Early biochemical studies defined 4 functional domains of the erythroid protein 4.1 (4.1R). From amino-terminal to carboxy-terminal, these are 30 kd, 16 kd, 10 kd, and 22/24 kd in size. Although the functional properties of both the 30-kd and the 10-kd domain have been demonstrated in red cells, no functional activities have been assigned to either the 16-kd or the 22/24-kd domain in these cells. We here describe new mutations in the sequence encoding the C-terminal 22/24-kd domain that are associated with hereditary elliptocytosis. An unusually mild phenotype observed in heterozygous and homozygous members of 1 family suggested heterogeneity in the pattern of expression of 4.1R deficiency. Using a variety of protein and messenger RNA (mRNA) quantification strategies, we showed that, regardless of the alteration in the C-terminal primary sequence, when the protein is produced, it assembles at the cell membrane. In addition, we found that alterations in red cell morphologic features and membrane function correlate with the amount of membrane-associated protein—and therefore with the amount of mRNA accumulated—rather than with the primary structure of the variant proteins. These data suggest that an intact sequence at exons 19 through 21 encoding part of the C-terminal 22/24-kd region is not required for proper protein 4.1R assembly in mature red cells.
Human erythroid protein 4.1 (4.1R) is encoded by the EPB41 gene, the genomic organization of which has been defined.1 The 4.1R pre-messenger RNA (mRNA) is processed in a tissue-specific and developmental-specific fashion, mainly through multiple splicing events.1-3 The erythroid 80-kd isoform is the best characterized to date. It results from 2 major pre-mRNA splicing events (Figure 1): the exclusion in early erythroblasts of a 17-nucleotide (NT)-sequence motif that contains the upstream initiation codon at the 5′ end of exon 2 and the incorporation at a later stage of exon 16, which encodes part of the 10-kd spectrin-actin binding domain.4 5
Schematic representation of 4.1R complementary DNA (cDNA) and functional domains.
The 17 nucleotides (NTs) at the 5′ end of exon 2 and exon 14, 15, 17a, and 17b sequences are absent from the erythroid mRNA isoform. Functional domains deduced from limited chymotryptic digestion are shown at the bottom of the cDNA depiction. The 4.1R interacts with many membrane or intracellular components. Some of these interactions (asterisks) were found in nonerythroid cells. PS indicates phosphatidylserine; and CaM, calmodulin. ▨, constitutive coding sequences;  , alternative coding sequences; ⊠, alternative non-coding sequence; □, untranslated regions.
, alternative coding sequences; ⊠, alternative non-coding sequence; □, untranslated regions.
Schematic representation of 4.1R complementary DNA (cDNA) and functional domains.
The 17 nucleotides (NTs) at the 5′ end of exon 2 and exon 14, 15, 17a, and 17b sequences are absent from the erythroid mRNA isoform. Functional domains deduced from limited chymotryptic digestion are shown at the bottom of the cDNA depiction. The 4.1R interacts with many membrane or intracellular components. Some of these interactions (asterisks) were found in nonerythroid cells. PS indicates phosphatidylserine; and CaM, calmodulin. ▨, constitutive coding sequences;  , alternative coding sequences; ⊠, alternative non-coding sequence; □, untranslated regions.
, alternative coding sequences; ⊠, alternative non-coding sequence; □, untranslated regions.
Four functional domains have been defined in 4.1R (Figure 1)6-8(for review, see Baklouti et al1 and Tang and Tang9). The first, the N-terminal 30-kd domain, interacts with a variety of proteins in erythroid and nonerythroid cells (Figure1). The second, the 16-kd domain, has no known function. The third, the 10-kd domain, tightens the spectrin-actin interaction at the red cell membrane. The fourth, the 22/24-kd domain, undergoes a deamidation (Asn 502).10 Although its function in erythrocytes is still unknown, this domain appears to play an important role in mitotic spindle formation in nucleated cells by means of a specific interaction with the nuclear mitotic apparatus.11 It also binds to ZO-2, a tight junction-specific protein.12
The aim of this study was to characterize the structural and functional aspects of mutations that affect the C-terminal region of 4.1R. Two of these mutations are associated with hereditary elliptocytosis (HE). In 3 variants, analysis of both the mutated mRNAs and the membrane-assembled polypeptides in mature red cells showed that the clinical and cellular severity of HE correlated with the mRNA metabolism (causing the reduced representation of other 4.1R domains) but not with protein alteration in the 22/24-kd domain. This finding suggests that an intact C-terminal region of protein 4.1R is probably not critical to the integrity of the red cell membrane.
Materials and methods
Protein electrophoresis and quantification
Red cell membrane proteins were purified and analyzed by using homogeneous 7% or 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as previously described.13Twenty micrograms of total membrane protein were loaded in each lane. The gels were stained with Coomassie blue, and the amount of protein 4.1R was determined with respect to other membrane proteins (primarily protein 4.2) after densitometric scanning of the gels. Densitometric measurements were performed by using ImageQuantMac software (version 1.2; Amersham Pharmacia Biotech AB, Uppsala, Sweden).
Western blot analyses
Western blot experiments were carried out as previously described14,15 by using 4 antibodies. In addition to a polyclonal antibody raised against the whole erythroid 80-kd isoform,14 3 new polyclonal antibodies were prepared and affinity-purified (VALBEX, IUT A, Université Lyon I, Villeurbanne, France). Ab10kd was raised against the spectrin-actin 10-kd binding domain. The region encoding this domain was amplified with polymerase chain reaction (PCR) using the following forward and reverse primers: 5′-GCGGATCCAAGAAAAAGAGAGAAAGACTAGAT-3′ and 5′-GGGGTACCTCAGAAGGGTGAGTGAGTGGATAA-3′, respectively (the boldfaced letters correspond to an added stop codon, and the sequences in italic letters contain restriction sites to facilitate cloning). The PCR fragment was cloned as aBamHI/KpnI insert into pQE-30 expression vector (Qiagen, Hilden, Germany). The fusion 10-kd protein was expressed as a poly-His fusion polypeptide in Escherichia coli JM109 (Promega, Madison, WI) and purified on a nickel-nitrilotriacetic acid-agarose column (Qiagen), according to the manufacturer's recommendations. The purity of the fusion protein was assessed by 15% SDS-PAGE.
AbCT and AbCO were directed against synthetic peptides CT and CO, respectively. The synthetic peptides were obtained from Synt:em (Nı̂mes, France). Peptide CT (NH2-CEAKEQHPDMSVTKV-CONH2) corresponds to the normal amino acid sequence at the C-terminal of 4.1R. The Cys residue (boldfaced letter) at the N-terminal was added for coupling purposes. Peptide CO (NH2-HVSDQGGRPPGDRDC-CONH2) is a missense amino acid sequence specific to the 4.1R Coimbra 1 truncated isoform. Both CT and CO peptides are encoded by exon 21. To raise the antibodies, the fusion 10-kd protein and the synthetic peptides were coupled to either bovine serum albumin (10 kd) or keyhole limpet hemocyanin (CT and CO) and injected into rabbits. After 4 weeks, the rabbits were given boosters of immunogens every 3 weeks, and serum samples were collected 8 days after the third or the fourth booster. All the immune antibodies were affinity-purified from serum by using either Sepharose 4B (Pharmacia, Uppsala, Sweden) and poly-His 10-kd fusion peptide as immobilized affinity ligand (Ab10kd) or HiTrap affinity column (Pharmacia) and uncoupled CT or CO peptides (AbCT and AbCO, respectively).
Ektacytometry
The deformability and stability of red cell membranes were assessed by osmotic gradient ektacytometry16 (Technicon ektacytometer, Tarrytown, NY). The deformability index was recorded at different osmolalities in samples from control subjects and from patients with 4.1R deficiency resulting from different genetic defects.
Secondary-structure predictions
Four different algorithms were used to determine the secondary structures of protein 4.1R variants (for review, see Bohm17): the GOR I and GOR II methods using the theory of information with conditional probability, the double prediction method, and the homologue method, which allows prediction of secondary structures by using not only sequence homology but also sequence similarity.
PCR methods and analysis of PCR products
Peripheral blood samples were used for simultaneous preparation of reticulocyte total RNA and genomic DNA. PCR amplification of genomic DNA was done with use of intronic primers surrounding each of the erythroid-coding exons, as previously described.15Reticulocyte RNA was reverse transcribed by using random hexamers as primers, and the complementary DNA (cDNA) was amplified by PCR according to previously described protocols.1,15 The PCR primers were designed to obtain overlapping fragments,15 which permits screening for changes at the sequences encompassing the primers, as well as screening for splicing alterations.
Two quantitative reverse transcriptase (RT)-PCR methods were used. In the experiment shown in Figure2B, a duplex PCR was performed by using 2 pairs of primers designed to simultaneously amplify 4.1R and protein 4.2 cDNA fragments (145 base pairs (bp) and 246 bp in size, respectively). Both the forward and the reverse 4.1R primers were designed within constitutive exons 11 and 12 to allow amplification of all the isoforms, deriving from either normal or shortened transcripts. Protein 4.2 primers were designed within exons 9 and 10. The RT step was performed as a standard reaction, that is, at 42°C for 20 minutes with use of random hexamers, and corresponded to a master RT reaction. The PCR reagents were then added, and the resulting master PCR mix was dispensed in ice into 5 time-course tubes. The PCR amplification was carried out for 20, 22, 24, 26, and 30 cycles. Each time-point PCR contained, as final concentrations, 200 nmol/L of each primer. When the desired number of cycles was completed, the corresponding time-point PCR was stopped by chilling the tube in dry ice. The PCR products were analyzed on agarose gels and quantified on a FluorImager 595 system (Molecular Dynamics, Sunnyvale, CA) after gel staining with Vistra Green dye (Amersham Pharmacia Biotech AB). The 4.1R mRNA level was measured as a ratio of 4.1R to 4.2 in each sample by using ImageQuantMac software (version 1.2).
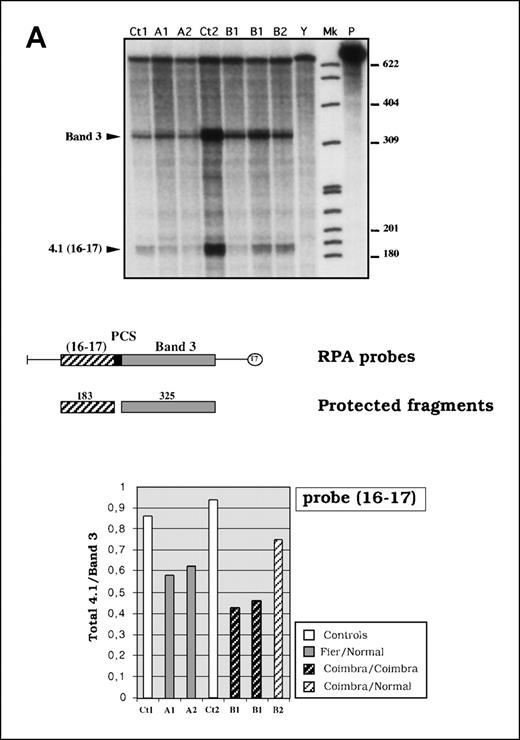
Quantitative analysis of reticulocyte 4.1R messenger RNA (mRNA).
(A) RNase protection assay. In the top panel is an autoradiograph showing phosphorus 32 (32P)-labeled protected fragments. Mk indicates size marker (32P end-labeled pBR 322 MspI fragments); P, undigested probe; Ct1 and Ct2, control mRNAs; and Y, yeast RNA used as negative control. The middle panel shows a schematic representation of the probe and the expected protected fragments. The bottom panel shows the 4.1R mRNA quantitation measured as a normalized ratio of total 4.1 to band 3. (B) Quantitative RT-PCR using protein 4.2 mRNA as the internal control. Three time points (20, 22, and 24 cycles) were considered. Mk indicates 100-base-pair size marker. Total 4.1R mRNA amounts are presented as ratios of 4.1R to 4.2.
Quantitative analysis of reticulocyte 4.1R messenger RNA (mRNA).
(A) RNase protection assay. In the top panel is an autoradiograph showing phosphorus 32 (32P)-labeled protected fragments. Mk indicates size marker (32P end-labeled pBR 322 MspI fragments); P, undigested probe; Ct1 and Ct2, control mRNAs; and Y, yeast RNA used as negative control. The middle panel shows a schematic representation of the probe and the expected protected fragments. The bottom panel shows the 4.1R mRNA quantitation measured as a normalized ratio of total 4.1 to band 3. (B) Quantitative RT-PCR using protein 4.2 mRNA as the internal control. Three time points (20, 22, and 24 cycles) were considered. Mk indicates 100-base-pair size marker. Total 4.1R mRNA amounts are presented as ratios of 4.1R to 4.2.
In the experiment shown in Figure 3, the forward primer was end-labeled by using T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and γ-phosphorus 32 (32P) adenosine triphosphate for 1 hour at 37°C, according to the manufacturer's instructions. After heat inactivation of the enzyme, the oligonucleotide was extracted with phenol and further purified through G-25 Sephadex columns (Boehringer Mannheim, Mannheim, Germany). The RT step was also performed as a standard reaction, and the master PCR mix was dispensed into 8 time-course tubes. The PCR amplification was carried out for 10, 13, 17, 20, 22, 25, 30, and 35 cycles. Each time-point PCR contained, as final concentrations, 200 nmol/L of radioactive 5′ terminals and 200 nmol/L of cold reverse primers. The PCR products were fractionated by PAGE and visualized and quantified by phosphorimaging on a Molecular Imager GS-525 (Bio-Rad, Hercules, CA).
Quantitative reverse transcription-polymerase chain reaction (RT-PCR) of CO.1 and CO.2 mRNA species.
The experiment was performed on reticulocyte mRNA obtained from the heterozygous individual B2 in family B. In the top panel is an autoradiograph showing the normal bands and the bands expected with CO.1 and CO.2. Longer exposure revealed the 3 specific bands at all the time points. The bottom panel shows phosphorimaging measurements of both the total CO mRNA levels compared with the normal 4.1R mRNA produced in trans (curve) and the relative amounts of CO.1 and CO.2 (bars).
Quantitative reverse transcription-polymerase chain reaction (RT-PCR) of CO.1 and CO.2 mRNA species.
The experiment was performed on reticulocyte mRNA obtained from the heterozygous individual B2 in family B. In the top panel is an autoradiograph showing the normal bands and the bands expected with CO.1 and CO.2. Longer exposure revealed the 3 specific bands at all the time points. The bottom panel shows phosphorimaging measurements of both the total CO mRNA levels compared with the normal 4.1R mRNA produced in trans (curve) and the relative amounts of CO.1 and CO.2 (bars).
Single-strand conformation polymorphism (SSCP) analysis, DNA cloning, and sequencing
PCR products resulting from DNA or RNA amplifications were analyzed by SSCP as previously described.15 Specific mutations were tested by restriction digestion of selected PCR fragments, according to the supplier's instructions. Nucleotide sequencing was performed either directly on purified PCR products or after DNA cloning into the TA-vector pCR II (Invitrogen, Carlsbad, CA).
RNase protection assay
Reticulocyte 4.1R mRNA output was assessed by chimeric probe-mediated RNase protection assay (RPA) as previously detailed.18 Three different probes were used. Two of them, 4.1(16-17)/band 3 and 4.1(20-22)/band 3, were previously described15,18; they encompass the 10-kd-domain–encoding and C-terminal-encoding regions, respectively, of the erythroid 4.1R isoform. The new probe, 4.1(18-20)/band 3, was designed to test the region of the mRNA from exon 18 to exon 20. Taking advantage of the cassette feature of the original recombinant plasmid,18 we cut out the 4.1(16-17) region from the plasmid and replaced it with the 4.1(18-20) region. In all these RPA experiments, phosphorimaging quantification was measured as a normalized ratio of 4.1R to band 3, where a band 3 protected fragment served as an internal control to quantify the 4.1R mRNA.
Results
Genealogic, cellular, and protein analyses
In this study, we investigated 3 families in which at least 1 member had HE. Family A (Figure 4) was mentioned (as family LA) in a previous report.19 Mother A1 and daughter A2 were studied in this work. Both had a typical 4.1(−) HE in the heterozygous state. The cell morphologic picture was characterized by nearly 100% smooth, elongated elliptocytes in the peripheral blood, with no cell fragmentation or hemolysis. Protein measurements revealed 30% and 31.5% deficiencies in membrane-assembled protein 4.1R in the mother and the daughter, respectively (Figure 5A).
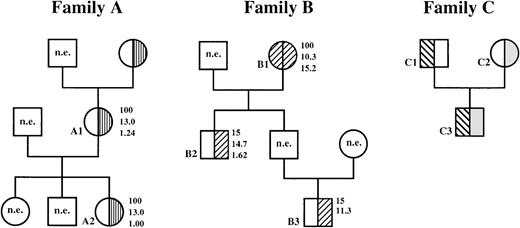
Hematologic variables and phenotypic expression of 4.1R deficiency in families A and B.
Note the unusually low elliptocyte counts in heterozygous members of family B compared with the typical 100% hereditary elliptocytosis HE in heterozygous members of family A; n.e. indicates not examined. Family C, which was described previously,23 24 is included for comparison. Numbers to right of circles and squares represent, in descending order, percent of elliptocytes; amount of hemoglobin, g/dL; and the reticulocyte count as a percentage of the whole blood cell population. □, normal 4.1R; ▥, 4.1R Fier; ▨, 4.1R Coimbra; ▧, uncharacterized 4.1R mutation;  , 4.1R Presles.
, 4.1R Presles.
Hematologic variables and phenotypic expression of 4.1R deficiency in families A and B.
Note the unusually low elliptocyte counts in heterozygous members of family B compared with the typical 100% hereditary elliptocytosis HE in heterozygous members of family A; n.e. indicates not examined. Family C, which was described previously,23 24 is included for comparison. Numbers to right of circles and squares represent, in descending order, percent of elliptocytes; amount of hemoglobin, g/dL; and the reticulocyte count as a percentage of the whole blood cell population. □, normal 4.1R; ▥, 4.1R Fier; ▨, 4.1R Coimbra; ▧, uncharacterized 4.1R mutation;  , 4.1R Presles.
, 4.1R Presles.
Quantitative and functional analysis of the C-terminal 4.1R variants.
Red cell membrane proteins were fractionated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), stained with Coomassie blue, and analyzed densitometrically. (A) Total protein 4.1R expression was measured as the ratio of protein 4.1R to protein 4.2. (B) The 2 truncated protein 4.1R variants (4.1R Coimbra and 4.1R Presles) and the normal 4.1R produced in trans in the heterozygous patients were estimated separately. WT and Mut indicate wild-type and mutated protein 4.1R, respectively. Note that the expression levels of both normal 4.1R are very similar (WT 4.1/4.2 bars), whereas 4.1R Coimbra is produced in a significantly lower amount than 4.1R Presles (Mut 4.1 bars). (C) Osmotic gradient ektacytometric curves. DI indicates deformability index; A, control; B, 4.1R Coimbra heterozygote (family member B2); C, 4.1R Algiers heterozygote 21; D, 4.1R Coimbra homozygote (patient B1); and E, 4.1R Algiers homozygote.21 Each 4.1R Coimbra allele yields a partial deficiency of 4.1R. Each 4.1R Algiers allele yields a complete deficiency of 4.1R.
Quantitative and functional analysis of the C-terminal 4.1R variants.
Red cell membrane proteins were fractionated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), stained with Coomassie blue, and analyzed densitometrically. (A) Total protein 4.1R expression was measured as the ratio of protein 4.1R to protein 4.2. (B) The 2 truncated protein 4.1R variants (4.1R Coimbra and 4.1R Presles) and the normal 4.1R produced in trans in the heterozygous patients were estimated separately. WT and Mut indicate wild-type and mutated protein 4.1R, respectively. Note that the expression levels of both normal 4.1R are very similar (WT 4.1/4.2 bars), whereas 4.1R Coimbra is produced in a significantly lower amount than 4.1R Presles (Mut 4.1 bars). (C) Osmotic gradient ektacytometric curves. DI indicates deformability index; A, control; B, 4.1R Coimbra heterozygote (family member B2); C, 4.1R Algiers heterozygote 21; D, 4.1R Coimbra homozygote (patient B1); and E, 4.1R Algiers homozygote.21 Each 4.1R Coimbra allele yields a partial deficiency of 4.1R. Each 4.1R Algiers allele yields a complete deficiency of 4.1R.
Family B (Figure 4) originated from Coimbra, Portugal. The propositus, the grandmother (B1), had chronic anemia, splenomegaly (spleen palpable 6 cm below the costal margin), and a massive elliptocytosis (100% elliptocytes) without fragmentation. Her son (B2) and grandson (B3) were asymptomatic and had an unusually low percentage of elliptocytes in circulating blood, with normal reticulocyte counts (B2). In B1, 4.1R was absent with a thicker 4.2 band, suggesting a truncated 4.1R comigrating with protein 4.2 (not shown). In B2 and B3, 4.1R was decreased and the 4.2 band was increased but to a lesser degree than in B1. These observations suggested a 4.1R mutation in the homozygous state in the grandmother and in the heterozygous state in the 2 other relatives.
Using 7% SDS-PAGE, we separated protein 4.2 and the variant 4.1R fractions. Protein quantitation showed a slight but significant decrease of about 17.5% of total 4.1R in the heterozygote B2 and ∼ 41% reduction in the homozygote B1 (Figure 5A). The shortened fractions in the membrane in B2 accounted for approximately 28.5% of the normal 4.1R produced in trans (Figure 5B). The homozygous and the heterozygous states were much milder than described previously,13 20-22 at both the cell-phenotypic and protein-expression levels. To further analyze the functional behavior of this unusual variant, we tested red cell deformability in both B1 and B2 patients (Figure 5C). Consistent with the cell-morphologic and protein-quantitation findings, the ektacytometric curves revealed a deformability index in B1 that was intermediate between that observed in a homozygous patient with complete absence of 4.1R and that observed in a heterozygous patient with typical 4.1(−) HE. Similarly, the deformability index was higher in B2 than in the heterozygous patient with typical 4.1(−) HE but lower than in the control.
Family C was described previously23,24 (Figure 4). The father (C1) had a typical 4.1(−) HE in the heterozygous state that was associated with a complete absence of 4.1R transcripts from the affected allele24 (and unpublished data). The mother (C2) carried 4.1R Presles, a shortened variant lacking exon 19-encoding amino acids. Yet, she had no clinical symptoms, and her red cells had normal morphologic features and normal mechanical properties (Feo et al, unpublished data). The son (C3) was a compound heterozygote for the 2 mutations and had the same phenotype as his father. Family C is included here for comparison. Consistent with our previous data,23 24 protein quantitation showed a 4.1R deficiency of ∼ 26.5% and 32% in the father and son, respectively, but no significant reduction in the mother, the 4.1R Presles carrier (Figure 5A). The shortened variant in the membrane appeared slightly reduced compared with the normal 4.1R produced in trans (Figure5B). This apparent reduction is most likely related to the 8-kd loss in the primary sequence of 4.1R Presles, which generates a proportional decrease in band intensity in the gel, rather than a decrease in the amount of 4.1 molecules per se.
Mutations at the gene level
Genomic DNA was first investigated by SSCP and restriction digestion of exon PCR products, as previously described.15 In family A, an abnormal SSCP pattern was found in exon 19 (not shown). Sequencing analysis revealed a dinucleotide frameshift deletion at position 2591-2592 (TA) or 2592-2593 (AT) (numbered according to Baklouti et al1), resulting in a premature termination codon (PTC) within the same exon (encoding the amino acids QTHIStop compared with QTITSE in the wild-type sequence). The mutation, defined as 4.1R Fier, neither created nor abolished a restriction site but was recognized as a smaller band on PAGE of restriction-digested PCR product (for example, AluI [Figure6A, left panel] or BsaI [not shown] enzymes).
Identification of 4.1R HE mutations.
(A) Identification of the genomic mutations by restriction digestion. In 4.1R Fier, the deletion neither creates nor abolishes a restriction site. (A1 + Ct) and (B1 + Ct) correspond to mixtures of patient and control DNAs. The restriction map is indicated at the bottom. Ct indicates control DNAs; and Mk, size markers. All sizes are in base pairs. (B) Reticulocyte 4.1R mRNA analysis. DdeI restriction digestion of RT-PCR product in 4.1R Fier revealed a single band that corresponds to the normal allele. In 4.1R Coimbra, RT-PCR amplification yielded 2 additional bands, CO.1 and CO.2, in the affected family members.
Identification of 4.1R HE mutations.
(A) Identification of the genomic mutations by restriction digestion. In 4.1R Fier, the deletion neither creates nor abolishes a restriction site. (A1 + Ct) and (B1 + Ct) correspond to mixtures of patient and control DNAs. The restriction map is indicated at the bottom. Ct indicates control DNAs; and Mk, size markers. All sizes are in base pairs. (B) Reticulocyte 4.1R mRNA analysis. DdeI restriction digestion of RT-PCR product in 4.1R Fier revealed a single band that corresponds to the normal allele. In 4.1R Coimbra, RT-PCR amplification yielded 2 additional bands, CO.1 and CO.2, in the affected family members.
In family B, an abnormal SSCP pattern was found at exon 20. Direct sequencing of the PCR products revealed a single base substitution at position 2720 (G→A), the last NT of exon 20. Both G and A residues were found in the grandson (B3), whereas only the A appeared in the grandmother (B1). The mutation, defined as 4.1R Coimbra, abolished a BslI restriction site. It was confirmed with different PCR products and was found in the heterozygous state in both the son and grandson and in the homozygous state in the grandmother (Figure 6A, right panel). In family C, the mutation responsible for allele 4.1R Presles remained elusive at the DNA level. So did the 4.1(−) HE mutation located in trans.24
Alterations at the mRNA level
Reticulocyte 4.1R mRNA was analyzed by RT-PCR. No shorter band was found on restriction digestion of RT-PCR products in family A, suggesting absence of the mutated transcripts (Figure 6B, left panel). The restriction fragments were also analyzed by SSCP; no abnormal pattern was observed. Direct sequencing of the PCR products showed a normal exon 19 sequence, further confirming the absence of mRNA species containing the dinucleotide deletion. In family B, RT-PCR amplification around exon 20 yielded 2 smaller bands in addition to the normal band in the heterozygous subjects (Figure 6B, right panel). This doublet was the exclusive product in the homozygous family member. Strikingly, the additional bands had a low intensity in comparison with the normal band in the heterozygous subjects. Nucleotide sequencing revealed that the upper band of the doublet (4.1R Coimbra 1, CO.1) contained only the first 10 NTs of exon 20, whereas the lower band (4.1R Coimbra 2, CO.2) was missing the whole exon 20 (Figure7). Reticulocyte RNA from the homozygote B1 was further analyzed by overlapping RT-PCR coupled to SSCP.15 No additional abnormalities were found.
The splicing defect in 4.1R Coimbra.
The G→A substitution precludes the splicing at the exon 20 donor splice site and partly activates an internal cryptic site 10 NTs downstream of the acceptor site. This splicing alteration leads to the production of CO.1 and CO.2 isoforms, with a partial and a total skipping of exon 20, respectively. CO.1 would possibly encode an out-of-frame C-terminal amino acid sequence. The predicted amino acid sequences of the truncated C-terminals are aligned to the wild-type sequence at the bottom.
The splicing defect in 4.1R Coimbra.
The G→A substitution precludes the splicing at the exon 20 donor splice site and partly activates an internal cryptic site 10 NTs downstream of the acceptor site. This splicing alteration leads to the production of CO.1 and CO.2 isoforms, with a partial and a total skipping of exon 20, respectively. CO.1 would possibly encode an out-of-frame C-terminal amino acid sequence. The predicted amino acid sequences of the truncated C-terminals are aligned to the wild-type sequence at the bottom.
mRNA quantification
The RT-PCR experiments suggested that the mutated 4.1R mRNA species were either absent (4.1R Fier) or reduced (4.1R Coimbra). We next analyzed 4.1R mRNA by RPA using a chimeric probe containing sequences upstream of the mutated region (probe 16-17; Figure 2A). Alternatively, we performed quantitative RT-PCR using protein 4.2 mRNA as an internal control. As in the previous experiments, the 4.1R mRNA was amplified at a constitutive region that is not affected by either of the mutations (Figure 2B). Both methods led to the following observations. In family A, the ratio of 4.1R to band 3 and the ratio of 4.1R to 4.2 showed a decrease in 4.1R mRNA in the heterozygous subjects, which we assumed to be exclusively associated with the 4.1R Fier allele. In family B, RNA quantitation experiments also showed a decrease in both 4.1R Coimbra heterozygous subjects and a more conspicuous decrease in RNA samples derived from 2 different blood samples from the propositus. These data confirmed the observations suggesting that exon 20 mutation was associated with a decrease in mRNA and protein levels. Total mRNA levels in the 4.1R Coimbra heterozygous subjects were higher than in the 4.1R Fier heterozygous subjects but were significantly lower than control levels. This difference was accounted for by the presence of the small amount of CO.1 and CO.2 mRNA species observed in the RT-PCR, whereas no shortened mRNA species were detectable in 4.1R Fier carriers (Figure 6B).
Two RPA probes encompassing exon 20 (probes 18-20 and 20-22; data not shown) were also used to quantitate the mRNA species. These experiments confirmed the virtually complete absence of normal-size mRNA in the homozygous 4.1R Coimbra patient. The 2 4.1R Coimbra heterozygous subjects had similar levels of normal allele mRNA. The reduction in the mRNA level was thus exclusively associated with the mutated allele.
To measure CO.1 and CO.2 mRNA accumulation, we performed a quantitative RT-PCR using an end-labeled primer (Figure 3). The sum of the shortened mRNA species averaged 30% of wild-type 4.1R mRNA produced intrans, which corresponded to 24% of total 4.1R in B2, again indicating a decrease in mRNA accumulation from the mutated allele. The relative expressions of CO.1 and CO.2 were reproducibly about 43% and 57%, respectively. In family C, previous work showed that mRNA from allele 4.1R Presles was present in normal amounts.24
Expression of 4.1R C-terminal variants in the red cell membrane
Total skipping of exon 20 did not affect the reading frame, and translation of CO.2 mRNA isoform generated a shortened protein lacking exon 20-encoding peptide but containing a C-terminal amino acid sequence identical to the wild-type sequence (Figure 7). In contrast, insertion of the first 10 NTs of exon 20 in CO.1 led to a frameshift and a potential 4.1R isoform with a missense C-terminal sequence of 29 amino acids and a PTC only 7 NTs upstream of the wild-type termination codon (Figure 7). Consistent with previous work by Conboy et al,8 secondary-structure analysis using 4 different prediction methods suggested an α-helical structure for the C-terminal end of 4.1R in the erythroid isoform, as well as in the shortened isoforms lacking either exon 19-encoding peptides (4.1R Presles) or exon 20-encoding peptides (CO.2). In contrast, the predicted secondary structure of CO.1 isoform with a missense C-terminal sequence indicated a β-sheet structure.
We assessed whether the shortened proteins, possibly translated from mRNAs with total or partial skipping of exon 20, were able to assemble to the red cell membrane. Western blot analysis of red cell membrane proteins using antibodies raised against the 80-kd erythroid isoform (Ab4.1) or against the 10-kd domain (Ab10 kd) revealed that, in addition to the normal 80-kd 4.1R, there was a shortened doublet in 4.1R Coimbra heterozygous subjects (Figure8, 1 and 7). The doublet was the exclusive product in the homozygous subject (6 in Figure 8). Ab10 kd recognized a peptide sequence upstream of the affected region in all 3 families. Consistent with the absence of mutated mRNA, 4.1R Fier red cell membrane did not contain any truncated form (5 in Figure 8). In contrast, 4.1R Presles was fully expressed in the membrane in the heterozygous parent (4 in Figure 8), as previously demonstrated.23
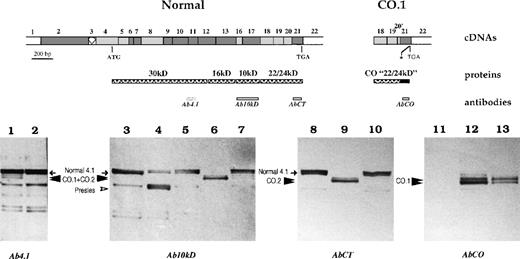
Western blot analysis of membrane-associated 4.1R.
Schematic representation of erythroid domains and corresponding cDNA structure for the normal 4.1R and CO.1 missense C-terminal domain is shown. The asterisk indicates the premature termination codon in CO.1. Red cell membrane proteins were resolved with SDS-PAGE and transferred to a nitrocellulose membrane. Each sample corresponds to 20 μg of total membrane protein. The antibodies used in this set of Western blot studies are depicted beneath the structural domains, except that Ab4.1 epitopes are not indicated precisely because this polyclonal antibody was raised against the whole 80-kd isoform. The tested samples were as follows: heterozygous 4.1R Coimbra (family member B2: 1, 7, 10, and 13), homozygous 4.1R Coimbra (family member B1: 6, 9, and 12), heterozygous 4.1R Fier (family member A1: 5), heterozygous 4.1R Presles (4), and control membrane samples with normal 4.1R (2, 3, 8, and 11).
Western blot analysis of membrane-associated 4.1R.
Schematic representation of erythroid domains and corresponding cDNA structure for the normal 4.1R and CO.1 missense C-terminal domain is shown. The asterisk indicates the premature termination codon in CO.1. Red cell membrane proteins were resolved with SDS-PAGE and transferred to a nitrocellulose membrane. Each sample corresponds to 20 μg of total membrane protein. The antibodies used in this set of Western blot studies are depicted beneath the structural domains, except that Ab4.1 epitopes are not indicated precisely because this polyclonal antibody was raised against the whole 80-kd isoform. The tested samples were as follows: heterozygous 4.1R Coimbra (family member B2: 1, 7, 10, and 13), homozygous 4.1R Coimbra (family member B1: 6, 9, and 12), heterozygous 4.1R Fier (family member A1: 5), heterozygous 4.1R Presles (4), and control membrane samples with normal 4.1R (2, 3, 8, and 11).
Membrane-assembled protein 4.1R species were further analyzed in 4.1R Coimbra samples by using specific AbCT and AbCO antibodies directed against the wild-type or the missense peptide sequences encoded by exon 21. Immunodetection with AbCT resulted in a pattern quite similar to that obtained with the common antibodies. However, the shortened forms that appeared in both the homozygous 4.1R Coimbra subject (9 in Figure8) and the heterozygous 4.1R Coimbra subject (10 in Figure 8) now corresponded specifically to the CO.2 isoform. Finally, when the antibody directed specifically against the missense sequence at the C-terminal of CO.1 (AbCO) was used, Western blot analysis showed a shortened doublet in 4.1R Coimbra, with a stronger reactivity in the homozygous subject (12 in Figure 8) than in the heterozygous subject (13 in Figure 8) and a complete absence of immunoreactivity in the control subject (11 in Figure 8).
Discussion
Impact of the mutations on mRNA metabolism
mRNA molecules containing a PTC either accumulate at a normal level or are targeted by nonsense-mediated mRNA decay. In many instances, nonsense mRNAs are also associated with aberrant splicing (for review, see Maillet et al15). Several models have been proposed in attempts to explain this discrepancy in the fate of nonsense mRNA.25-27 In 4.1R Fier, the frameshift deletion generates a PTC within exon 19, which most likely leads to degradation of nonsense mRNA, as suggested by qualitative and quantitative mRNA analyses (Figure 2 and Figure 6B). This interpretation is consistent with the recently described rule for termination-codon position.27 28 The rule stipulates that stop codons, including PTC, mediate a reduction in mRNA abundance only when they are located more than about 50 NTs upstream of the last exonexon junction.
In 4.1R Coimbra, the splicing mutation is associated with a reduced amount of total (CO.1 plus CO.2) mRNA. One possible explanation for this finding is that the reduction occurs exclusively at the expense of the nonsense mRNA species CO.1. However, the PTC in this case appears only 7 NTs upstream of the wild-type stop codon (ie, 16 NTs upstream of the exon 21-exon 22 junction) and should therefore elicit a normal expression of the nonsense mRNA according to the rule for termination-codon position. Alternatively, the low mRNA expression could possibly derive from an additional molecular defect that is inherent to the 4.1R Coimbra allele, such as a promoter mutation. Accordingly, we would assume that both CO.1 and CO.2 mRNAs are affected and, consequently, that the percentage of CO.1 mRNA species relative to total 4.1R Coimbra mRNA (∼ 43%) reflects the efficiency of use of the internal cryptic site (Figure 7).
Limited role for the C-terminal domain in mature red cells
To assess the role of the C-terminal region of 4.1R in red cells, we examined the effects of 3 different mutations on both the cell morphologic features and the 4.1R membrane assembly. The 3 mutations led or would lead to 4 altered C-terminal domains. Using specific antibodies, we showed that the 3 altered 4.1R proteins that are actually produced in the red cells (Presles, CO.1, and CO.2) assemble to the cytoskeletal membrane where they are probably functional owing to the presence of unaltered 30-kd and 10-kd domains.
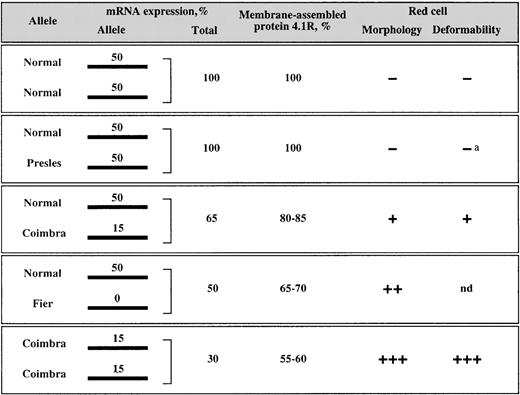
Figure 9 summarizes the data obtained from the mRNA and protein measurements. Assuming that all the normal 4.1R alleles in these families have roughly the same mRNA expression level, we can consider that they would produce 50% of the mRNA molecules in normal diplotypes. In the families with HE, if we assign the same rate (50) to each of the normal 4.1R chromosomes, the contribution of the mutated alleles in trans would accordingly be 50 for 4.1R Presles, 5 for 4.1R Coimbra, and 0 for 4.1R Fier (Figure 9). The allele expression rate of 15 in Coimbra corresponds to 30% of the normal allele expression intrans, as shown in Figure 3.
Correlation of mRNA expression with protein assembly and membrane functional properties.
Minus signs indicate the absence of membrane defects; plus signs, the presence and degree of functional alterations; a, Feo et al (unpublished data); and nd, not determined. The 4.1R Fier is a typical 4.1R deficiency, in which only 1 haploid set of 4.1R is expressed. Therefore, the red cell membrane properties in family A must be comparable to those described for most of the heterozygous 4.1R deficiencies reported to date, including heterozygous 4.1R Algiers20 21 (Figure 5).
Correlation of mRNA expression with protein assembly and membrane functional properties.
Minus signs indicate the absence of membrane defects; plus signs, the presence and degree of functional alterations; a, Feo et al (unpublished data); and nd, not determined. The 4.1R Fier is a typical 4.1R deficiency, in which only 1 haploid set of 4.1R is expressed. Therefore, the red cell membrane properties in family A must be comparable to those described for most of the heterozygous 4.1R deficiencies reported to date, including heterozygous 4.1R Algiers20 21 (Figure 5).
Previous studies have emphasized that protein 4.1R is synthesized in excess in maturing red cells.29,30 Therefore, a unique functional allele can partly compensate for the absence of protein synthesis in trans, the resulting protein deficiency being restricted to approximately 30% instead of 50%.13 14Consistent with these observations, subjects heterozygous for 4.1R Fier had 65% to 70% total 4.1R in red cell membranes (Figure 5 and Figure9). The efficient use of the excess protein also applies to the 4.1R Coimbra allele; the amount of membrane-assembled 4.1R was higher than would be expected on the basis of the mRNA quantitations (Figure 5 and Figure 9).
Collectively, the data shown in Figure 9 suggest that the membrane functional defect correlates with the available amount of protein 4.1R as represented by the mRNA level rather than with the integrity of the primary sequence of the protein. More specifically, in 4.1R Presles, the mRNA level was normal, the truncated protein was present in the membrane in a normal amount, the blood smear was devoid of elliptocytes, and the mechanical properties of the membrane were normal. In 4.1R Fier, only mRNA from the normal allele in transwas detected, and (consistently) no truncated protein was present in the membrane. The massive elliptocytosis was due simply to the presence of 1 haploid set of 4.1R. However, it is likely that the truncated protein, if produced, would also assemble. Previous work showed that the presence of the 10-kd domain alone is sufficient for binding to the spectrin-actin cytoskeletal complex.31,32 Finally, 4.1R Coimbra showed an intermediate pattern at all mRNA, protein, and functional levels. These findings suggest that the 4.1R C-terminal domain has a limited functional importance for protein 4.1R assembly in mature red cells. In the light of recent findings, however, it will be interesting to examine the behavior of the altered proteins during erythroid differentiation and, more generally, in nucleated cells, in which the role of protein 4.1 is currently being elucidated.11
Acknowledgments
We thank Dr C. Martin and Dr L. Roda for referring family A; Dr A. Yvorra for performing the secondary structure prediction analysis; Dr N. Alloisio, Dr G. Brunet, Dr G. Tchernia, and Dr L. Morlé for support and fruitful discussions; and M.-Th. Ducluzeau for technical assistance.
Supported by the Centre National de la Recherche Scientifique, Université Claude Bernard Lyon 1, the Institut National de la Santé et de la Recherche Médicale, the Association pour la Recherche sur le Cancer, the Association Française contre les Myopathies, the Assistance Publique-Hôpitaux de Paris, and the Faculté de Médecine Paris-Sud.
Reprints:Faouzi Baklouti, Centre de Génétique Moléculaire et Cellulaire, Bât 741, CNRS UMR 5534, Université Lyon 1, 43, Boulevard du 11 Novembre 1918, 69622 Villeurbanne Cedex, France; e-mail: baklouti@univ-lyon1.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.