Abstract
Hypotransferrinemic (Trfhpx/hpx) mice have a severe deficiency in serum transferrin (Trf) as the result of a spontaneous mutation linked to the murine Trf locus. They are born alive, but before weaning, die from severe anemia if they are not treated with exogenous Trf or red blood cell transfusions. We have determined the molecular basis of the hpx mutation. It results from a single point mutation, which alters an invariable nucleotide in the splice donor site after exon 16 of the Trf gene. No normalTrf messenger RNA (mRNA) is made from the hpx allele. A small amount of mRNA results from the usage of cryptic splice sites within exon 16. The predominant cryptic splice site produces aTrf mRNA carrying a 27-base pair (bp), in-frame deletion. Less than 1% of normal levels of a Trf-like protein is found in the serum of Trfhpx/hpx mice, most likely resulting from translation of the internally deleted mRNA. Despite their severe Trf deficiency, however,Trfhpx/hpx mice initially treated with transferrin injections can survive after weaning without any further treatment. They have massive tissue iron overload develop in all nonhematopoietic tissues, while they continue to have severe iron deficiency anemia. Their liver iron burden is 100-fold greater than that of wild-type mice and 15- to 20-fold more than that of mice lacking the hemochromatosis gene, Hfe. Trfhpx/hpx mice thus provide an additional model with a defined molecular defect for the study of genetic iron disorders.
Iron is necessary for a wide spectrum of biologic functions, including oxygen transport, mitochondrial electron transfer, and DNA synthesis. However, because of its insolubility at physiologic pH and its ability to generate free radicals, iron in biologic systems must be tightly complexed by proteins. The plasma glycoprotein Trf accounts for the majority of the iron-binding capacity in blood. Trf not only serves a protective role, but also facilitates the delivery of iron to tissues via the Trf cycle (reviewed by Andrews1). Iron-loaded Trf specifically binds the Trf receptor (Trfr), initiating receptor-mediated endocytosis. Acidification of specialized Trf cycle endosomes aids in the dissociation of iron from Trf, and potentiates the apo-Trf-Trfr interaction. Iron exits the endosome through a transmembrane transporter, DMT1 (formerly Nramp2 or DCT1).2The apo-Trf-Trfr complex is recycled to the plasma membrane, where neutral pH favors the release of apo-Trf back into the circulation.
Hypotransferrinemic (Trfhpx/hpx) mice carry a spontaneous mutation linked to the Trflocus.3 With the use of immunologic methods, circulating Trf levels have been measured to be about 1% of normal. NewbornTrfhpx/hpx mice are viable, albeit profoundly anemic, and can survive for up to 2 weeks after birth without red blood cell transfusions or Trf replacement. When treated, however, their development is normal, apart from subtle architectural changes in the central nervous system and iron overload in multiple organs.4-6 The degree of anemia and rate of iron absorption are inversely proportional to the extent to which the animals are treated.7 A similar iron-overloaded anemic phenotype has been observed in human patients with congenital atransferrinemia.8-11 However, the molecular basis of human atransferrinemia has not been defined.
It is likely that the Trf cycle is essential for normal erythropoiesis because it provides a mechanism of iron uptake sufficient to support high levels of hemoglobin production. The receptor for Trf, Trfr, is expressed at levels that vary with the physiologic and developmental needs of each tissue. Developing erythroid cells express very high levels of Trfr. Our laboratory recently generated mice carrying a disrupted Trfr allele, and showed that homozygous mice lacking Trfr (Trfr−/−) die in utero from severe anemia.12 This result was surprising, because it indicated that mice lacking Trfr are more severely affected thanTrfhpx/hpx mice lacking Trf. We considered 2 possible explanations. First, we speculated that, inTrfr−/− mice, iron bound to circulating Trf might be sequestered from the developing erythron. In the absence of the Trf cycle, erythroid cells might not be able to dissociate iron from Trf for uptake through other pathways. Alternatively, it is possible thatTrfhpx/hpx mice express a small amount of Trf that is sufficient for erythroid iron delivery via the Trf cycle, allowing homozygous animals to survive past birth. To investigate these possibilities, we sought to determine the molecular basis of the Trfhpx/hpx phenotype.
The genetic defect in Trfhpx/hpxmice was previously unknown. However, linkage to the Trf locus on chromosome 9 suggested that it involved the Trfgene.3 A previous report postulated a defect in Trfmessenger RNA (mRNA) splicing, because nuclear RNA fromTrfhpx/hpx brain contained a 5-kilobase (kb) Trf transcript that inappropriately retained the last 2 introns of the Trf gene.13 Nonetheless, the molecular details of theTrfhpx/hpx mutation were not described. We have now identified the Trfhpxmutation: a G-to-A transition at the +1 position of the splice donor site of the last intron in the Trf gene. When this mutation is present, a small amount of a near-normal–sized transcript can be detected by Northern blot analysis, but this transcript results from use of a cryptic splice donor site 27-base pair (bp) upstream from the normal intron 16 splice donor site. Western blot analysis shows a small amount of Trf protein in the plasma ofTrfhpx/hpx mice, which is likely the result of translation of this abnormal Trf mRNA.
Materials and methods
Animals
Animals produced from matings of heterozygous BALB/cJ-Trfhpx/+ mice were used for these studies. Dr Jerry Kaplan (University of Utah) generously provided founder animals for this colony. For RNA analysis,Trfhpx/hpx animals were treated with weekly intraperitoneal injections of 6 mg human Trf (Boehringer Mannheim, Indianapolis, IN) until 8 weeks of age, and then killed at 10 weeks of age. For Western blot analysis of serum and tissue iron studies, animals were given 4 weekly doses of 6 mg of human Trf and subsequently maintained without treatment until humanely killed at 8 to 9 months of age. All mice were fed standard rodent chow and water ad libitum.
RNA isolation and analysis
Total RNA was prepared from frozen livers of 10-week-old animals using RNA STAT-60 (Leedo Medical Laboratories, Houston, TX), according to the manufacturer's instructions. 20 μg of total RNA was electrophoresed on 1% agarose, 0.7% formaldehyde gels and blotted onto Hybond N (Amersham Pharmacia Biotech, Piscataway, NJ). The Northern blot was hybridized to 106 cpm/mL of a32P-labeled probe in QUIKHYB (Stratagene, LaJolla, CA), according to the manufacturer's instructions, and washed under high stringency. The probe was a polymerase chain reaction (PCR) product generated from a template containing the 5′ end of the mouseTrf complementary DNA (cDNA) (−52 bp to 470 bp relative to the ATG start codon), using primers 5′-GAAGCGGGTCGGTCTGTACTCCCC-3′ and 5′-GCTTACAGAAGA GCAAGCCAATGG-3′. The blot was stripped and rehybridized with a mouse β-actin probe.
Reverse transcriptase-polymerase chain reaction and Southern blot analysis
Reverse transcriptase (RT)-PCR was performed by using 1 μg total RNA. cDNA samples were prepared fromTrfhpx/hpx,Trfhpx/+, andTrf+/+ mice using oligo-dT and random hexamers and the Superscript Preamplification kit (GIBCO/BRL, Bethesda, MD), according to the manufacturer's instructions. PCR of the full-length Trf transcript was performed by using primers 5′-GAAGCGGGTCGGTCTGTACTCCCC-3′ and 5′-CTGTCTCCACCACAGT GGCAACCC-3′. By homology to the human and rabbit Trf genes,14 15 we inferred the intron/exon boundaries for exons 13 to 17 of the murine Trfgene. The presence of introns at the expected locations was confirmed by comparing the results of PCR from genomic DNA and cDNA from wild-type animals. RT-PCR across the splice junction of intron 15 was performed by using primers 5′-CCCAAGCTCCAAAC CATGTTGTGG-3′ and 5′-GTGGTACCCTCTGGAAGTTTAACG-3′. PCR across the splice junction of intron 16 was performed by using primers 5′-CGTTAAACTTCCAGAGGGTACCAC-3′ and 5′-CTGTCTCCACCACAGTGGCAACC C-3′. All PCR reactions were carried out for 40 cycles using an annealing temperature of 55°C. DNA sequences were determined by direct PCR (cycle) sequencing of the PCR products by using an automated ABI sequencer (Howard Hughes Medical Institute Biopolymer Facility at Harvard Medical School). Aliquots of PCR products were further analyzed by agarose gel electrophoresis and Southern blot hybridization. Blots were hybridized with 106cpm/mL of a 32P–end-labeled oligonucleotide probe in QUIKHYB solution (Stratagene, LaJolla, CA) at 49°C, and washed to 0.1X SSC, 0.1% SDS at 45°C. The oligonucleotide probes used were 24 nucleotides in length: probe A: 5′-GCTCAACCTCACGACTCCTG GAAG-3′; probe B: 5′-TGCAATCTGTCGGACTCCTGGAAG-3′.
Genomic DNA analysis
Genomic DNA was amplified by PCR with primers flanking the mutation: 5′-CGTTAAACTTCCAGAGGGTACCAC-3′ and 5′-CTGCCTTAGTATCCTGGGTCTGC G-3′. PCR products were subcloned using the TOPO-TA kit (Invitrogen, Carlsbad, CA), according to manufacturer's instructions. The DNA sequence of each subclone insert was determined with the primer 5′-CTGGAATGGTAGTTACAAGAACTC-3′ by using the35S-dideoxy-nucleotide method with the Sequenase version 2.0 (United States Biochemical, Cleveland, OH), according to the manufacturer's instructions. DNA sequence analysis was carried out on 11 subclones from an obligateTrfhpx/+ animal. Single subclones were sequenced from PCR reactions by using genomic DNA from 3 phenotypically affected Trfhpx/hpxanimals, as well as 19 additional inbred strains: A/J, AKR/J, BALB/cJ, BALB/cByJ, BUB/BnJ, C3H/HEJ, C57BL/6J, CAST/Ei, CBA/J, CFO, DBA/2J, FL/1ReJ, FL/4ReJ, LP/J, SEC/1ReJ, SPRET/Ei, ST/bJ, WB/1ReJ, and YBR/Ei. The genomic PCR products were also analyzed for evidence of single strand conformational polymorphisms (SSCPs) using a nondenaturing gel system. Radiolabeled PCR reaction mixtures, generated with a32P–end-labeled oligonucleotide, were loaded onto a 5% polyacrylamide, 0.5X TBE gel and fractionated at 30 W at 4°C for approximately 6 hours. The gel was exposed to x-ray film for visualization of conformational polymorphisms.
Genotyping
Genotype determinations for animals used in these studies were performed by sequence analysis of genomic PCR products, SSCP analysis, measurement of serum Trf (TIBC), or a combination of these methods. There were no inconsistencies in the results. TIBC levels were measured with the Iron and Iron-Binding Capacity kit (Sigma, St Louis, MO), according to the manufacturer's instructions.
Western blot analysis
Mouse Trf-specific antibodies were affinity purified from a sheep antimouse Trf antiserum (Chemicon International, Temecula, CA) with a SulfoLink Coupling Gel column (Pierce, Rockford, IL) coupled with 5 mg of purified mouse Trf (Chemicon International), according to the manufacturer's instructions. For Western blots, 0.5 μL of mouse serum containing approximately 25 μg of total protein was separated on a 7.5% acrylamide SDS-PAGE minigel. Purified mouse (0.05 μg) and human (1 μg) Trfs were used as controls. The protein was transferred onto a PVDF membrane (Amersham Pharmacia Biotech) with the use of a Semi-Phor apparatus (Hoefer Scientific, San Francisco, CA) with transfer buffer (25 mmol/L Tris.HCl, 192 mmol/L glycine, 10% methanol). The membrane was blocked in 3% bovine serum albumin (BSA) before incubating with the affinity purified primary antibody in TBS-T (100 mmol/L Tris, 150 mmol/L NaCl, 0.1% Tween 20, pH 8.0), containing 0.5% BSA for 1 hour at room temperature. Excess primary antibody was removed by washing in TBS-T before incubation with a 1:20 000 dilution of a donkey antisheep HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) in TBS-T, followed by washing in TBS-T. Visualization was accomplished by using ECL+Plus (Amersham Pharmacia Biotech) chemiluminescent detection agent, according to the manufacturer's instructions.
Histology and tissue iron analysis
Mouse tissues were fixed in phosphate-buffered formaldehyde (3.7%) before dehydration and paraffin embedding using standard methodology. Sections were cut at 4 μm and stained with hematoxylin and eosin or Perl's stain.16 Iron determination was performed on fresh frozen liver samples as previously described.17
Results
The Trf hpx/hpx mutation dramatically reduces Trf mRNA levels
On the basis of previous reports,3 13 it seemed likely that the mutation responsible for mouse hypotransferrinemia lay within the Trf gene. To explore this hypothesis, we reevaluated the expression of TrfmRNA in BALB/cJ-Trfhpx mice. Total RNA was extracted from livers of 10-week-old BALB/cJ-Trfhpx/hpx, -Trfhpx/+, and -Trf+/+ mice. A Northern blot containing 20 μg of total RNA was hybridized to a probe from the 5′ end of the mouse Trf cDNA. All samples showed hybridization to a 2.5-kb band, consistent with the size of the full-length Trf transcript (Figure1). The signal detected by using RNA fromTrf+/+ mice was comparable to that of the BALB/cJ controls. In contrast, phosphorimager analysis (Molecular Dynamics, Sunnyvale, CA) showed that the signal from theTrfhpx/+ animals was approximately 50% as strong as the controls, whereas inTrfhpx/hpx animals, the 2.5-kb species was reduced to approximately 5% of wild-type levels. No larger transcripts were detected in total liver RNA from the mutant animals. These data findings reconfirmed that theTrfhpxallele significantly abrogates the expression of Trf mRNA.
Northern blot analysis of Trf expressed by wild-type and mutant animals.
Total liver RNA from homozygous, heterozygous, and wild-type animals was analyzed by Northern blot with a probe derived from the 5′ end of mouse Trf cDNA. All samples show hybridization to a 2.5-kb message, as expected for the full-length wild-type Trftranscript. The Trfhpx/+ animal shows approximately 50% as much signal as wild-type animals. However, very little Trf transcript was detected inTrfhpx/hpx mice. The lower panel shows the same blot after being stripped and reprobed with radiolabeled β-actin cDNA to demonstrate equal loading of the lanes.
Northern blot analysis of Trf expressed by wild-type and mutant animals.
Total liver RNA from homozygous, heterozygous, and wild-type animals was analyzed by Northern blot with a probe derived from the 5′ end of mouse Trf cDNA. All samples show hybridization to a 2.5-kb message, as expected for the full-length wild-type Trftranscript. The Trfhpx/+ animal shows approximately 50% as much signal as wild-type animals. However, very little Trf transcript was detected inTrfhpx/hpx mice. The lower panel shows the same blot after being stripped and reprobed with radiolabeled β-actin cDNA to demonstrate equal loading of the lanes.
The hypotransferrinemia phenotype is due to a splice donor site mutation
Huggenvik et al13 described a nuclear RNA species fromTrfhpx/hpx brain that, when analyzed by Northern hybridization, appeared to contain introns 15 and 16 of theTrf gene. To evaluate whether introns 15 and 16 were not properly excised from Trfpre-mRNAs inTrfhpx/hpx animals, RT-PCR was performed on total liver RNA from BALB/cJ-Trfhpx/hpx and BALB/cJ-Trf+/+ mice. Similar to the Northern blot analysis, full-length RT-PCR products from homozygous mutant and control RNAs produced single bands approximately 2.2 kb in length that were indistinguishable by agarose gel electrophoresis (data not shown). Sequencing plasmid subclones of these products, however, showed that they were different: theTrfhpx/hpx subclone contained a 27-bp deletion (coding nucleotides 2032-2058) located within exon 16 (Figure 2).
Partial sequence of mouse Trf mRNA produced by wild-type and hpx alleles.
The coding sequence and predicted translation are shown for murine Trf exon 16 (pink) and the coding portion of exon 17 (blue). The boxed sequence is deleted in cDNA prepared fromTrfhpx/hpx mice. The deletion does not change the protein reading frame.
Partial sequence of mouse Trf mRNA produced by wild-type and hpx alleles.
The coding sequence and predicted translation are shown for murine Trf exon 16 (pink) and the coding portion of exon 17 (blue). The boxed sequence is deleted in cDNA prepared fromTrfhpx/hpx mice. The deletion does not change the protein reading frame.
To confirm this small deletion, and to determine whether there was any wild-type mRNA present, RT-PCR across the splice junctions of introns 13, 14, 15, and 16 was performed. UsingTrfhpx/hpx RNA, the intron 13 and 14 splice junction primers yielded single products of the expected sizes, indicating that the mRNA had been spliced appropriately (data not shown). However, RT-PCR across the splice junction for intron 15 inTrfhpx/hpx RNA produced 1 major and 1 minor DNA band using this primer pair (Figure3 upper panel, hpx/hpx lane 2). Direct sequencing of these products showed the smaller fragment to be the normally spliced mRNA, and the larger product to contain the entire 1.4-kb sequence of intron 15. This latter product was not seen in wild-type animals. A third, minor band of intermediate-molecular weight was variably present. RT-PCR across the intron 16 splice junction also produced 1 major and 1 minor DNA band (Figure 3 upper panel,hpx/hpx lane 3). Sequence analysis of the smaller product showed the same 27-bp deletion seen in the full-length clone. The larger product contained the entire 1.6-kb sequence of intron 16. It is likely that the 1.4-kb and 1.6-kb products detected with intron 15 and 16 splice junction primers, respectively, are derived from incompletely spliced RNAsretaining introns 15 and 16 described by Huggenvik and colleagues.13
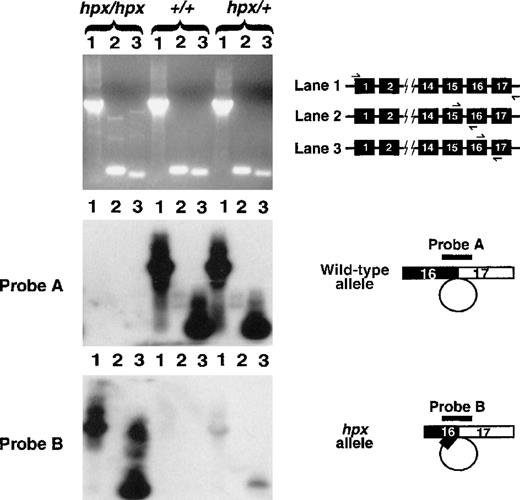
RT-PCR and Southern hybridization analysis of TrfmRNA.
The cartoon in the top right corner illustrates the placement of primers in the Trf gene, which were used for RT-PCR. For each animal, lane 1 represents full-length RT-PCR, lane 2 represents RT-PCR across the splice junction for intron 15, and lane 3 represents RT-PCR across the splice junction for intron 16. The agarose gel in the top panel was analyzed by Southern blot analysis. Probe A is specific for cDNA reverse transcribed from correctly spliced Trf mRNA, completely excising intron 16. Strong hybridization signals are shown in lanes from animals carrying a wild-type allele and are absent inTrfhpx/hpx mice. Probe B hybridizes to cDNA reverse transcribed from Trf mRNA resulting from use of the cryptic splice donor site in exon 16, causing excision of intron 16 plus 27 bp of exon 16. Probe B hybridizes specifically to RT-PCR products from the Trfhpx allele.
RT-PCR and Southern hybridization analysis of TrfmRNA.
The cartoon in the top right corner illustrates the placement of primers in the Trf gene, which were used for RT-PCR. For each animal, lane 1 represents full-length RT-PCR, lane 2 represents RT-PCR across the splice junction for intron 15, and lane 3 represents RT-PCR across the splice junction for intron 16. The agarose gel in the top panel was analyzed by Southern blot analysis. Probe A is specific for cDNA reverse transcribed from correctly spliced Trf mRNA, completely excising intron 16. Strong hybridization signals are shown in lanes from animals carrying a wild-type allele and are absent inTrfhpx/hpx mice. Probe B hybridizes to cDNA reverse transcribed from Trf mRNA resulting from use of the cryptic splice donor site in exon 16, causing excision of intron 16 plus 27 bp of exon 16. Probe B hybridizes specifically to RT-PCR products from the Trfhpx allele.
Comparison of the sequence of intron 16 derived from hpx cDNA and that of wild-type genomic DNA revealed that the first nucleotide of the hpx intron was an adenosine rather than a guanosine. A guanosine at the +1 position is highly conserved among mammalian splice donor sites. Given this, it seemed likely that this G-to-A transition was the mutation responsible for the hypotransferrinemia phenotype.
To test this hypothesis, we undertook analysis of genomic DNA. with the use of a forward primer in exon 16 and a reverse primer in intron 16, PCR was performed on genomic DNA from 2Trfhpx/hpx mice. The PCR products were subcloned and sequenced. Our results confirmed the presence of the G-to-A transition in the +1 position of the splice donor site of intron 16 in each subclone. The same genotyping procedure was performed on genomic DNA from an obligateTrfhpx/+ female mouse. Sequence analysis of 11 genomic PCR subclones revealed 6 clones containing the wild-type allele and 5 clones containing the mutant allele (data not shown). Although the mutation arose and is maintained on BALB/cJ, we also considered the possibility that this nucleotide change might be a polymorphism present in other strains that was introduced by genetic contamination of the BALB/cJ-Trfhpxstrain. To test this, we analyzed genomic DNA from the mutant strain, as well as 19 other wild-type inbred strains of mice, for this change by SSCPs. We found abnormal SSCP bands only in mice that were heterozygous or homozygous for the hypotransferrinemia trait (data not shown). The lack of this polymorphism in wild-type strains was further confirmed by sequence analysis of genomic PCR clones.
The Trf hpx/hpx mutation completely eliminates normal Trf mRNA splicing
With the use of primers spanning the intron 16 splice junction, discrete RT-PCR products of similar size could be amplified from liver RNA from Trfhpx/hpx and wild-type mice. However, sequence analysis indicated that theTrfhpx/hpx product contained the 27 nucleotide deletion. To search for low levels of normal TrfmRNA in Trfhpx/hpxmice, we carried out a combined PCR-Southern blot experiment. Three separate amplification reactions were performed on first-strand cDNA from total liver RNA from Trfhpx/hpx,Trfhpx/+, andTrf+/+ animals. Primers flanking the entire coding region of the Trfgene and 2 other primer sets flanking the splice junctions of introns 15 and 16 were used for 40 cycles of PCR amplification. We analyzed these PCR products by gel electrophoresis and Southern blot hybridization (Figure 3). To distinguish different splicing products, hybridization was performed using 24-mer oligonucleotide probes that bridged splice junctions. Each oligonucleotide was designed to contain 12 nucleotides of homology on each side of the splice site, so that the probes would hybridize only to DNA amplified from mRNA with a specific splice pattern under moderately stringent conditions. Probe A was designed to hybridize to amplification products derived from mRNAs in which intron 16 had been correctly removed. This probe detected amplification products from all animals, except those with theTrfhpx/hpx phenotype. Evidence of correct excision of intron 16 was not detected even after prolonged exposures of the blot. Probe B was designed to identify transcripts made by using the cryptic GT splice donor site 27-bp upstream of the normal exon 16/intron 16 boundary. Probe B hybridizes strongly to PCR products amplified from homozygous mutant mice and weakly to bands from heterozygous mice. Mice carrying only wild-type alleles of Trfdid not utilize the cryptic splice site.
On further investigation, we discovered transcripts from theTrfhpx allele that resulted from the use of at least 1 additional, cryptic splice donor site in exon 16 (data not shown). However, by far the most abundant transcript is that described above. In summary,Trfhpx/hpx mice have defectiveTrfmRNA splicing, and all detectable mRNA results from misspliced transcripts that use cryptic donor splice sites. Even by using a very sensitive RT-PCR–Southern blot detection method, we could not detect any correctly spliced TrfmRNA in the livers of the homozygous mutant mice.
Trf hpx/hpx mice survive without therapy after weaning, but develop massive hemosiderosis
Without serum, purified Trf injections or red blood cell treatments, newborn Trfhpx/hpx mice die of complications of anemia. Mice maintained on routine treatment protocols have been reported to live up to 15 weeks,3 but the survival and phenotype of adult animals from which protein or blood treatments have been withdrawn have not been described. We reasoned that, by the time of weaning,Trfhpx/hpx mice should have passed through the period of maximal growth and iron demand. To investigate the possibility that they might no longer be dependent on treatments for viability, we reared a cohort of mice with no hematologic support after 4 weeks of age. In the absence of exogenous Trf or transfusions, these animals continued to grow although they remained runted and pale compared with their littermates. One of 5Trfhpx/hpx mice died in the first 2 months after weaning. By 6 months of age, all the remaining animals developed severe kyphosis of the cervical and thoracic spine and a distinct red-brown hue to exposed areas of skin, particularly the ears. When killed at 8 to 9 months of age, all lacked appreciable body fat and most organs, including skeletal muscle, had taken on a rusty tinge. The liver and pancreas were particularly noteworthy for their dark brown color. Peripheral blood smears showed extremely hypochromic red cells. Tissues were examined histologically for nonheme iron deposition. As shown in Figure 4, there was massive iron accumulation in the liver, kidney, and heart ofTrfhpx/hpx mice compared with wild-type controls. Liver iron deposition was seen in both Kupffer cells and hepatocytes. Kidney iron deposition localized to a distinct band at the corticomedullary junction, likely representing deposition in a specific population of tubular epithelial cells. Cardiac iron deposition was seen in both macrophages and cardiac myocytes. Abnormal iron accumulation was also found in the adrenal medulla and the exocrine pancreas, but the pancreatic islets were spared (Figure 5). Strikingly, in no tissue was there histologically detectable fibrosis (data not shown). Despite the marked iron accumulation in many tissues, the spleen contained very scant stainable iron (data not shown).
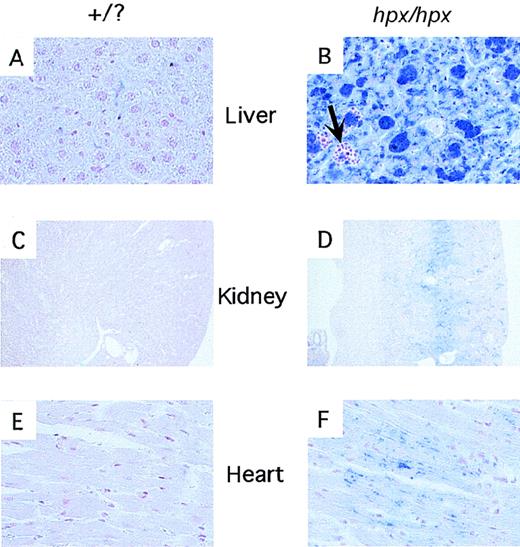
Iron deposition in tissues from wild-type andTrfhpx/hpx mice.
Histologic sections were stained with the Perl's stain for iron. Panels A, C, and E are sections of liver, kidney, and heart (respectively) from nonanemic mice (Trfhpx/+ orTrf+/+ genotype). Panels B, D, and F are sections of liver, kidney, and heart fromTrfhpx/hpx mice. Iron deposits stain blue. The arrow in panel B highlights an area of extramedullary hematopoiesis within the liver of the mutant animal.
Iron deposition in tissues from wild-type andTrfhpx/hpx mice.
Histologic sections were stained with the Perl's stain for iron. Panels A, C, and E are sections of liver, kidney, and heart (respectively) from nonanemic mice (Trfhpx/+ orTrf+/+ genotype). Panels B, D, and F are sections of liver, kidney, and heart fromTrfhpx/hpx mice. Iron deposits stain blue. The arrow in panel B highlights an area of extramedullary hematopoiesis within the liver of the mutant animal.
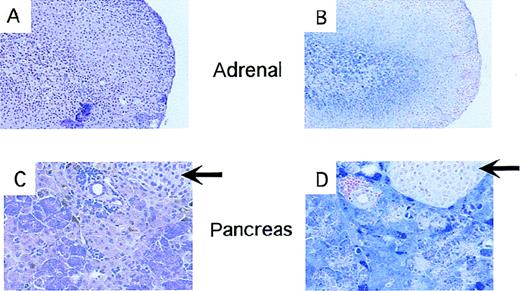
Iron deposition in endocrine tissues fromTrfhpx/hpx mice.
All panels show tissues fromTrfhpx/hpx mice last treated with Trf 7 to 8 months earlier. Panels A and C are stained with hematoxylin and eosin; panels B and D are stained with the Perl's stain for iron. Panels A and B show adrenal tissue; panels C and D show pancreas. Iron deposits stain blue. Arrows indicate pancreatic islets.
Iron deposition in endocrine tissues fromTrfhpx/hpx mice.
All panels show tissues fromTrfhpx/hpx mice last treated with Trf 7 to 8 months earlier. Panels A and C are stained with hematoxylin and eosin; panels B and D are stained with the Perl's stain for iron. Panels A and B show adrenal tissue; panels C and D show pancreas. Iron deposits stain blue. Arrows indicate pancreatic islets.
Quantitative analysis of liver iron revealed massive accumulation, averaging 16 027 μg/g wet weight (n = 4, range = 15 340 to 17 485 μg/g), which was nearly 100-fold increased compared with their wild-type and heterozygous control littermates (n = 12, average = 167 μg/g wet weight, range 87-223 μg/g). It is also 15- to 20-fold greater than liver iron levels in mice lacking the hereditary hemochromatosis gene, Hfe,17 of similar age (J. E. Levy, L. K. Montross and N. C. A., unpublished data).
Untreated Trf hpx/hpx mice have a limited amount of immunoreactive Trf
Our genetic and expression analysis indicated thatTrfhpx/hpx mice do not produce detectable amounts of correctly spliced TrfmRNA. To look for Trf-related protein in their sera, approximately 25 μg of total serum protein was tested for the presence of immunoreactive Trf by Western blot analysis that used an antimouse Trf antibody.Trfhpx/hpx mice used in this analysis were last treated with human Trf 6 to 8 months earlier. By Western blot, sera from homozygous mice contained a small amount of a Trf-like protein, whose electrophoretic migration was not discernibly different from the protein present in wild-type animals (Figure6). With dilutions of serum from aTrf+/+ animal as a reference,Trfhpx/hpx animals have between 0.5% and 1% the normal amount of immunoreactive transferrin.
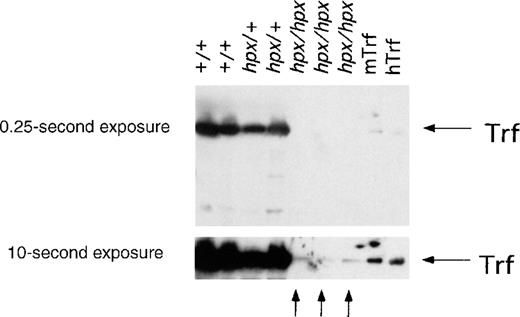
Western analysis of Trf protein in wild-type and mutant animals.
Sera from Trfhpx/hpx mice, last treated with human Trf 6 to 8 months earlier, were analyzed for the presence of immunoreactive Trf by Western blotting using an antimouse Trf antibody. Two exposures of the same blot are shown here, with 50 ng of mouse Trf (mTrf) and 1 μg of human Trf (hTrf) as control lanes on the right. Trf was abundant in serum from wild-type and heterozygous mice, as seen in the first 4 lanes. In contrast, little Trf was detected in serum from Trfhpx/hpxmice. A band that migrated similarly to Trf was visible after a prolonged exposure, as shown by the arrows at the bottom. There was far less of this species present inTrfhpx/hpx mice than Trf in wild-type mice.
Western analysis of Trf protein in wild-type and mutant animals.
Sera from Trfhpx/hpx mice, last treated with human Trf 6 to 8 months earlier, were analyzed for the presence of immunoreactive Trf by Western blotting using an antimouse Trf antibody. Two exposures of the same blot are shown here, with 50 ng of mouse Trf (mTrf) and 1 μg of human Trf (hTrf) as control lanes on the right. Trf was abundant in serum from wild-type and heterozygous mice, as seen in the first 4 lanes. In contrast, little Trf was detected in serum from Trfhpx/hpxmice. A band that migrated similarly to Trf was visible after a prolonged exposure, as shown by the arrows at the bottom. There was far less of this species present inTrfhpx/hpx mice than Trf in wild-type mice.
Discussion
We have shown that the spontaneous mutation responsible for murine hypotransferrinemia disrupts a splice donor site at the end of exon 16 of the Trfgene. This mutation can fully account for the observed abnormalities in Trf mRNA levels and splicing seen in these animals. The mutation precludes normal excision of intron 16, resulting in the usage of a cryptic splice donor site 27-bp upstream of the normal splice junction (Table 1). No mRNA with intron 16 correctly excised can be detected. The low levels ofTrfmRNA seen by Northern blot may reflect the inefficient use of the nonconsensus, cryptic splice site. Alternatively, it may be that the internally deleted mRNA is less stable than the wild type. In addition to this aberrant splice, we and others13 identified a subset of transcripts containing introns 15 and/or 16. Although the mutation directly affects the donor site for exon 16, it could also be expected to result in retention of intron 15 in some RNAs, as mutations in 5′ splice donor sites can lead to inefficient splicing of the preceding intron.18 In any case, only a trace amount of a Trf-related protein can be detected by Western blot analysis of serum fromTrfhpx/hpx mice. Considering the fact that no normal mRNA is detectable, yet the electrophoretic migration of the Trf-related protein is indistinguishable from that of the wild-type protein, it appears that the slightly smaller, deleted message is translated. We infer that an aberrant protein, containing a 9 amino acid internal deletion removing residues 679 to 687 near the carboxyl-terminus of the 697 residue Trf protein (including the leader sequence), circulates in Trfhpx/hpxmice.
We have not yet determined whether the mutated form of Trf can bind iron. It seems likely that it can, as deletion of amino acids 679 to 687 does not remove any of the residues known to be important for binding iron atoms.19 It is also unclear whether Trf produced from the Trfhpxallele binds to the transferrin receptor. Regardless, the amount of mutant Trf protein is no more than 1% of wild-type levels. Whether or not it can function effectively, there is clearly too little Trf to provide an amount of iron sufficient for erythropoiesis, as approximately 109atoms of iron are required for each developing red cell.
The phenotype of Trfhpx/hpx mice, deficient in Trf, is less severe than that ofTrfr−/− mice,12which do not produce any functional Trf receptor. Initially, the difference between mice lacking ligand and receptor seemed surprising. However, although the only proven function of Trfr is the binding and receptor-mediated endocytosis of ferric-Trf, Trf has an additional function. It serves to complex and sequester iron, attenuating the reactivity of the metal ion. In the absence of a functional Trf cycle in Trfr−/− mice, iron is bound to normal Trf, but there is no endosomal process for iron release.
Considering these facts, there are 2 plausible explanations for the difference in phenotype betweenTrfr−/− mice andTrfhpx/hpx mice. First, it is possible that the small amount of aberrant Trf protein produced byTrfhpx/hpx mice provides a sufficient amount of iron to support erythropoiesis at a level that allows the animals to survive. In embryos, a small amount of maternal transferrin might also cross the placenta, allowing the animals to survive to birth and briefly thereafter. Alternatively, it may be that non-Trf–bound iron (NTBI) is more available for non-Trf cycle iron uptake pathways than Trf-bound iron. It is indisputable that such pathways must exist; Trfhpx/hpx mice develop massive iron deposition in most tissues even though they have little if any Trf cycle iron uptake activity. However, the persistence of severe anemia in adultTrfhpx/hpx mice indicates that very little iron enters erythroid precursors through non-Trf cycle pathways. At present, we cannot rule out either hypothesis.
This is the first report of survival ofTrfhpx/hpx mice for a period of months after Trf administration was discontinued. Our findings are striking for several reasons. First, although earlier studies indicated that Trf treatment was necessary for survival ofTrfhpx/hpx mice to the time of weaning,3 it appears that little, if any, Trf is absolutely necessary after weaning. AdultTrfhpx/hpx mice are severely anemic, yet viable off treatment. Second, untreated adultTrfhpx/hpx mice demonstrate massive iron loading. This apparently results from increased intestinal iron absorption secondary to an as yet unknown regulatory signal from the iron-deficient erythron to intestinal absorptive cells.20Iron deposits in nearly all tissues, including the expected targets for iron overload (liver, heart, endocrine tissues) as well as the kidney. Remarkably, Trfhpx/hpx mice have very little stainable iron in splenic macrophages, despite iron overload elsewhere. This may be because very little iron enters splenic macrophages, because the iron that does enter splenic macrophages leaves rapidly, or a combination of both.
The amount of tissue iron inTrfhpx/hpx mice greatly exceeds that found in mice carrying targeted mutations in the hemochromatosis gene,Hfe.17,21 When matched for age, the amount of iron in the livers of mature Trfhpx/hpxmice is 15- to 20-fold greater than that ofHfe−/− mice (N. C. Andrews, L. K. Montross, and J. E. Levy, unpublished data). This indicates thatHfe−/− mice must retain some regulation of intestinal iron absorption, despite the fact that they produce no Hfe protein. There must be other regulatory mechanisms, in addition to that involving Hfe, that govern the amount of dietary iron entering the body. Interestingly, although they accumulate enormous iron burdens, the tissues ofTrfhpx/hpx mice do not develop the secondary changes, such as cirrhosis and pancreatic fibrosis, seen in human iron overload disorders. This is despite the amount of iron seen in Trfhpx/hpx livers is greater than the amount typically associated with the development of cirrhosis in humans with hereditary hemochromatosis (HH).22In addition, similar to humans with HH, iron accumulates largely in acinar cells of the pancreas, preferentially sparing the islets.23 The lack of secondary sequelae may be due to the fact these changes require long-standing iron deposition. Alternatively, mice may be intrinsically more resistant to the development of cirrhosis and pancreatic fibrosis.
Trfhpx/hpx mice offer a unique model in which to study both iron deficiency and iron overload. Although hematopoietic cells are severely deprived of iron, most, if not all, other tissues develop marked iron overload. These animals complement other models, including microcytic anemia (mk) mice,24 Belgrade (b) rats,2 sex-linked anemia (sla) mice,25Trfr-/-mice12, and Hfemutant mice17 21 for the study of disorders of iron homeostasis.
Acknowledgments
We thank Dr Jerry Kaplan for providingTrfhpx/hpx mice and Dr Adriana Donovan for assistance with the preparation of the figures. We thank Dr Robin Reed for discussing splicing aberrancies with us. C.C.T. is currently a medical student at the University of Tennessee, Memphis.
Supported by the Howard Hughes Medical Institute and grant number HL51057 from the National Institutes of Health to N.C.A.
Reprints:Nancy C. Andrews, Children's Hospital, Enders 720, 300 Longwood Ave, Boston, MA 02115; e-mail:nandrews@rascal.med.harvard.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.