Abstract
Interferons (IFNs) mediate their diverse biologic activities through induction of the expression of multiple genes. Whereas the mode of action of certain of these IFN-regulated genes has been well characterized, most of the molecular and cellular events underlying the constellation of biologic responses to the IFNs remain unresolved. This study showed that the newly identified PLSCR1 gene for phospholipid scramblase, previously implicated in remodeling of plasma membrane phospholipids, is regulated at the transcriptional level by IFN-. Analysis of 5′ flanking genomic sequence in reporter constructs showed that transcriptional control of PLSCR1 was entirely regulated by a single IFN-stimulated response element located in the first exon. A similar induction of PLSCR1 by IFN-2a was also observed in a variety of other human tumor cell lines as well as in human umbilical vein endothelial cells. In these cell lines, the marked IFN-2a–induced increase in PLSCR1 protein expression, ranging as high as 10-fold above basal levels, was not accompanied by increased cell surface exposure of phosphatidylserine, suggesting that remodeling of the cell surface requires both exposure to IFN and a second yet-to-be identified event to stimulate plasma membrane phospholipid scramblase activity and to mobilize phosphatidylserine to the cell surface.
Interferons (IFNs)1 are a family of cytokines that exert antiviral, antiproliferative, antitumor, and immune modulatory functions through transcriptional induction of specific IFN-stimulated genes (ISGs).1,2 On binding to their cell surface receptors, IFNs initiate a signaling cascade involving the JAK family of tyrosine kinases and signal transducers and activators of transcription (STATs) that ultimately results in ISG transcription. The cellular proteins encoded by ISGs largely mediate the actions of IFNs. The best characterized ISGs include certain proteins that are thought to contribute to the antiviral properties of IFNs, most notably the double-stranded RNA-activated protein kinase (PKR), RNase L, the 2′-5′ oligoadenylate (2-5A) synthetases, and the Mx proteins, as well as those ISGs that directly participate in transcriptional regulation of ISG and IFN-gene expression, including the STATs and IFN regulatory factor (IRF) families of transcription factors.1,3,4 The biologic functions of many other ISGs remain unclear and the mechanisms by which the IFNs exert their diverse range of biologic activities remains largely unresolved.1,2 5
Phospholipid scramblase 1 (PLSCR1; GenBank Accession Numbers AF098 642, AF153 715) encodes an endofacial plasma membrane protein that is implicated to mediate an accelerated transbilayer movement of membrane phospholipids under conditions of elevated calcium or acidification of the cytosol.6-8 The properties of this protein suggest that it may contribute to the rapid transbilayer movement of plasma membrane phospholipids that is observed in activated platelets and injured or apoptotic cells that are exposed to elevated intracellular [Ca++].9-12 Whereas PLSCR1 is detected in a wide range of cells and tissues, it has been noted that there can be marked cell-to-cell variability in the level of expression of messenger RNA (mRNA) or protein.7 When membranes differing in PLSCR1 content were compared, the level of PLSCR1 protein generally correlated with the observed sensitivity of membrane phospholipids to the effects of elevated Ca++at the endofacial surface.8
Differential oligonucleotide array screening of ISG expression in the IFN-responsive fibrosarcoma cell line HT1080 suggested a nearly 10-fold increase of PLSCR1 mRNA in cells treated with either IFN-α or IFN-β, and to lesser degree in response to IFN-γ.5 This study demonstrates that the amount of PLSCR1 expressed in the plasma membrane of a variety of cells is under transcriptional control by an IFN-stimulated response element located in the untranslated first exon, and we consider how this up-regulation of PLSCR1 expression may relate to the biologic activities of IFN.
Materials and methods
Materials
Recombinant human interferon-α2a (IFN-2a) 3 × 106 IU/mL was from Roche Laboratories (Nutley, NJ). Bovine prothrombin, factor Va, and factor Xa were obtained from Haematologic Technologies (Essex Junction, VT). Chromogenic thrombin substrate CBS 34.47 was from Diagnostica Stago, Asnières, France. RPMI-1640, Dulbecco's modified Eagle's medium (DMEM), modified Eagle's medium essential (MEME), and OPTI-MEM were from GIBCO-BRL (Grand Island, NY). Murine monoclonal antibody V237 specific for the light chain of factor Va was a gift from Dr Charles T. Esmon (The Oklahoma Medical Research Foundation, Oklahoma City, OK).13Murine monoclonal antibody 4D2 was raised against purified recombinant human PLSCR1. Cell lines: Daudi, Raji, HeLa, and Jurkat cells were from American Type Culture Collection (Rockville, MD); human umbilical vein endothelial cells and CS-C medium were from Cell Systems Co. (Kirkland, WA); fibrosarcoma cell lines HT1080 and STAT1-null U3A cells were a gift from Dr George R. Stark (Cleveland Clinic Foundation, Cleveland, OH).14
Cell culture
The Burkitt's B-cell lymphoma cell lines, Daudi and Raji, and Jurkat T-cell line were cultured in RPMI-1640 complete medium. Human fibrosarcoma HT1080 cells and U3A cells were cultured in DMEM, HeLa cells in MEME, and human umbilical vein endothelial cells in CS-C medium. All culture media were supplemented with 10% fetal bovine serum (FBS; 20% in case of Daudi cells) and 100 U/mL of penicillin and 100 μg/mL of streptomycin, and all cells were maintained at 37°C in 5% CO2.
Northern blotting
Cells were washed twice in phosphate-buffer saline (PBS) and the total RNA was extracted with Trizol reagent (GIBCO-BRL). RNAs, 20 μg/lane, were separated in 1.2% agarose, 2.2 mol/L formaldehyde gels and transferred to Nylon membranes (Amersham, Piscataway, NJ) for 18 to 20 hours. RNA was cross-linked to the membrane, incubated in prehybridization solution at 42°C for 16 hours, and probed with an EcoRI fragment of PLSCR1 complementary (cDNA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA, or β-actin cDNA labeled with 32P-dCTP by random priming with the Prime-a-gene labeling system (Promega, Madison, WI). Membranes were washed and used to expose x-ray film.
Western blotting
For Western blotting, 106 cells were harvested and lysed in 30 μL of cell lysis buffer (2% NP-40 in PBS containing 5 mM EDTA, 50 mM benzamidine, 50 mM N-ethyl maleimide, 1 mM phenylmethylsulfonyl fluoride, and 1 mM leupeptin) at 4°C for 1 hour. Cell lysate was centrifuged at 250,000g for 30 minutes at 4°C, and the supernatants denatured (100°C, 5 minutes) in 10% (w/v) sodium dodecyl sulfate (SDS) sample buffer containing 2% β-mercaptoethanol. Following SDS-polyacrylamide gel electrophoresis (0.23 μg total protein per lane) and transfer to nitrocellulose, the membrane was blocked with 4% low-fat milk and incubated for 1 hour at room temperature in the presence of 2 μg/mL of 4D2, a monoclonal antibody raised against PLSCR1. The blots were incubated with horseradish peroxidase–conjugated goat antimouse IgG (Sigma, St Louis, MO) for 1 hour at room temperature, developed by SuperSignal ULTRA Chemiluminescence (Pierce, Rockford, IL), and analyzed on a Kodak Image Station 440CF (Eastman Kodak, Rochester, NY). PLSCR1 antigen detected by Western blotting was quantified using Kodak's 1D Image Analysis Software version 3.0.
Protein concentrations
The protein concentration of cell lysates was determined by bicinchoninic acid (BCA) assay. In brief, 150 μL of 1:90 diluted cell lysate was mixed with 150 μL of BCA reagent (Pierce, Rockford, IL) and incubated at 37°C for 30 minutes. Absorbance at 562 nm was measured, and protein concentration calculated using bovine serum albumin (BSA) as standard.
Confocal fluorescence microscopy
Cells were subcultured on glass cover slips and treated with 1000 IU/mL of IFN-α2a for 0 to 18 hours at 37°C. All following procedures were performed at room temperature. Cells were washed in PBS and fixed with 2% paraformaldehyde in PBS for 30 minutes. After permeabilization by 0.005% saponin in PBS for 5 minutes, cells were incubated in 2% whole goat serum in PBS for 30 minutes, followed by incubation with mab 4D2 (20 μg/mL in 2% goat serum in PBS) for 1 hour. Cells were stained with fluorescein isothiocyanate (FITC)-goat antimouse IgG (2μg/mL in PBS) for 1 hour, followed by nuclear counterstain with propidium iodide (0.1 μg/mL in PBS) for 10 minutes. Cover slips were mounted on glass slides and samples analyzed on a Bio-Rad MRC1024 laser scanning confocal microscope attached to a Zeiss Axiovert S100TV microscope with Infinity Corrected Optics (40 × oil immersion objective). Images were collected using Bio-Rad's LaserSharp (v3.2) software. Specificity of staining observed for mab 4D2 was evaluated by cell staining with the identical concentration of an isotype-matched antibody raised against complement C9, substituting for mab 4D2.
Molecular cloning of 5′ flanking region of PLSCR1gene and construction of deletions
Human PLSCR1 gene was cloned from a BAC-human genomic library (Genome System, St. Louis, MO) using full-length PLSCR1 cDNA for hybridization, and 4.12 kb of 5′ flanking region was sequenced (GenBank AF153 715). The 4.18-kb DNA consisting of the 5′ flanking region (−1 to −4120) and the first 60 bp of the first exon of the gene (+1 to +60) was amplified by polymerase chain reaction (PCR) using Advantage DNA polymerase mix (CLONTECH Laboratories, Palo Alto, CA), and PCR products were cloned into pGL3-basic-luciferase reporter vector (Promega, Madison, WI). Analysis of the 5′ flanking region for the presence of putative binding sites for transcription factors was performed using MatInspector V2.2. The 4 putative binding sites for ISGF3 or IRFs (Figure 5) were deleted by PCR-mediated deletion. All DNA sequencing was performed on an ABI DNA Sequencer Model 373 Stretch (Applied Biosystems, Foster City, CA) using PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin Elmer, Foster City, CA).
Transfection of Daudi cells
Daudi cells were harvested in exponential growth phase, washed twice, and suspended to 1.35 × 107/mL in OPTI-MEM. To 800 μL of cell suspension in a 0.4-cm electroporation cuvette, 20 μg of pGL3-5′ flanking region (or deletions) of PLSCR1 and 20 μg of pSV-β-galactosidase (Promega) were added, and the mixture was incubated for 10 minutes on ice. Electroporation was performed at 380 V and 500 μF using a Bio-Rad Gene Pulser II (Bio-Rad Laboratories, Hercules, CA). Following incubation for 10 minutes at 37°C, the cells were plated in 10 mL RPMI-1640 complete medium onto tissue culture plates and cultured for 24 hours. Cells were then cultured for an additional 18 hours in the presence or absence of 1000 IU/mL of IFN-α2a, and harvested for luciferase and β-galactosidase assay.
Luciferase and β-galactosidase assay
Luciferase activity was measured using a Luciferase Assay Kit (Promega). In brief, Daudi cells were harvested, washed with PBS, and lysed for 15 minutes with reporter lysis buffer. Cell lysates were vortexed for 15 seconds and centrifuged at 12,000g for 2 minutes at 4°C. In a 96-well plate, 20-μL aliquots of lysate (18 μg protein) were mixed with 100 μL of luciferase assay buffer by automated injection using a MicroLumatPlus microplate luminometer (EG&G Berthold, Gaithersburg, MD), and luminescence was measured for a period of 30 seconds. β-Galactosidase activity was determined with o-nitrophenyl-β-d-galactopyranoside (OPNG) as a substrate. A 100-μL aliquot of cell lysate (90 μg protein) was incubated with 100 μL of 4.4 mM ONPG for 1 hour at 37°C, and absorbance was read at 420 nm. Luciferase activity was expressed in arbitrary light units and corrected for transfection efficiency of β-galactosidase.
Evaluation of cell surface–exposed phosphatidylserine in adherent cells
The cell surface exposure of phosphatidylserine resulting from treatment with IFN induction and ionophore treatments of adherent cell lines HT1080 and human umbilical vein endothelial cells was evaluated by expression of membrane catalytic function in the prothrombinase enzyme reaction. Cells were grown to about 80% confluence in a 48-well culture plate and induced overnight (18 hours) with either 0 or 1000 IU/mL IFN-α2a. After 3 washes, cells were incubated at 37°C in the presence of either 5 μM (HT1080) or 10 μM (human umbilical vein endothelial cells) A23187 in Hanks' balanced salt solution (HBSS) containing 2 mM Ca++, 0.8 mM Mg++, and 0.1% BSA for the time periods indicated. Controls omitting A23187 received identical 1% (final volume) solvent DMSO. During the last 2 minutes of treatment with A23187, the prothrombinase reaction was initiated by addition of factor Va (2 nM), factor Xa (1 nM), and prothrombin (1.4 μM). Thrombin generation was terminated by dilution of cell supernatants into HBSS containing 0.1% BSA and 20 mM EDTA, and samples were stored on ice. Aliquots were transferred to a 96-well plate, and thrombin generated was assayed in HBSS containing 0.1% BSA in the presence of 150 μM chromogenic substrate CBS 34.47 by monitoring time-dependent changes in absorbance at 405 nm using a Thermomax plate reader (Molecular Devices, Sunnyvale, CA).
Flow cytometry
Interferon- and A23187-induced cell surface exposure of phosphatidylserine was evaluated in the suspension cell lines Daudi and Raji using flow cytometric detection of bound factor Va (light chain) as previously described.8 Following 18 hours of induction with either 0 or 1000 IU/mL IFN-α2a, cells were washed once with RPMI and suspended (3 × 106 cells/mL) in RPMI containing 0.1% BSA, 4 mM Ca++. After 2 minutes at 37°C, A23187 (0 or 1 μM) was added. At each time point, the reaction was stopped by addition of 10 mM EGTA, and cells incubated with bovine factor Va (10 μg/mL, 15 minutes at room temperature) and bound factor Va were detected with mab FITC-V237 specific for the light chain.13Cells staining positive for bound factor Va were analyzed by flow cytometry (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ). Data were expressed as percentage of gated factor Va–positive cells in the total cell population.
Results
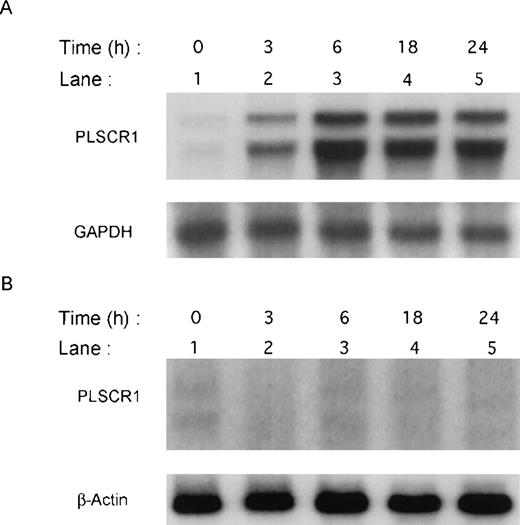
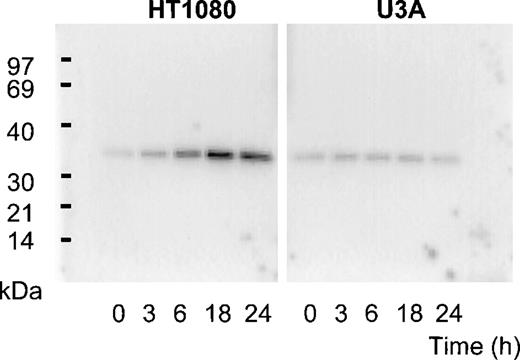
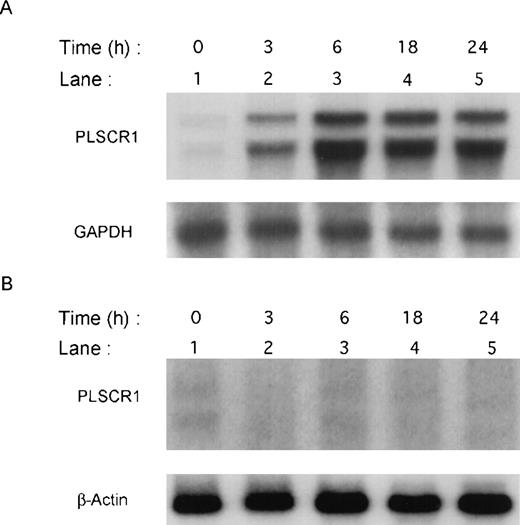
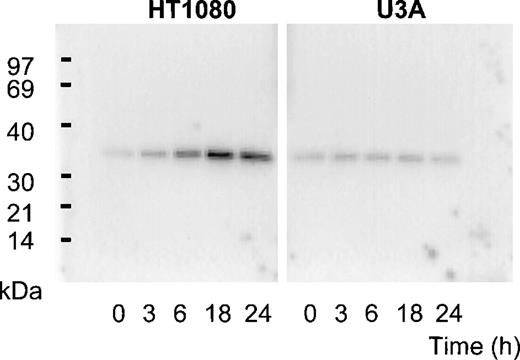
Previous screening by high-density oligonucleotide microarrays provided evidence of an induction of PLSCR1 mRNA in HT1080 cells 6 hours after exposure to IFN-α, β, or γ.5 These findings were extended by demonstrating the time-dependent induction of both PLSCR1 mRNA and protein by IFN-α2a in Northern and Western blots (Figures 1and 2, respectively). Increased PLSCR1 mRNA was detected by 3 hours after IFN-α2a (1000 IU/mL) addition, with protein expression increasing to approximately 10-fold above basal levels at 18 hours. Peak expression of PLSCR1 mRNA was observed at 6 hours, the same length of IFN treatment as in the microarray analysis.5 By contrast to this response observed in the IFN-responsive HT1080 cells, treatment with IFN-α2a had no effect on PLSCR1 expression in mutant U3A cells, an HT1080 derivative cell line deficient in STAT1 transcription factor required for signaling through IFN-receptors.14
Induction of PLSCR1 mRNA by IFN-2a.
Northern blotting of human PLSCR1 in HT1080 cells (panel A) and in STAT-defective U3A cells (panel B) was performed using full-length cDNA for PLSCR1. Times indicate hours after addition of 1000 IU/mL IFN-α2a (see “Materials and methods”). Also shown are results obtained when same blots were probed with control cDNAs for GAPDH and β-actin.
Induction of PLSCR1 mRNA by IFN-2a.
Northern blotting of human PLSCR1 in HT1080 cells (panel A) and in STAT-defective U3A cells (panel B) was performed using full-length cDNA for PLSCR1. Times indicate hours after addition of 1000 IU/mL IFN-α2a (see “Materials and methods”). Also shown are results obtained when same blots were probed with control cDNAs for GAPDH and β-actin.
Expression of PLSCR1 in HT1080 cells.
Western blotting for PLSCR1 expressed in HT1080 (left panel) and U3A (right panel) cells after treatment with 1000 IU/mL IFN-α2a under conditions of Figure 1 was performed with mab 4D2 (see “Materials and methods”). The IFN-induced increase in PLSCR1 antigen detected in HT1080 (normalized to t = 0) was 1.5-fold (t = 3 hours), 4.3-fold (t = 6 hours), 8.3-fold (t = 18 hours), and 6.8-fold (t = 24 hours). In U3A, the increase was 1.4-fold (t = 3 hours), 1.3-fold (t = 6 hours), 1.3-fold (t = 18 hours), and 1.2-fold (t = 24 hours). Data of single experiment, representative of 3 so performed.
Expression of PLSCR1 in HT1080 cells.
Western blotting for PLSCR1 expressed in HT1080 (left panel) and U3A (right panel) cells after treatment with 1000 IU/mL IFN-α2a under conditions of Figure 1 was performed with mab 4D2 (see “Materials and methods”). The IFN-induced increase in PLSCR1 antigen detected in HT1080 (normalized to t = 0) was 1.5-fold (t = 3 hours), 4.3-fold (t = 6 hours), 8.3-fold (t = 18 hours), and 6.8-fold (t = 24 hours). In U3A, the increase was 1.4-fold (t = 3 hours), 1.3-fold (t = 6 hours), 1.3-fold (t = 18 hours), and 1.2-fold (t = 24 hours). Data of single experiment, representative of 3 so performed.
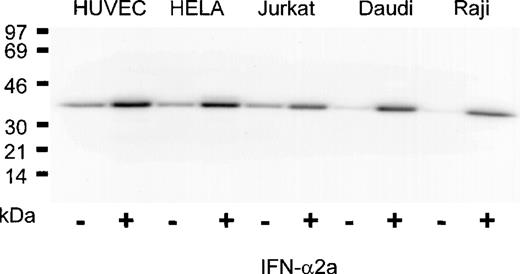
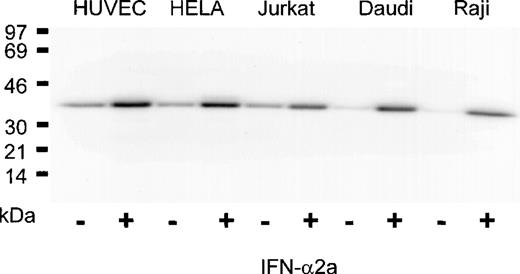
The IFN-α2a dependence of PLSCR1 expression observed in HT1080 cells was confirmed in a variety of other transformed cell lines as well as in cultures of human umbilical vein endothelial cells and nontransformed peripheral blood mononuclear cells isolated from whole blood (Figure 3 and data not shown). In all cases, incubation with IFN-α2a caused a marked increase in PLSCR1 protein expression, ranging to as high as 10-fold above basal levels in the Raji and Daudi cell lines. These data indicate that thePLSCR1 gene is highly up-regulated by IFN-α2a treatment in a variety of normal and transformed cell types.
Induction of PLSCR1 by IFN-2a in various human cell lines.
Western blotting for PLSCR1 was performed in cells indicated after 18 hours of treatment with 0 (−) or 1000 IU/mL (+) IFN-α2a. HUVEC denotes human umbilical vein endothelial cells. The IFN-induced increase in PLSCR1 antigen detected was 2-fold (Jurkat), 3-fold (HUVEC, HELA), and 10-fold (Daudi, Raji). Data of single experiment, representative of at least 3 similar experiments so performed on each cell line.
Induction of PLSCR1 by IFN-2a in various human cell lines.
Western blotting for PLSCR1 was performed in cells indicated after 18 hours of treatment with 0 (−) or 1000 IU/mL (+) IFN-α2a. HUVEC denotes human umbilical vein endothelial cells. The IFN-induced increase in PLSCR1 antigen detected was 2-fold (Jurkat), 3-fold (HUVEC, HELA), and 10-fold (Daudi, Raji). Data of single experiment, representative of at least 3 similar experiments so performed on each cell line.
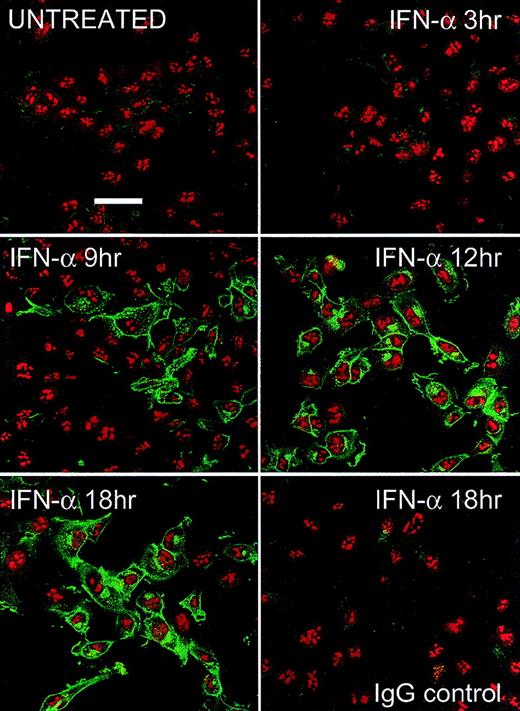
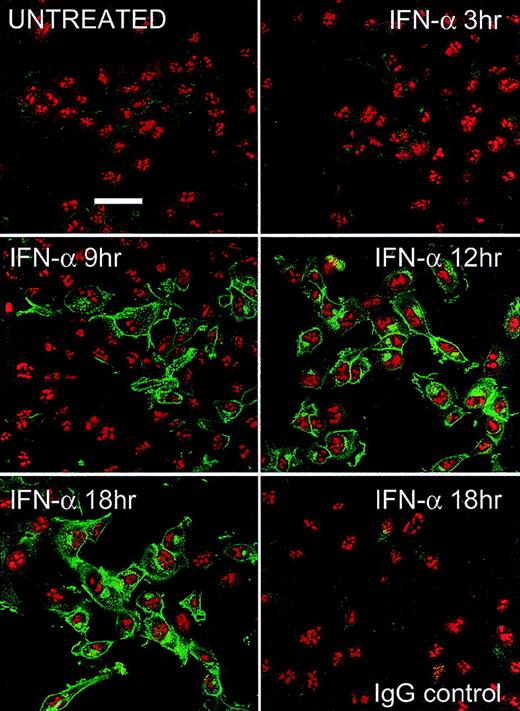
After IFN treatment, newly synthesized PLSCR1 was detected in the plasma membrane where it appeared to concentrate in membrane protrusions. In addition to plasma membrane, PLSCR1 antigen also appeared to be distributed in a variety of other intracellular membranous structures, suggestive of Golgi and endoplasmic reticulum (Figure 4).
Fluorescence microscopy.
HT1080 cells were incubated with 1000 IU/mL IFN-α2a for various times indicated. After fixation and permeabilization, PLSCR1 antigen (green fluorescence) was detected using mab 4D2 (see “Materials and methods”). Cell nuclei are counterstained with propidium iodide (red fluorescence). Control IgG refers to IFN-induced cells that were identically treated and stained with IgG1 murine antibody to irrelevant antigen (complement C9) substituting for mab 4D2. Data representative of 2 independent experiments so performed. White bar indicates 50 μ scale.
Fluorescence microscopy.
HT1080 cells were incubated with 1000 IU/mL IFN-α2a for various times indicated. After fixation and permeabilization, PLSCR1 antigen (green fluorescence) was detected using mab 4D2 (see “Materials and methods”). Cell nuclei are counterstained with propidium iodide (red fluorescence). Control IgG refers to IFN-induced cells that were identically treated and stained with IgG1 murine antibody to irrelevant antigen (complement C9) substituting for mab 4D2. Data representative of 2 independent experiments so performed. White bar indicates 50 μ scale.
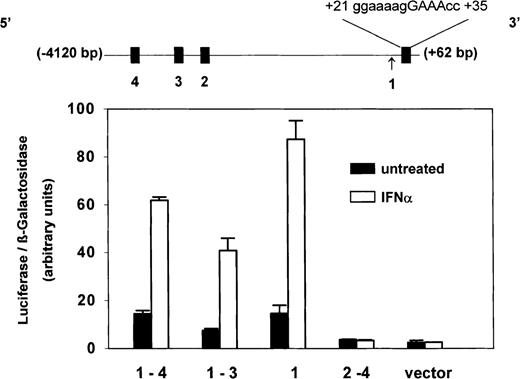
Inspection of PLSCR1 genomic sequence revealed 3 potential IFN-regulated sites within the first 4 kb of 5′ flanking sequence; a potential binding site for IRF-2 at (−3815)gaaaagaGAATcc(−3800); potential binding sites for ISGF3 at (−2733)acaaaaaGAAAgc(−2721) and at (−2519)aaaaacaGAAAcc(−2497), and a single consensus interferon-stimulated response element (ISRE) in the untranslated exon 1 at (+21)gggaaaagGAAAccg(+35) (Figure 5). To identify which of these 4 putative regulatory sites actually contributed to the observed IFN-inducible expression of PLSCR1, luciferase reporter constructs incorporating 5′ untranslatedPLSCR1 gene sequence spanning 1 or more of the putative sites were expressed in Daudi cells, and the response of the transfected cells to IFN-α2a was determined. As shown in Figure 5, these experiments revealed that the IFN-inducible expression of PLSCR1 appears to be controlled by the single ISRE that is located in exon 1. The close proximity of this ISRE to the PLSCR1 transcriptional start site may account for the observed potency of IFN-α2a in inducing PLSCR1 expression.
Identity of ISRE in PLSCR1 genomic sequence.
Four putative ISRE-like elements (filled boxes) located between −4120 bp and +60 bp of the 5′ flanking region and first untranslated exon of PLSCR1 gene are depicted in linear map (top): #4 = (−3815)gaaaagaGAATcc(−3800); #3 = (−2733)acaaaaaGAAAgc(−2721); #2 = (−2519aaaaacaGAAAcc(−2497); #1 = (+21)gggaaaagGAAAccg(+35). Arrow denotes transcription initiation site. Sequence spanning these various putative ISRE-like elements were selectively deleted by PCR and the truncated PLSCR1 DNA cloned into pGL3-luciferase reporter vector as described in “Materials and methods.” Daudi cells were then cotransfected with β-galactosidase-pSV (as transfection efficiency control) and these PLSCR1-pGL3-luciferase plasmids containing the following insertions of PLSCR1 5′ genomic DNA: −4120 bp to +60 bp (spanning #1-4); −3307 bp to +60 bp (spanning #1-3); −2277 bp to +60 bp (spanning #1 only); −4120bp to +18 bp (spanning #2-4); and pGL3 vector without insert (vector). After 24 hours of transfection, either 0 (solid bars) or 1000 IU/mL (open bars) IFN-α2a was added to the cell cultures, and 18 hours later, the cells were harvested for measurement of luciferase and β-galactosidase activities (see “Materials and methods”). Bar graph reports ratio of luciferase/β-galactosidase activities measured at 18 hours. Error bars denote mean ± SEM (n = 3). Data of single experiment, representative of 3 experiments so performed. The average IFN-induced increase (mean ± SD) obtained for each reporter construct from the combined data of all 3 experiments was 3.9 ± 0.4 (insert spanning #1-4); 5.3 ± 1.0 (insert spanning #1-3); 5.2 ± 0.8 (insert spanning #1); 0.8 ± 0.1 (insert spanning #2-4); 0.8 ± 0.2 (vector control).
Identity of ISRE in PLSCR1 genomic sequence.
Four putative ISRE-like elements (filled boxes) located between −4120 bp and +60 bp of the 5′ flanking region and first untranslated exon of PLSCR1 gene are depicted in linear map (top): #4 = (−3815)gaaaagaGAATcc(−3800); #3 = (−2733)acaaaaaGAAAgc(−2721); #2 = (−2519aaaaacaGAAAcc(−2497); #1 = (+21)gggaaaagGAAAccg(+35). Arrow denotes transcription initiation site. Sequence spanning these various putative ISRE-like elements were selectively deleted by PCR and the truncated PLSCR1 DNA cloned into pGL3-luciferase reporter vector as described in “Materials and methods.” Daudi cells were then cotransfected with β-galactosidase-pSV (as transfection efficiency control) and these PLSCR1-pGL3-luciferase plasmids containing the following insertions of PLSCR1 5′ genomic DNA: −4120 bp to +60 bp (spanning #1-4); −3307 bp to +60 bp (spanning #1-3); −2277 bp to +60 bp (spanning #1 only); −4120bp to +18 bp (spanning #2-4); and pGL3 vector without insert (vector). After 24 hours of transfection, either 0 (solid bars) or 1000 IU/mL (open bars) IFN-α2a was added to the cell cultures, and 18 hours later, the cells were harvested for measurement of luciferase and β-galactosidase activities (see “Materials and methods”). Bar graph reports ratio of luciferase/β-galactosidase activities measured at 18 hours. Error bars denote mean ± SEM (n = 3). Data of single experiment, representative of 3 experiments so performed. The average IFN-induced increase (mean ± SD) obtained for each reporter construct from the combined data of all 3 experiments was 3.9 ± 0.4 (insert spanning #1-4); 5.3 ± 1.0 (insert spanning #1-3); 5.2 ± 0.8 (insert spanning #1); 0.8 ± 0.1 (insert spanning #2-4); 0.8 ± 0.2 (vector control).
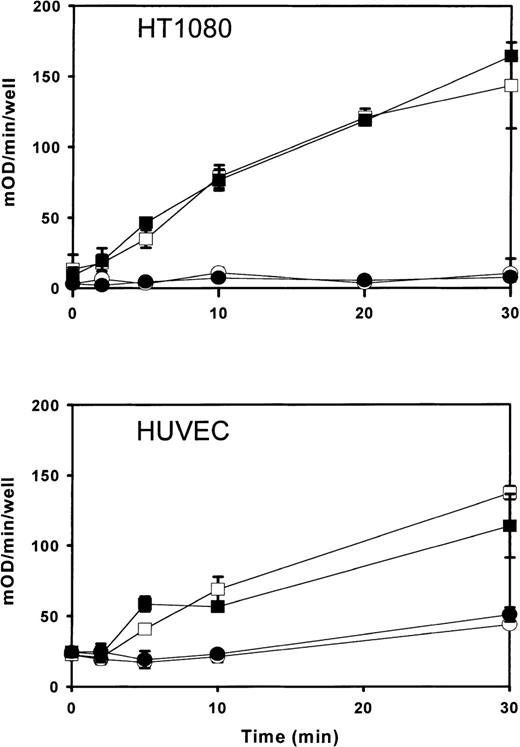
In reconstituted proteoliposomes, PLSCR1 has been shown to mediate accelerated transbilayer migration of membrane phospholipids in the presence of Ca++ or under acidic conditions.6,7,9,15 Furthermore, the level of expression of this protein was generally found to correlate with the extent to which phosphatidylserine was exposed at the cell surface following calcium ionophore treatment, suggesting that PLSCR1 participates in the remodeling of plasma membrane phospholipids in activated platelets and injured or apoptotic cells exposed to increased intracellular Ca++concentration.8 A potential influence of PLSCR1 on either cell proliferation or cell clearance in vivo was also suggested by the observation of altered transcription—including alternative splicing—of a murine PLSCR1 orthologue in leukemogenic versus nonleukemogenic cell clones.16 17 We therefore considered whether the marked up-regulation of PLSCR1 induced by IFN-α2a is also accompanied by changes in the plasma membrane that might increase the likelihood of phosphatidylserine becoming exposed at the cell surface. Despite the presumed activity of PLSCR1 in mediating accelerated transbilayer movement of plasma membrane phospholipids leading to transfer of phosphatidylserine to the outer leaflet, we were not able to detect any change in the IFN-α2a–treated cells indicative of increased surface exposure of plasma membrane phosphatidylserine (Figures 6and 7).
Prothrombinase assay for cell surface phosphatidylserine exposed in IFN-2a–induced cells.
Cells were incubated 18 hours with either 0 (open symbols) or 1000 IU/mL (closed symbols) IFN-α2a. After induction with IFN-α2a, cells were challenged with calcium ionophore (open and closed squares) and prothrombinase activity measured as a function of time after addition of A23187 (abscissa). Open and closed circles denote results for identically matched cells omitting A23187. Upper panel shows results for cell line HT1080 receiving 5 μM A23187; lower panel shows results for human umbilical vein endothelial cells receiving 10 μM A23187. Results (mean ± SD) are plotted from a single experiment representative of similar experiments performed at least 3 times with each cell type.
Prothrombinase assay for cell surface phosphatidylserine exposed in IFN-2a–induced cells.
Cells were incubated 18 hours with either 0 (open symbols) or 1000 IU/mL (closed symbols) IFN-α2a. After induction with IFN-α2a, cells were challenged with calcium ionophore (open and closed squares) and prothrombinase activity measured as a function of time after addition of A23187 (abscissa). Open and closed circles denote results for identically matched cells omitting A23187. Upper panel shows results for cell line HT1080 receiving 5 μM A23187; lower panel shows results for human umbilical vein endothelial cells receiving 10 μM A23187. Results (mean ± SD) are plotted from a single experiment representative of similar experiments performed at least 3 times with each cell type.
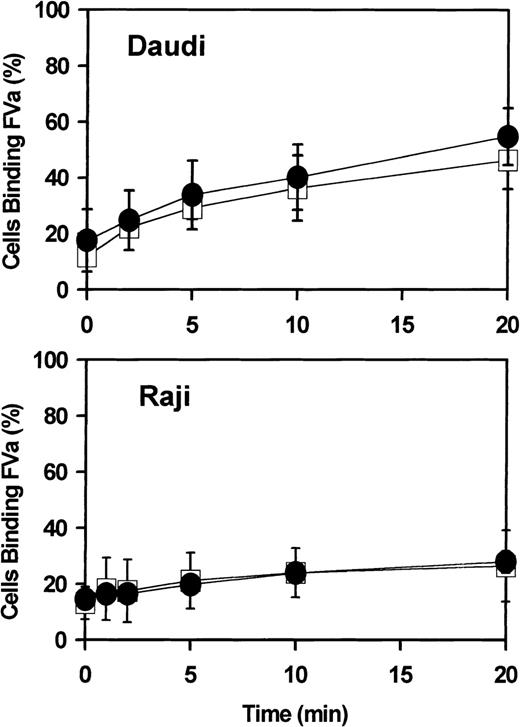
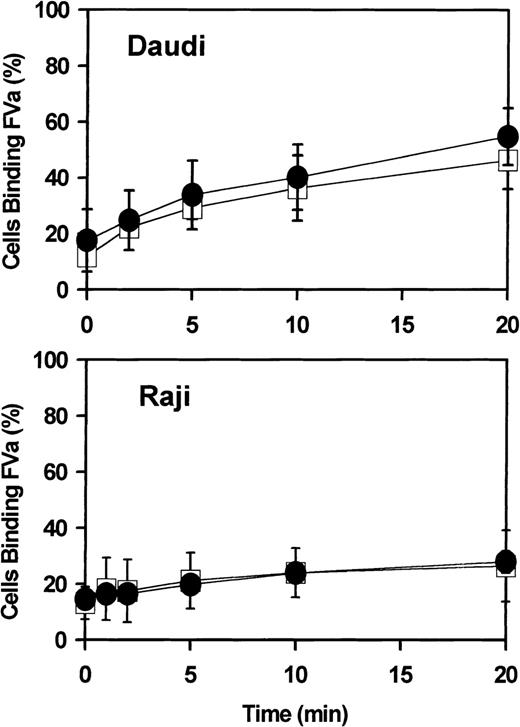
Flow cytometry.
The suspension cell line Daudi (upper panel) or Raji (lower panel) was incubated 18 hours with either 0 (open squares) or 1000 IU/mL (closed circles) IFN-α2a and then analyzed for cell surface–exposed phosphatidylserine, as detected by the binding of coagulation factor Va (FVa; see “Materials and methods”). Ordinate represents percent of cells analyzed by flow cytometry detected as positive for bound FVa. Error bars represent mean and range of combined results of 2 independent experiments. Similar data were obtained for Jurkat cells (not shown).
Flow cytometry.
The suspension cell line Daudi (upper panel) or Raji (lower panel) was incubated 18 hours with either 0 (open squares) or 1000 IU/mL (closed circles) IFN-α2a and then analyzed for cell surface–exposed phosphatidylserine, as detected by the binding of coagulation factor Va (FVa; see “Materials and methods”). Ordinate represents percent of cells analyzed by flow cytometry detected as positive for bound FVa. Error bars represent mean and range of combined results of 2 independent experiments. Similar data were obtained for Jurkat cells (not shown).
Discussion
These experiments demonstrate that the PLSCR1 gene is a member of the IFN-stimulated gene family requiring JAK/STAT signaling for optimal expression. The locus of the controlling ISRE in the untranslated first exon of the PLSCR1 gene represents a putative binding site for the ISGF3 transcription factor complex. Whereas most known ISREs map to flanking sequence that is 5′ to the transcriptional start site, the location of an active ISRE in untranslated exonic sequence has also previously been described for the p202 gene, another IFN-stimulated gene regulated through ISGF3.18 As has been noted in other IFN-stimulated genes, the marked effect of IFN-α2a in up-regulating PLSCR1 expression is consistent with the relatively close proximity of this single active ISRE to the transcriptional start site.
Certain of the IFN-stimulated genes regulated through ISREs are thought to be involved in the apoptotic, antiproliferative, and tumor suppressive activities of IFN-α, although the precise roles of the downstream effector genes actually responsible for these activities remain to be resolved.19-21 Induction of apoptosis of malignant cells or virus-infected cells by IFNs, and clearance of these apoptotic cells by the reticuloendothelial system, are widely assumed to underlie the therapeutic response to IFN treatment. In light of the putative role of the PLSCR1 gene product in catalyzing movement of phospholipids between plasma membrane leaflets, it is of particular interest that one of the most prominent changes observed in apoptotic cells is a remodeling of the topology of plasma membrane phospholipids, with surface exposure of phosphatidylserine and other aminophospholipids that are normally sequestered to the inner leaflet. Such cell surface exposure of phosphatidylserine has been implicated in promoting clearance of injured or apoptotic cells by the reticuloendothelial system.10,11 22
Phospholipid scramblase is an endofacial-oriented plasma membrane protein that has been proposed to contribute to accelerated movement of phospholipids between plasma membrane leaflets in activated platelets as well as in injured and apoptotic cells that are exposed to local elevations in Ca++ concentration or to acidification affecting the inner plasma membrane leaflet.9-12 This activity of the PLSCR1 gene product in promoting Ca++ and pH-dependent movement of phospholipids between membrane leaflets was demonstrated in reconstituted proteoliposomes containing this protein, and the level of cellular expression of PLSCR1 was previously found in general to correlate with the observed extent of transfer of phosphatidylserine to the cell surface in response to induced elevations of cytoplasmic Ca++ concentration.Nevertheless, the exact role of this protein in promoting transbilayer movement of phosphatidylserine and other plasma membrane phospholipids and the actual mechanism of activation of the phospholipid scramblase pathway in situ remains to be clarified. As noted above, induction of PLSCR1 by IFN-α2a leads to a marked increase in concentration of phospholipid scramblase that is expressed in the plasma membrane of the IFN-treated cells, but we were unsuccessful in detecting either a corresponding increase in surface-exposed phosphatidylserine or increased sensitivity of the plasma membranes of these cells to subsequent treatment with calcium ionophore. These data suggest that the mobilization of phosphatidylserine to the cell surface cannot simply be attributed to the level of expression of the PLSCR1 gene product as was previously assumed, but is likely to require additional factors, including potentially another protein, that either acts directly on the plasma membrane or that interacts with PLSCR1 to accelerate transbilayer movement of phospholipids in the plasma membrane. Alternatively, the endogenous level of PLSCR1 expressed in the plasma membrane of these cells before IFN treatment may itself be sufficient to mediate maximal response to calcium ionophore under the conditions of these experiments, masking more subtle changes in phospholipid trafficking between plasma membrane leaflets arising from the IFN-induced increase in plasma membrane concentration of the protein. It also remains to be determined whether the marked increase in PLSCR1 expression induced by IFN promotes remodeling of plasma membrane phospholipids or cell clearance in vivo, under physiologic conditions relevant to tumor growth and viral infection.
The conclusions of the present experiments are consistent with recent observations relating to the possibility that mutation affecting PLSCR1 was responsible for the cellular defect underlying Scott syndrome, a rare inherited bleeding disorder characterized by reduced mobilization of phosphatidylserine to the surface of activated platelets and other blood cells.23,24 When PLSCR1 expressed in Scott blood cells was analyzed, the deduced amino acid sequence and levels of expressed mRNA and protein were normal.25 Furthermore, phospholipid scramblase derived from Scott cell membranes showed apparently normal function once extracted in detergent and reconstituted in proteoliposomes.26 Thus, despite the aberrant properties of the plasma membranes of the Scott syndrome cells suggesting an inherited defect or deficiency affecting the plasma membrane phospholipid scramblase pathway, no abnormality of PLSCR1 per se was detected. This raised the likelihood that another gene is aberrantly expressed or mutated in patients with Scott syndrome, affecting either another as yet unidentified protein with such activity, or alternatively, a regulatory cofactor required for normal PLSCR1 function in situ.
The IFNs have been used for more then 10 years in the treatment of various diseases in humans, including chronic viral syndromes and various malignant and nonmalignant cell proliferative disorders.1 The clinical outcome of IFN treatment is variable. The IFNs induce remission in some patients who have good response to these cytokines and can lead to improvement in these patients. On the other hand, the same IFN therapy results in no benefit at all in some patients who have poor response to IFNs and such treatment can result in severe side effects.27 28 The mechanisms underlying the antitumor or antivirus effects of IFNs and the reason for resistance to IFN therapy in some patients are poorly understood. The discovery of a new IFN-stimulated gene with induced mRNA and protein that can be readily detected in IFN-responsive cells may provide a new tool for quantifying or predicting response to IFN therapy, and promises to shed new insight into the molecular and cellular events that underlie the diverse biologic activities of these cytokines.
Acknowledgments
The authors acknowledge the gift of cell line U3A from Dr George R. Stark (The Cleveland Clinic Foundation) and antibody V237 from Dr Charles T. Esmon (The Oklahoma Medical Research Foundation). The assistance of Dr Malcolm Wood (The Scripps Research Institute) with confocal microscopy is gratefully acknowledged as is the superb technical assistance of Lilin Li, Hongfan Peng, and Yolanda Montejano.
Supported by grants HL36946 (P.J.S.), HL61200 (T.W.), and HL63819 (P.J.S.) from the Heart, Lung, and Blood Institute, and by grant CA44059 (R.H.S.) from the National Cancer Institute, National Institutes of Health, and by the Stein Endowment Fund.
Reprints:Peter J. Sims, Departments of Molecular and Experimental Medicine and Department of Vascular Biology, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: psims@scripps.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.