Abstract
Hereditary hemochromatosis (HC) is one of the most common single-gene hereditary diseases. A phenotypic hallmark of HC is low iron in reticuloendothelial cells in spite of body iron overload. Most patients with HC have the same mutation, a change of cysteine at position 282 to tyrosine (C282Y) in the HFE protein. The role of HFE in iron metabolism and the basis for the phenotypic abnormalities of HC are not understood. To clarify the role of HFE in the phenotypic expression of HC, we studied monocytes–macrophages from subjects carrying the C282Y mutation in the HFE protein and clinically expressing HC and transfected them with wild-type HFE by using an attenuated Salmonella typhimurium strain as a gene carrier. The Salmonella system allowed us to deliver genes of interest specifically to monocytes–macrophages with high transduction efficiency. The accumulation of 55Fe delivered by55Fe-Tf was significantly lower in macrophages from patients with HC than from controls expressing wild-type HFE. Transfection of HC macrophages with the HFE gene resulted in a high level of expression of HFE protein at the cell surface. The accumulation of 55Fe delivered by 55Fe-Tf was raised by 40% to 60%, and this was reflected by an increase in the55Fe-ferritin pool within the HFE-transfected cells. These results suggest that the iron-deficient phenotype of HC macrophages is a direct effect of the HFE mutation, and they demonstrate a role for HFE in the accumulation of iron in these cells.
Hereditary hemochromatosis (HC) is one of the most common single-gene hereditary diseases.1 A cornerstone of HC genetics was laid in 1996, with the isolation of the hemochromatosis gene, now called HFE.2 Most (83% to 100% in different series) patients with HC carry the same mutation, resulting in a change from cysteine at position 282 to tyrosine (C282Y) in the HFE protein.2-4 The human HFE protein is closely related to the family of major histocompatibility complex class I molecules. The C282Y mutation disrupts a critical disulfide bond in the α3 domain of the HFE protein and abrogates binding of the mutant HFE protein to β2-microglobulin (β2M). This results in reduced transport to and expression on the cell surface.5,6Some light has been shed on the role of HFE in iron metabolism by the observation that wild-type HFE makes a stable complex with transferrin receptor (TfR)7 and that HFE, TfR, and transferrin (Tf) form a ternary complex, with binding of HFE to TfR strictly dependent on the pH.8 In spite of the growing data on the molecular interactions of TfR and HFE in transfected cell lines, little is known about the biologic effect of this interaction and the phenotypic consequences of a mutated HFE in HC.
In patients with HC9 and in murine models of HC (ie, HFE and β2M knockout mice)10,11 iron preferentially accumulates in parenchymal cells; little metal is stored in reticuloendothelial (RE) cells. This indicates an abnormal regulation of iron metabolism in RE cells in HC.9Macrophages are the central portal of entry for Salmonellainfection.12 We took advantage of the fact that an attenuated Salmonella typhimurium carrier strain harboring eukaryotic expression vectors can mediate gene transfer in vivo.13 14 Differentiated, nondividing macrophages from patients with HC carrying the C282Y mutation in HFE were transfected with wild-type HFE. This allowed us to study the effect of a functional HFE on specific parameters of iron metabolism in cells from patients with HC.
Materials and methods
Isolation and culture of monocytes
Patients with HC were characterized through full clinical evaluation and HFE genotyping, as described.15 Monocytes from 8 patients (2 women, 6 men; mean age, 47 ± 10 years) with untreated HFE-associated HC (homozygous for the C282Y genotype) and clinically expressing the disease were obtained by gradient centrifugation of peripheral white blood cells as previously described.9 For the iron incorporation experiments, cells from HFE wild-type control subjects (3 women, 3 men; mean age 40 ± 14 years) without abnormalities of iron metabolism, were used. Yield, purity, viability, and recovery (usually between 95% and 98% as assessed by specific markers) of monocytes were as previously reported.9 In some experiments contaminating lymphocytes were depleted by complement-mediated lysis using a monoclonal antibody to CD3 (clone HIT3a; PharMingen, San Diego, CA) and rabbit complement (Cederlane, Hornby, Canada). To induce differentiation to macrophages, monocytes (approximately 2.5 × 106cells) were maintained in culture for 10 to 12 days in RPMI 1640 medium containing 2 mmol/L glutamine, antibiotics, and 20% heat-inactivated human serum and kept in 5% CO2 at 37°C. The medium and all reagents were endotoxin free.
Bacterial strains, plasmids, and media
The auxotrophic S typhimurium aroA− strain SL7207 (S typhimurium 2337-65 derivative hisG46, DEL407-aroA::Tn10[Tc-s]) was kindly provided by B. A. D. Stocker (Stanford University School of Medicine, Stanford, CA). Bacterial strains were routinely grown at 37°C in brain–heart infusion broth or agar (BHI; Sigma, Milan, Italy), supplemented with 100 μg/mL ampicillin when required. The eukaryotic pCMV-βgal and the pCMV-GFP vectors that contains the β-galactosidase (βgal) or the green fluorescent protein (GFP) gene, respectively, under the control of the immediate early promoter of cytomegalovirus (CMV) (Clontech, Palo Alto, CA), were used to characterize gene transfer and transgene expression. Full-length HFE cDNA was generated by reverse transcription–polymerase chain reaction (RT-PCR) amplification of human lung RNA, and the PCR product was sequenced and subcloned into the pCDNA3.1 expression vector (Invitrogen, Groningen, The Netherlands) that contains a c-myc epitope to produce an HFE/c-mycmRNA product.
In vitro assay for gene transfer and HFE expression
Human monocytes–macrophages were exposed in vitro to recombinantSalmonella to test for transfection efficiency.Salmonella infection was preferentially carried out after 10 to 12 days of in vitro culture. Medium was replaced with antibiotic free medium containing recombinant Salmonella at a 10:1 to 50:1 ratio (bacteria/cell). Infection was carried out for 30 minutes at 37°C. Wells were then carefully washed and refilled with gentamicin-containing fresh medium to kill any residual extracellularSalmonella. Gene transfer was determined 24 to 48 hours after infection by cytofluorometric analysis. HFE was detected by staining with anti-HFE monoclonal antibody (HFE-JB1)16 followed by biotinylated antimouse immunoglobulin and streptavidin–phycoerythrin (PharMingen). Appropriate isotype-matched antibodies were used as negative controls. Cells have also been stained with anti-CD14PE or fluorescein isothiocyanate (PharMingen) and with anti-CD71PE for the detection of TfR. Expression of βgal, TfR, and HFE was determined on CD14-positive gated cells using a FACScalibur Instrument (Becton Dickinson, San Jose, CA) and cellQuest software. Cells were processed for βgal activity by X-gal staining, as previously specified.13 HFE protein was also detected by Western blot analysis. Briefly, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed under denaturing conditions, followed by transfer to a polyvinylidene difluoride membrane (Amersham, Little Chalfont, UK). Immunostaining was performed by incubation with a rabbit polyclonal antibody to the α3 domain of HFE. This antibody was made by immunizing a Sandy half-lop rabbit with 0.3-mg peptide (KDKQPMDAKEFEPKD) glutaraldehyde cross-linked to keyhole limpet hemocyanin. The serum was taken after 3 more immunizations of antigen, and the antiserum was purified using a specific peptide-affinity column.17 Peroxidase-conjugated goat antirabbit antibody (Sigma) was then used and was followed by chemiluminescence detection. Total RNA was extracted from in vitro differentiated macrophages by cesium chloride gradient and subjected to RT-PCR. First-strand DNA was generated from 1 μg RNA using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) for 1 hour at 42°C. The cDNA was amplified by PCR with Taq DNA polymerase (Promega) using primers specific for β2m (direct, 5′-CTCGCGCTACTCTCTCTTTCTGG-3′; reverse, 5′-GCTTACATGTCTCGATCCCACTTAA-3′), HFE (direct, 5′-CTTGCTGCGTTCACACTCTCTG-3′; reverse, 5′-GGCTTGAAATTCTACTGGAAACCC-3′), βgal (direct, 5′-CGTGACGTCTCGTTGCTGCAT-3′; reverse, 5′-CACCATCGTCTGCTCATCCATG-3′), and HFE/c-myc(direct, 5′-GGGGAAGAGCAGAGATATACGTGC-3′; reverse, 5′-TGAGATGAGTTTTTGTTCGGGC-3′). The HFE/c-myc cDNA encompasses the last HFE exon and the c-myc epitope. PCR was carried out in a 30-μL volume (1 μmol/L primers, 1 U Taq DNA polymerase, 2 mmol/L MgCl2, 25 μmol/L dNTPs) for 30 cycles (1 minute denaturation at 94 °C, 1 minute annealing at 60°C, 45-second extension at 72°C) using a thermal cycler (PCT-200 Peltier Thermal Cycler; MJResearch). The PCR products were then analyzed on an 8% to 10% polyacrylamide gel and stained by ethidium bromide.
Transferrin-iron incorporation
Human (apo)Tf (Sigma) was passed over an S-200 (Pharmacia, Little Chalfont, UK) column to remove any aggregated protein, and it showed a single peak on high-pressure liquid chromatography. Saturation of Tf with iron was performed as follows: a stock solution of 0.2 mmol/L 55FeCl3, > 5 mCi/mg Fe (Amersham), and citric acid (molar ratio, 1:2) in Tris-HCl 100 mmol/L, pH 8.8, was prepared and added to a solution of Tf dissolved in 10 μmol/L NaHCO3 (molar ratio Fe-Tf, 2:1) and incubated at room temperature for 60 minutes. The 55Fe-Tf solution was dialyzed against phosphate buffer, checked by nondenaturing PAGE, and added to the culture medium at 0.5-μmol/L final Tf concentration. Cells were kept for 24 hours after infection in fresh culture medium before the addition of 55Fe-Tf. Cell-associated radioactivity was measured at different incubation times by using liquid scintillation counting (Beckman, Palo Alto, CA). In addition to monitoring the incorporation of radioactive iron in the macrophages, cell lysates were subjected to nondenaturing PAGE coupled with autoradiography and densitometry. The main cellular radioactive protein comigrated with control purified human ferritin (kindly provided by P. Arosio, Milan, Italy). Relative quantities of ferritin in cell lysates were normalized to cell protein content.
Statistical analysis
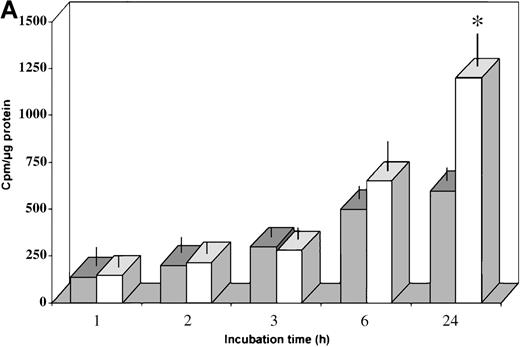
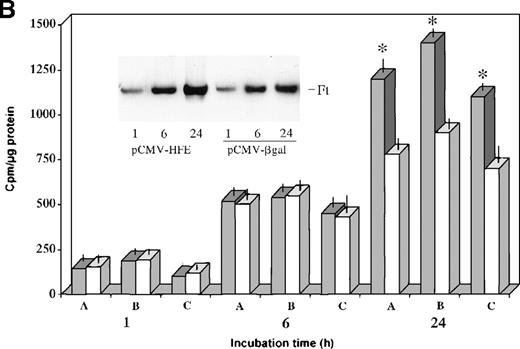
The values shown in Figure 4 are expressed as means ± SD. Significant differences between normal and HC macrophages (Figure 4A) or HC macrophages transduced with the control vector or the HFE vector (Figure 4B) were evaluated by Student t test using the Stat View 4.0 program (Abacus Concept, Berkeley, CA).
Results
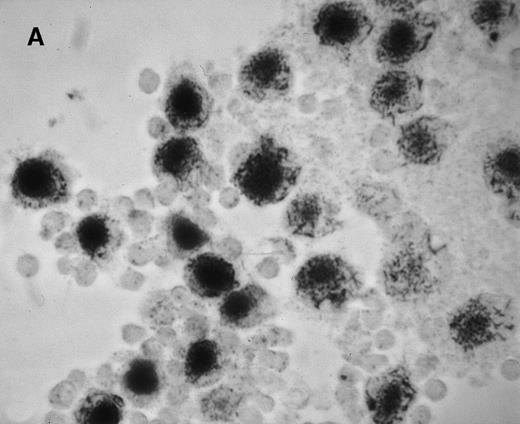
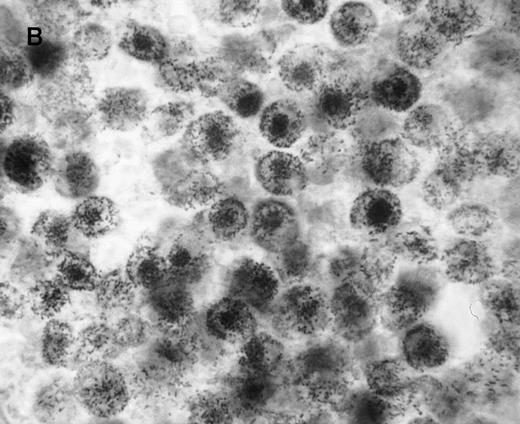
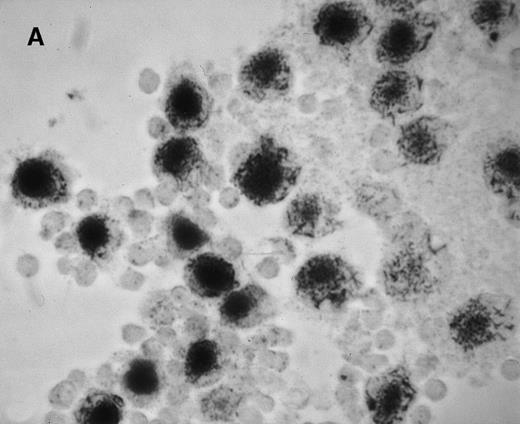
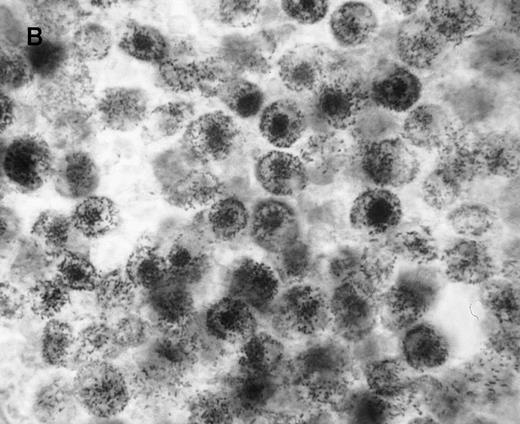
We first assessed the specificity of gene delivery in HC cells by using an attenuated S typhimurium aroA− SL7207 strain harboring the pCMV-βgal vector that contains the βgal gene under the control of the immediate early promoter of CMV. After infection of peripheral blood mononuclear cells, we found that βgal expression was restricted to macrophages and spared lymphocytes (Figure1A). Figure 1B indicates that after lymphocyte depletion, most of the remaining cells were infected and expressed the transgene. Transduction efficiency after infection ranged from 85% to 95%, as assessed by cytofluorometric analysis.
Gene delivery in HC monocytes–macrophages bySalmonella carrier.
In vitro differentiated macrophages from patients with HC were infected with Salmonella harboring the pCMV-βgal plasmid. (A) βgal expression in peripheral blood mononuclear cells was restricted to macrophages (large cells) and spared lymphocytes (small cells). (B) A duplicate cell preparation of A after complement-lysis of lymphocytes showing high βgal expression in most cells.
Gene delivery in HC monocytes–macrophages bySalmonella carrier.
In vitro differentiated macrophages from patients with HC were infected with Salmonella harboring the pCMV-βgal plasmid. (A) βgal expression in peripheral blood mononuclear cells was restricted to macrophages (large cells) and spared lymphocytes (small cells). (B) A duplicate cell preparation of A after complement-lysis of lymphocytes showing high βgal expression in most cells.
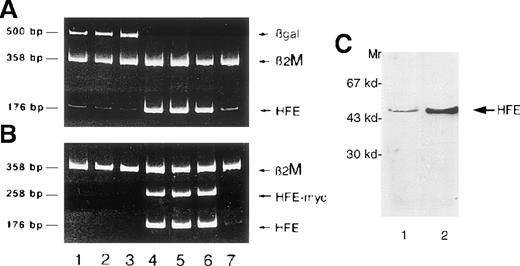
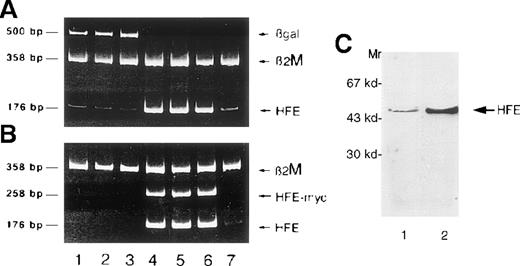
Once the DNA delivery system was optimized, we infected HC macrophages with Salmonella carrying the pCMV-βgal plasmid or a human HFE cDNA under the control of the CMV promoter. At 24 hours after infection with the pCMV-HFE construct, we found a significant increase in HFE expression by RT-PCR that was maintained up to 48 hours, as with cells transduced with the pCMV-βgal plasmid (Figure2A-B). We also showed that the up-regulation of HFE expression correlated with the detection of RT-PCR product from the c-mycepitope linked to the HFE cDNA, proving transgene expression (Figure2B). Up-regulation of HFE in transduced cells was confirmed by Western blot analysis (Figure 2C) with a polyclonal antibody to a peptide derived from the α3 domain.
Salmonella mediates HFE gene transfer in HC macrophages.
Expression of different genes in HC macrophages as detected by RT-PCR at 24 hours (lanes 1 and 4), 30 hours (lanes 2 and 5), and 48 hours (lanes 3 and 6) after Salmonella infection. Lanes 1 to 3, macrophages transfected with βgal; lanes 4 to 6, macrophages transfected with HFE; lane 7, noninfected HC macrophages. The same cDNA preparations were divided and amplified in duplicate test tubes with primers for either HFE and βgal (A) or HFE and HFE/c-myc (B), using β2M primers as an internal control for each reaction. The presence of the transgene is demonstrated by the detection of HFE/c-myc RNA. (C) Western blot of proteins from infected cells. Proteins (30 μg) from cell homogenates were separated by SDS-PAGE, blotted, and analyzed by Western blot using the anti-HFE polyclonal antibody. A significant increase of HFE expression was detected in cells infected with Salmonella carrying the pCMV-HFE plasmid (lane 2) compared with those infected with the pCMV-βgal plasmid (lane 1).
Salmonella mediates HFE gene transfer in HC macrophages.
Expression of different genes in HC macrophages as detected by RT-PCR at 24 hours (lanes 1 and 4), 30 hours (lanes 2 and 5), and 48 hours (lanes 3 and 6) after Salmonella infection. Lanes 1 to 3, macrophages transfected with βgal; lanes 4 to 6, macrophages transfected with HFE; lane 7, noninfected HC macrophages. The same cDNA preparations were divided and amplified in duplicate test tubes with primers for either HFE and βgal (A) or HFE and HFE/c-myc (B), using β2M primers as an internal control for each reaction. The presence of the transgene is demonstrated by the detection of HFE/c-myc RNA. (C) Western blot of proteins from infected cells. Proteins (30 μg) from cell homogenates were separated by SDS-PAGE, blotted, and analyzed by Western blot using the anti-HFE polyclonal antibody. A significant increase of HFE expression was detected in cells infected with Salmonella carrying the pCMV-HFE plasmid (lane 2) compared with those infected with the pCMV-βgal plasmid (lane 1).
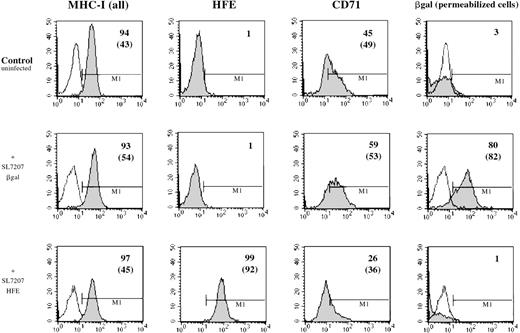
To demonstrate expression of the HFE protein on the cell surface afterHFE gene transduction in HC macrophages, cells were stained with the HFE-specific monoclonal antibody HFE-JB116 and were analyzed by FACS. As a control, Salmonella carrying a pCMV-βgal plasmid was used to infect the same cell preparation. Results of a representative experiment are reported in Figure3. HC macrophages were efficiently transduced with pCMV-βgal, and HFE gene transduction increased HFE expression in HC macrophages. Interestingly,Salmonella infection with HFE wild type led to a slight reduction of the cell surface expression of TfR, as identified by specific antibody to CD71.
Cell surface expression of HFE in transfected macrophages.
Human macrophages from a patient with HC were infected in vitro withSalmonella strain SL7207 carrying either the pCMV-βgal (middle panels) or the pCMV-HFE (lower panels) plasmid. Results from a representative experiment in which cells were analyzed for gene expression by FACS 48 hours after infection using monoclonal antibodies against major histocompatibility class I, HFE, CD71, and βgal followed by a PE-conjugated anti-murine IgG polyclonal antibody. Appropriate isotype-matched monoclonal antibody controls have been used for instrument settings (empty histograms). Detection of βgal expression was performed after saponin-mediated permeabilization of monocytes. Percentage of positive cells and mean values of fluorescence (in brackets) are reported for each single histogram. Basal expression level for each marker in untreated HC–monocytes is also shown (upper panel).
Cell surface expression of HFE in transfected macrophages.
Human macrophages from a patient with HC were infected in vitro withSalmonella strain SL7207 carrying either the pCMV-βgal (middle panels) or the pCMV-HFE (lower panels) plasmid. Results from a representative experiment in which cells were analyzed for gene expression by FACS 48 hours after infection using monoclonal antibodies against major histocompatibility class I, HFE, CD71, and βgal followed by a PE-conjugated anti-murine IgG polyclonal antibody. Appropriate isotype-matched monoclonal antibody controls have been used for instrument settings (empty histograms). Detection of βgal expression was performed after saponin-mediated permeabilization of monocytes. Percentage of positive cells and mean values of fluorescence (in brackets) are reported for each single histogram. Basal expression level for each marker in untreated HC–monocytes is also shown (upper panel).
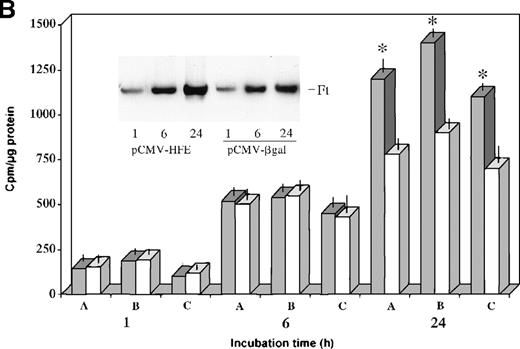
In vitro differentiated HC macrophages showed a significantly lower accumulation of iron than macrophages from normal controls (Figure 4A). To evaluate the effect of HFE reconstitution on iron metabolism in HC macrophages 24 hours after Salmonella infection, HC cells were incubated for an additional 24 hours in the presence of 55Fe-Tf. The HFE-transduced HC cells showed a 40% to 60% increase in the accumulation of 55Fe delivered by55Fe-Tf compared with HC cells transduced with the control vector (Figure 4B). Reconstitution of HC macrophages with wild-type HFE also led to expansion of the iron–ferritin pool, which is inappropriately low in HC macrophages9(Figure 4B, inset). Densitometric evaluation of radioactive ferritin bands showed a 30% to 45% increase in 4 separate experiments after normalization to protein content. No differences in cell growth rate (and protein content) were detected in HFE-transduced cells compared with cells transduced with the control plasmid.
Transferrin–iron incorporation in HC macrophages transfected with HFE.
(A) HC (solid bars) and normal (empty bars) macrophages were incubated for 24 hours in the presence of 55Fe-Tf and cell-associated radioactivity measured at the indicated time points. Data are mean ± SD from 6 separate experiments performed in triplicate.*P < .001 versus HC macrophages. (B) HC macrophages transduced with the pCMV-HFE (solid bars) or the pCMV-βgal (empty bars) plasmids were incubated for 24 hours in the presence of 55Fe-Tf. Cell-associated radioactivity was measured at the indicated incubation time points. Data are mean ± SD of experiments in macrophages from 3 subjects with HC (A, B, C) performed in triplicate, representative of 8 separate experiments in HC macrophages. *P < .001 versus HC macrophages transduced with control vector. (Inset) Time-course of intracellular55Fe-ferritin accumulation in HC macrophages transduced with the pCMV-HFE or the pCMV-βgal plasmid as evaluated by nondenaturing PAGE of pooled cell lysates coupled with55Fe-autoradiography. Ft, ferritin.
Transferrin–iron incorporation in HC macrophages transfected with HFE.
(A) HC (solid bars) and normal (empty bars) macrophages were incubated for 24 hours in the presence of 55Fe-Tf and cell-associated radioactivity measured at the indicated time points. Data are mean ± SD from 6 separate experiments performed in triplicate.*P < .001 versus HC macrophages. (B) HC macrophages transduced with the pCMV-HFE (solid bars) or the pCMV-βgal (empty bars) plasmids were incubated for 24 hours in the presence of 55Fe-Tf. Cell-associated radioactivity was measured at the indicated incubation time points. Data are mean ± SD of experiments in macrophages from 3 subjects with HC (A, B, C) performed in triplicate, representative of 8 separate experiments in HC macrophages. *P < .001 versus HC macrophages transduced with control vector. (Inset) Time-course of intracellular55Fe-ferritin accumulation in HC macrophages transduced with the pCMV-HFE or the pCMV-βgal plasmid as evaluated by nondenaturing PAGE of pooled cell lysates coupled with55Fe-autoradiography. Ft, ferritin.
Discussion
Reticuloendothelial cells play a central role in iron metabolism. They process more than 80% of the iron entering the plasma each day and act as the main storage compartments for iron.18Although erythrocyte phagocytosis accounts for most of the iron acquired by these cells in vivo, TfR-mediated uptake may also occur in culture19 and in vivo.20 Iron metabolism in RE cells is altered in HC. In patients and in murine models of HC, the metal accumulates preferentially in parenchymal cells, and little iron is stored in RE cells despite the rise in total body iron.10,11 Indeed, inappropriately high activity of the iron regulatory protein, an intracellular sensor for iron, has been documented in macrophages and in duodenal cells of patients with HC, which suggests that these cells are actually iron deficient.9,21 It is of interest that the macrophages of the liver (Kupffer cells) and intestinal crypt cells are sites of strong HFE expression in tissue-staining experiments with HFE-specific antibodies (including JB-1, used in this study).16 22 The mechanism that underlies this defect in iron storage by RE cells in HC is unknown and led us to examine the effect of expressing wild-type HFE in macrophages from patients with HC. We found that expression of HFE enhanced iron and ferritin accumulation in these cells, detected after exposure to 55Fe-Tf. In addition, we observed a small reduction in surface expression of TfR consistent with iron accumulation within the cells.
Recent work7,8,23,24 has shown that HFE can bind to TfR and inhibit the binding of Fe-Tf. HFE might, therefore, be expected to inhibit iron uptake by cells. Experiments involving the transfection of HFE into HeLa cells in vitro23 certainly confirm that under these conditions, the expression of HFE results in reduced uptake of Fe-Tf and in a detectable reduction in cellular ferritin content. This is the exact opposite of our results with HC macrophages, and it suggests that HFE may serve a function in specialized RE cells different from that detected in established cell lines in vitro.
The biochemical basis for the effect of HFE that we observed is as yet unknown. We can speculate that it could be mediated by either enhanced uptake of iron through the TfR cycle or by reduced egress of iron from the cell. Our observation that an appreciable effect on iron accumulation was seen only after a 24-hour exposure of macrophages to55Fe-Tf would be more compatible with an inhibitory effect of HFE on the egress of iron. This is also consistent with the enhanced release of iron observed in macrophages isolated from patients with HC.25 It has been estimated that macrophages in vivo deliver 10 million atoms of iron each minute to circulating transferrin,18 but the mechanism for this movement of iron out of the macrophage is unknown. Recently, a novel transporter of Fe2+ ions, Ferroportin1, was shown to be expressed in mature duodenal enterocytes, where it may mediate the transport of iron across the basolateral membrane.26 Ferroportin1 was also expressed in Kupffer cells (macrophages) in the liver,26 a known site of iron storage and HFE expression,16 where it may mediate iron egress. It is tempting to speculate that a function of HFE or the HFE-TfR complex is to counterbalance (or to directly inhibit?) the function of Ferroportin1 in RE macrophages and thereby promote iron storage. The loss of HFE expression in HC would result in an RE system incontinent of iron. In support of this conclusion, spleens from HFE knock-out mice are relatively resistant to dietary iron loading, possibly reflecting decreased accumulation of Tf-bound iron by the HFE−/−splenic macrophages.27 Because the iron content of RE cells may have a role in controlling intestinal iron absorption through a negative feedback mechanism exerted on enterocytes,18 20 the chronically iron-deficient RE cells in HC may determine or contribute to enhanced intestinal iron absorption.
Acknowledgment
We thank Prof Alain Townsend for reading the manuscript and offering helpful comments.
Supported by grants E609 and A102 from Comitato Telethon Fondazione Onlus and by a grant from Cofinanziamento MURST.
Reprints:Antonello Pietrangelo, Unit for the Study of Disorders of Iron Metabolism, Department of Internal Medicine, Policlinico, Via del Pozzo 71, 41100 Modena, Italy; e-mail:pietra@unimo.it.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.