused signal was assigned to a marker metacentric chromosome. This element seems to exclusively contain BCR/ABL sequences, since it appeared totally painted in red and green (Figure 7D, E). The single fused signals were mapped to a small acrocentric chromosome resembling the standard Philadelphia chromosome. The fluorescence intensity of bcr/ablsignals was evaluated using IPLab Spectrum software. Forty-eight individual signals and 22 marker elements were segmented and quantitatively analyzed. The sum of the pixel values within the segmented targets (total intensity) was obtained for each target and averaged for the individual fusion and for the marker. These procedures were performed separately for the red and the green signals. The mean intensity of the marker was then compared to the mean intensity of the individual fusions. For the green signals, the fluorescence in the marker was found to be 14 times more intense than in the individual fusion; for the red signals, the marker intensity was found to be 13.2 times higher than the individual fusion.
FISH analysis of LAMA84 and LAMA84R cell lines.
(A) Interphase nuclei from LAMA84 displaying 2, 3, and 5 copies of the fused BCR/ABL signal indicating the BCR/ABL chimeric gene. (B) Metaphase spreads in the same cell line showing the modal number of 4 copies of BCR/ABL per cell. In contrast to the single fused signals observed in LAMA84, the resistant LAMA84R line additionally displayed an amplified signal containing multiple copies of BCR/ABL. (C) Interphase nuclei from LAMA84R showing the amplified signal plus single copies of the fused signal. (D) A metaphase sample of LAMA84R shows a similar signal compatible with the amplified element, shown at higher magnification in (E).
FISH analysis of LAMA84 and LAMA84R cell lines.
(A) Interphase nuclei from LAMA84 displaying 2, 3, and 5 copies of the fused BCR/ABL signal indicating the BCR/ABL chimeric gene. (B) Metaphase spreads in the same cell line showing the modal number of 4 copies of BCR/ABL per cell. In contrast to the single fused signals observed in LAMA84, the resistant LAMA84R line additionally displayed an amplified signal containing multiple copies of BCR/ABL. (C) Interphase nuclei from LAMA84R showing the amplified signal plus single copies of the fused signal. (D) A metaphase sample of LAMA84R shows a similar signal compatible with the amplified element, shown at higher magnification in (E).
In conclusion, our findings suggest that LAMA84 contains multiple copies of the chimeric BCR/ABL gene, with the average of 4 per cell. By contrast, LAMA84R cells have been demonstrated to contain, in addition to single copies, approximately 14 copies of the chimeric gene in a specific marker chromosome, thus indicating the presence of further bcr/abl gene amplification in the resistant cell line.
Cross resistance to doxorubicin and expression of the MDR1 gene
Since increased bcr/abl expression was shown to be associated with resistance to various cytotoxic drugs,19 20the in vitro sensitivity of LAMA84 and LAMA84R to doxorubicin was studied. Cell survival of LAMA84 and LAMA84R cells after exposure to various doses of doxorubicin is presented in Figure8. LAMA84R cells showed a relative resistance to doxorubicin with an IC50 of 100 ± 9 (SE) ng/ml, while LAMA84 showed an IC50 of 20 ± 1 (SE) ng/ml. Remarkably, these data suggest that the resistant cell line presented 5-fold less sensitivity to the cytotoxic effect exerted by doxorubicin, when compared to the parental sensitive cell line.
Sensitivity of LAMA84 and LAMA84R to doxorubicin.
The graph shows the sensitivity of LAMA84 (diamonds) and LAMA84R (squares) to doxorubicin, calculated by cell count.
Sensitivity of LAMA84 and LAMA84R to doxorubicin.
The graph shows the sensitivity of LAMA84 (diamonds) and LAMA84R (squares) to doxorubicin, calculated by cell count.
To further investigate whether the overexpression of the Multi Drug Resistant 1 (MDR1) gene, a phenomenon often found in antracycline-resistant neoplastic cells, was responsible for the increased resistance to doxorubicin observed in the LAMA84R cells, the 2 experimental cell lines were assayed for the expression of theMDR1 gene. Reverse transcription and PCR of MDR1and β-actin were simultaneously undertaken to obtain a semiquantitative analysis of the MDR1 gene. In both cell lines, the intensity of the PCR product representing theMDR1 gene was compared to that of the β-actin gene, and the ratio obtained in the 2 lines were compared. In the parental LAMA84 cell line, the MDR1/β-actin ratio was 1.72, whereas in the resistant LAMA84R line this ratio was 1.26 (data not shown). Based on these data, an amplification of the MDR1 gene in the LAMA84R line was excluded, and doxorubicin-resistance was attributed tobcr/abl overexpression.
Discussion
The generation of drug resistance represents an important point in the development of new compounds. STI571 represents an innovative new drug for leukemia treatment. This molecule is in fact operationally specific for neoplasias caused by the oncogenic tyrosine kinasebcr/abl, and holds promise for minimal toxicity to normal cells when compared to now available cytotoxic drugs.5 Study of the possible development of resistance to this and similar compounds is rare at present. To the best of our knowledge, the data reported here represent the first example of a successful generation and selection ofbcr/abl positive cells (LAMA84) that show a significant and specific resistance to STI571 effects. Here, we show that this newly selected resistant cell line, designated LAMA84R, shows about 4-fold increased expression of bcr/abl at the protein level.
As already established, STI571 was shown to selectively inhibit theabl tyrosine phosphorylation, most likely by competitive binding in the ATP-binding pocket of the abl kinase domain.5 This observation prompted us to ask whether the inhibitory drug had the same effect on the bcr/abl tyrosine phosphorylation and kinase activity of the newly generated resistant cell line. Our data revealed that higher concentrations of STI571 were required to inhibit bcr/abl tyrosine phosphorylation and to reduce the kinase activity in the LAMA84R cells than in the parental cell line. Nevertheless, no point mutations within the ATP-binding site of the bcr/abl kinase domain, that might be responsible for the observed resistance, was found in the resistant cell line. In addition, both tyrosine-phosphorylation and kinase activity experiments show that STI571 concentrations active against LAMA84R were also able to inhibit the function of thebcr/abl protein in the same cells. Thus, our findings let us conclude that no apparent intrinsic resistance to STI571 appears to be present in the LAMA84R cell line.
Based on our previous results, which showed that STI571 commitsbcr/abl positive cells to apoptosis,6 we sought to compare the effect of the inhibitory drug on the induction of the caspase 3–like activity and the subsequent induction of the programmed cell death in both our experimental systems. Our data show that the resistant LAMA84R cell line presented resistance to apoptosis induced by concentrations of STI571 active on LAMA84 cells, and that no activation of the caspase 3 pathway, which was instead triggered in the sensitive cell line, was induced. Taken together, these findings provide additional evidence for a specific resistance to the STI571 effects present in the LAMA84R cell line.
Since a higher content of the bcr/abl protein in the resistant cell line was found, we sought to determine the origin of such overexpression. Our results clearly indicate that the bcr/ablmRNA is a 4.6-fold overtranscribed in the LAMA84R cells, compared to the sensitive cell line. This finding indicates that bcr/abltranscriptional activity is increased in LAMA84R.
As previously observed, the amplification of target genes is a frequently used mechanism to generate drug resistance in neoplastic cells.18 This observation prompted us to ask whetherBCR/ABL gene amplification developed in the resistant cell line. Thus, we used a FISH approach to investigate the gene amplification rate both in the sensitive and in the resistant LAMA84. We found that the LAMA84R cells contain more than 15 copies of the chimeric gene per cell. The ratio between the total number of copies of the BCR/ABL gene found in LAMA84R cells (14 + 4) and in LAMA84 (4), parallels the observed 4- to 5-fold increase in protein and mRNA levels.
It is known that bcr/abl delivers a strong “survival signal” that antagonizes many inducers of apoptosis, including cytotoxic drugs and radiation19 20; the higher IC50 for doxorubicin noted in LAMA84R cells is consistent with this hypothesis. LAMA84R cells did not show intrinsic resistance to the inhibition operated by STI571; rather, LAMA84R cells contain higher baseline levels of functional Bcr/Abl protein. It is therefore possible that at STI571 concentrations (0.1-1 μM) sufficient to drop the activity ofbcr/abl below the threshold needed for survival in sensitive LAMA84 cells, this limit is not reached in LAMA84R cells, where higher STI571 concentrations are needed. These results are compatible with a model in which bcr/abl needs to deliver a certain “amount” of signaling, to keep leukemic cells alive. In LAMA84 cells, the inhibition operated by STI571 at 0.1 to 0.6 μM is sufficient to decrease the amount of bcr/abl signaling below the level required for survival. In LAMA84R cells, higher baseline levels of bcr/abl activity are present; STI571 produces an inhibition of bcr/abl activity similar in percentage to LAMA84, but the total “amount” of residual signal is still sufficient for cell survival.
These data also contain important information on strategies to avoid the selection of resistant cells. In fact, selection of resistant cells occurred on several months of exposure to STI571, starting at a very low concentration. In addition, even when using this selection procedure, the induction of resistance was not simple and could not be achieved in all the cell lines tested. Therefore, clinical protocols should aim at quickly achieving STI571 concentrations higher than the IC50, and should avoid the exposure of the leukemic cells to marginally active STI571 concentrations.
The generation of a LAMA84 subline resistant to STI571 still represents an example of in vitro selection, and its relevance to resistance possibly developing in vivo21 remains to be investigated in clinical trials. It is known however that progression of CML to blast crisis is associated with bcr/abl overexpression, and that in some patients multiple copies of BCR/ABL can be observed during blast crisis (Gambacorti-Passerine, unpublished). It will be important to determine whether patients treated with STI571 will show increased amounts of phosphorylated bcr/abl associated with disease relapse. In our case, the presence of a marker chromosome containing several copies of the BCR/ABL gene facilitated the characterization of the phenotype observed; it is likely that different mechanisms should be operative in other lines generated in our laboratory.17 These data, coupled with the results of ongoing clinical trials, will be useful to select treatment schedules designed to maximize therapeutic effects and to avoid the selection of resistant subpopulations of leukemic cells.
Acknowledgments
We thank Enrico Garatini, MD, for helpful discussion and support.
This work was supported in part by the Italian Association for Cancer Research (AIRC 420.198.662); the Italian Research Council (95.00842,9600225.CT04); Istituto Superiore di Sanità (881A/10); BIOMED-2 grant BMH4-CT96-0848; EU-TMR grant BMH4-CT96-5006; and NCI P30 CA 46934.
P.L.C. and E.T. are equally contributing authors.
Reprints:Carlo Gambacorti, Department of Experimental Oncology, Istituto Nazionale Tumori, Via Venezian 1, 20133 Milan, Italy; e-mail: gambacorti@istitutotumori.mi.it.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.