Abstract
Polymerase chain reaction genotyping of 32 unrelated Jknull individuals originating predominantly from Polynesia and Finland indicated that all were homozygous for the JK*Bpolymorphism and that 17 of 32, including the 14 Polynesians, carried a 3′-acceptor splice site mutation of intron 5 that resulted in the skipping of exon 6 (called mutation JkΔ6). The remaining 15 Jknull donors from Finland were homozygous for a new T871C transition resulting in a S291P amino acid substitution at a consensusN-glycosylation site of the Jk polypeptide. Transcription-translation assays revealed that the Jk(S291P) mutant was translated into a glycosylated component as efficiently as the wild-type Jk polypeptide (wt Jk)] in the presence of microsomes, thus indicating that the S291P mutation has no effect on theN-glycosylation pattern of the Jk protein. Expression studies in Xenopus oocytes revealed that the Jk(S291P) polypeptide functions as a urea transporter, but the transport activity and the membrane expression level of the mutant protein was reduced to a similar extent. A substantial fraction of the mutant protein was retained intracellularly suggesting that the transit to the plasma membrane was reduced, presumably because of the S→P mutation. After transfection in erythroleukemia K562 cells the wild-type, but not the mutant, protein was efficiently expressed at the cell surface. Because the Jk(S291P) mutant polypeptide was not present in human red cells from Jknull individuals, expression data in the erythroid context clearly indicates that the S→P mutation is the molecular basis of the Finnish Jknull phenotype.
Introduction
The Kidd (JK) blood group antigens are carried by a multispanning red blood cell (RBC) membrane glycoprotein of 45-kd protein recently identified as the erythroid urea transporter,1,2 although it is also expressed in the kidney.3,4 The 2 major codominant alleles of theJK gene, JK*A and JK*B, have a similar frequency in Caucasian populations (0.51 and 0.49, respectively)5,6 and define the 3 common phenotypes Jk(a+b−), Jk(a−b+), Jk(a+b+). A rare null phenotype, called Jk(a−b−) or Jknull, in which the RBCs lack all Jk antigen and the Jk protein,5 was first described by Pinkerton and colleagues.7 The Jknull phenotype may result from 2 different genetic backgrounds: (1) homozygous inheritance of a “silent” Jk allele at the JK locus and (2) inheritance of a dominant inhibitor gene In(Jk), unlinked to the JK locus.5 6
Many individuals of Asian and Polynesian extraction have been identified as Jknull.8 Other populations representing this rare phenotype include ethnic groups from Brazil,9 India,10 and Japan.11However, the Jknull phenotype is strikingly absent from Caucasians although cases have been found in France,12Australia,13 and Finland where it was first described in 1984.14
The Jknull phenotype is not associated with any obvious clinical syndrome, except for a urine-concentrating defect,15 which probably results from the absence of Kidd/urea transporter protein on endothelial cell of the vasa recta in the inner and outer medulla of the kidney.3,4 Although it has been postulated that the erythrocyte Kidd/urea transporter might prevent the swelling of RBCs when they leave the hyperosmotic medulla,16 no hemolysis has been reported in Jknull individuals. Jknull RBCs, however, are easily identified by an increased resistance to lysis in aqueous 2 M urea solution,17 a property widely used to screen for individuals of Jknull phenotype.3 4
Recently, we have shown that the JK gene is composed of 11 exons spreading over 30 kb of DNA19 and that theJK*A/JK*B polymorphism resulted from a G838A transition, causing a D280N amino acid substitution in the 3rd extracellular loop of the Jk polypeptide.20 Moreover, 2 different splice-site mutations were identified in 2 unrelated Jknull individuals homozygous for the JK*B polymorphism. In the Chinese variant B.S., the mutation affected the invariant G residue of the 3′-acceptor splice site of intron 5, the Jk(Δ6) mutation, whereas in the French variant L.P., the mutation affected the invariant G residue of the 5′-donor splice site of intron 7, the Jk(Δ7) mutation.19The Jk(Δ6) and Jk(Δ7) mutations cause the skipping of exons 6 and 7 of the JK gene, respectively, and the corresponding truncated proteins neither mediated a facilitated urea transport inXenopus oocytes nor were they expressed on the oocyte's plasma membrane, thus providing a rational explanation for the lack of Kidd/urea transporter protein on the RBC surface.19
In this report, we have further analyzed 32 samples of unrelated Jknull donors originating mainly from Polynesia and Finland to determine their molecular basis. All Polynesian samples exhibited the Jk(Δ6) mutation, whereas each of the Finnish samples exhibited an undescribed missense mutation of the Jk polypeptide, the expression and function of which were analyzed in Xenopus.
Materials and methods
Blood samples and reagents
The Jknull blood samples in this study were obtained from Polynesian blood donors who reside in Auckland (New Zealand), from the Finnish Red Cross (Helsinki, Finland), from the Irwin Memorial Blood Center (San Francisco, CA) and from the Centre National de Référence sur les Groupes Sanguins (CNRGS, Paris, France). Some of these samples were initially identified by 2 M urea lysis screening,17 but all were identified as Jk(a−b−) by routine serology. Blood samples of common Jk phenotypes came from the Institut National de la Transfusion Sanguine (Paris, France) and all samples were collected on EDTA or heparin. The human serum anti-Jk3 was obtained from an immunized Jk(a−b−) individual. Human polyclonal anti-Jka or anti-Jkbsera and monoclonal anti-Jka (IgM, clone MS15) and anti-Jkb (IgM, clone MS8) were from from Biotest AG (Dreieich, Germany). Restriction endonucleases and modifying enzymes came from New England Biolabs (Hertfordshire, UK). Radiolabeled nucleotides, l-[35S]-methionine (1.85 GBq/mmole) and [14C]-urea (1.96 GBq/mmol) were purchased from Amersham/Pharmacia Biotech (Bucks, UK) and [3H]-raffinose (188.7 GBq/mmole) from Dupont-NEN (Boston, MA). The Taq polymerase, the Expand High Fidelity PCR and Titan 1 tube RT-PCR systems from Boehringer-Mannheim/Roche Diagnostics (Germany) were used for polymerase chain reaction (PCR) amplification. Nucleotide sequences were determined on both strands by the dideoxychain termination method (Sanger) with ThermoSequenase fluorescent-labeled primer cycle sequencing Kit from Amersham Pharmacia Biotech using 5′(Cy5)-primers (Genset, France).
Amplification by reverse transcription coupled with PCR (RT-PCR)
Total blood RNA extracted by the acid-phenol-guanidinium method21 was used in the Titan 1 tube RT-PCR system (50°C for 30 minutes [1 cycle]; 94°C for 30 seconds, 64°C for 30 seconds, 68°C for 2 minutes [30 cycles]) between primers SP-1 and AS-2 (Table 1) according to the manufacturer's instructions. The second PCR was performed with 1/25 of the first PCR reaction in the same conditions except the first step, using primers SP-1 and AS-3, and enzyme mix (Taq and Pwo DNA polymerase) using the Expand High Fidelity system. The final PCR product was identified by Southern blot analysis, subcloned into theEcoRV-digested pT7TS plasmid (kindly provided by P. Krieg, Austin, TX), and sequenced on both strands using an automated Alf-Express sequencer (Parmacia, Uppsala, Sweden).
Genomic DNA analysis by PCR
The PCR genotyping assays were developed to detect the mutations that cause the basis of Jknull phenotypes. To identify the Jk(Δ6) variant, an allele-specific PCR reaction was performed between primers SP-4, specific of the mutated allele, and AS-5 under stringent conditions (94°C for 2 minutes [1 cycle]; 94°C for 30 seconds, 64°C for 30 seconds, 72°C for 30 seconds [30 cycles]; 72°C for 2 minutes [1 cycle]) using Taq polymerase. The 221-bp PCR product was electrophoresed on a 1.8% (w/v) agarose gel stained with ethidium bromide. To identify the Jk(Δ7) variant, a PCR-restriction fragment length polymorphism (PCR-RFLP) was carried out by a hemi-nested PCR reaction (94°C for 2 minutes [1 cycle]; 94°C for 30 seconds, 54°C for 30 seconds, 72°C for 20 seconds [30 cycles]; 72°C for 2 minutes [1 cycle]) using the Expand High Fidelity system. The first amplification was performed between primers SP-6 and AS-8 and the second between primers SP-7 and AS-8 with 1/50 of the first reaction in the same experimental conditions. The 76-bp PCR product was digested with 5 U of Mse I restriction enzyme and electrophoresed on a 15% acrylamide gel stained with ethidium bromide. To identify the Finnish Jk(S291P) variants, a PCR-RFLP was performed between primers SP-9 and AS-10 in the same conditions as above except that annealing was performed at 58°C. The 81-bp PCR product was digested with 5 U ofMnl I restriction enzyme and analyzed as previously on a 15% acrylamide gel. Each PCR reaction was performed in a total volume of 50 μL containing 250 ng genomic DNA extracted from whole blood cells using a Wizard Genomic DNA Purification kit from Promega (Madison, WI). All primers used are given in Table 1.
Site directed mutagenesis of the Jk complementary DNA (cDNA)
A nucleotide substitution A632T (resulting in a N211I substitution in the Jk polypeptide) was introduced into the wt Jk cDNA using a 2-step PCR approach with common SP-1 and AS-3 primers, in addition to sequence specific mutagenic primers overlapping each other, AS-11 and SP-12. Annealing was done at 60°C for the second PCR step. After subcloning into the EcoRV-digested pT7TS plasmid, the mutant construct was sequenced on both strands to confirm that the correct mutation has been obtained.
Transcription-translation assays
Polypeptides corresponding to wt Jk, Jk(S291P), and Jk(N211I) were in vitro synthesized from each pT7TS cDNA constructs (see below) using the transcription-translation coupled reticulocyte lysate kit from Promega in the presence ofl-[35S]-methionine and with or without canine pancreatic microsomal membranes as recommended by the manufacturer. One tenth of the reaction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 15% separating gel on a discontinuous buffer system,22 followed by enlightening treatment (NEN, Boston, MA) and autoradiography.
Oocyte urea flux measurements and immunocytochemistry
After linearization of the pT7TS-wt Jk or −Jk(S291P) cDNA constructs with SmaI restriction enzymes, capped sense RNAs were synthesized using T7 RNA polymerase from the mCAP mRNA capping kit (Stratagene, La Jolla, CA). Expression studies were carried out by microinjection of complementary RNAs (cRNAs) (0.1 ng/oocyte) in collagenase-treated Xenopus laevi oocytes23 and functional tests were realized 3 days after injection as described previously.24 In parallel, groups of 3 to 6 oocytes without chorionic membrane were embedded in paraffin as described.25 Sections (7 μm thickness) were stained overnight at 4°C with 10 μg/mL of affinity purified polyclonal antibody directed against the N-terminal region (residues 8-22) of the Kidd/urea transporter protein, described previously,1,3 and visualized with fluorescein-conjugated goat antirabbit IgG (1:100 dilution) for 1 hour at room temperature.26 Immunostaining of the oocyte plasma membrane was imaged and quantified using a Nikon Eclipse TE300 microscope with epifluorescence illumination (Nikon, Paris, France) coupled to a Biocom computer system of image integration (Visiolab 2000 program, Biocom, Les Ulis, France).
Northern blot analysis
Total RNAs isolated from 6 injected oocytes, according to Chomezynski and Sacchi,27 were resolved by electrophoresis on 6% (w/v) formaldehyde, 1% (w/v) agarose gel, and transferred to nylon filters (Zeta-probe GT, Bio-Rad, Hercules, CA). Hybridization with a [32P]-labeled Jk cDNA probe (1.2 kb) using the random primed DNA labeling kit (Boehringer-Mannheim, Germany) was carried out at 65°C in 0.25 mol/L Na2HPO4,7% (wt/vol) SDS. Stringent washes were performed in 0.02 mol/L Na2HPO4, 1% (wt/vol) SDS at 65°C for 30 minutes and exposed to Biomax-MR film with intensifying screen at −80°C.
Cell culture, transfection, and flow cytometry
Human erythroleukemic K562 cells were obtained from the American Type Culture Collection (Manassas, VA) and were grown in Iscove's modified Dulbecco medium with Glutamax-1 (IMDM, Gibco BRL, Eragny, France) supplemented with penicillin-streptomycin and 10% fetal calf serum (FCS). To express wt Jk and Jk(S291P) polypeptides in K562 cells, wt Jk and Jk(S291P) cDNAs obtained by PCR between SP-1 and AS-3 primers (Table 1) were subcloned into pCEP4 episomal expression vector (Invitrogen, Leek, The Netherlands). After transfection using the Lipofectin-Reagent according to the manufacturer's instructions (Life Technologies, Gaithersburg, MD), stable K562 transfectants resistant to hygromycin (0.3 mg/mL) were selected for Jk protein expression by immunomagnetic separation using human anti-Jk3 antiserum and Biomag goat antihuman-IgG (Perseptive Biosystems, Connecticut, MA). The expression of Kidd/urea transporter polypeptides was examined by flow cytometry (FACScan, Becton Dickinson, San Jose, CA) using cultured cells (3-5 × 105) incubated 60 minutes at 22°C with human polyclonal or monoclonal anti-Jkb and anti-Jka at saturating concentration. The cells were then washed and stained with 100 μL of phycoerythrin (PE)-conjugated F(ab′)2 fragments of goat antihuman IgG diluted 1:40 (Coulter/Immunotech, Marseille, France). After another washing step, 20 nmol/L of TO-PRO-1, molecular probes (Interchim, Monluçon, France), was added to the cell suspension 15 minutes before analysis, to exclude dead cells (TO-PRO positive cells).
Western blot analysis
The RBC membranes from donors of known phenotypes were prepared by hypotonic lysis.28 For Western blot analysis, RBC membrane proteins were separated by discontinuous SDS-PAGE, transferred to nitrocellulose sheets (Schleicher and Schull, Keene, NH; 0.1 μm) and incubated for 90 minutes at 37°C with 10 μg/mL of an affinity-purified rabbit antibody raised against the Kidd/urea transporter protein (see above). Bound antibodies were visualized with alkaline phosphatase-labeled goat antirabbit IgG 1:800 and the alkaline phosphatase substrate kit (BioRad).
Results
Detection of mutations in JK gene from Jknulldonors
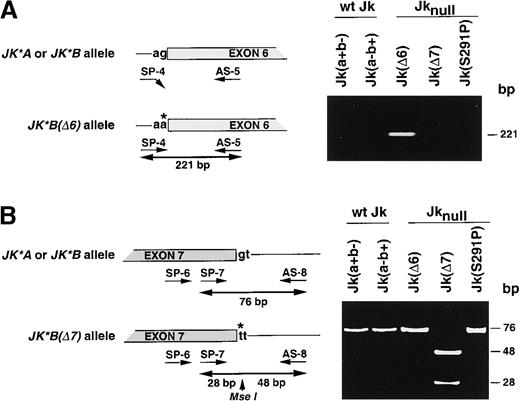
To determine the molecular defect occurring in Jknullphenotypes of 32 unrelated donors, among whom 14 originated from Polynesia, 15 from Finland, and 3 from diverse origins (Australia, Europe, and United States), we first used PCR-RFLP assays to screen for the JK*A/JK*B polymorphism20 and developed PCR genotyping assays for the 2 splice-site mutations previously identified in 2 Jknull individuals.19 To detect the Jk(Δ6) mutation, an allele specific-PCR (AS-PCR) assay was performed between the SP-4 primer, specific for the mutated JK*Ballele, and the common antisense AS-5 primer (Figure1A). Homozygosity for this mutation was determined by sequencing 5′-intron exon 6 boundaries as previously described.19 In parallel, a hemi-nested PCR-RFLP assay was developed to detect the Jk(Δ7) mutation (Figure 1B). The results of the genotyping are given in Table 2.
Identification of JK gene mutations in Jknull variants.
(A) Left, schematic representation of the Jk(Δ6) DNA genotyping assay by allele-specific PCR using primers SP-4 and AS-5. The invariant G nucleotide at the acceptor splice-site of intron 5 in JK*Aor JK*B alleles is replaced by an A in the Jk(Δ6)null allele. Right, the 221-bp PCR product was amplified from the Jk(Δ6) mutant but not from the wt Jk, Jk(a+b−) or Jk(a−b+), Jk(Δ7), and Jk(S291P) samples demonstrating the specificity of the assay. (B) Left, schematic representation of the Jk(Δ7) DNA genotyping assay by PCR-RFLP using primers SP-6, SP-7, and AS-8. The invariant G nucleotide at the donor splice-site of intron 7 inJK*A or JK*B alleles is replaced by a T in theJk(Δ7) null allele, which creates a Mse I restriction site. Right, the 76-bp PCR product encompassing the splice-site mutation was cleaved by Mse I into 48- and 28-bp fragments in the Jk(Δ7) mutant. As expected the PCR products from wt Jk, Jk(a+b−) or Jk(a−b+), Jk(Δ6), and Jk(S291P) samples were uncut. Single base substitution and intronic sequence are shown by a star and in lower case, respectively. Size of fragments (bp) are given on the right.
Identification of JK gene mutations in Jknull variants.
(A) Left, schematic representation of the Jk(Δ6) DNA genotyping assay by allele-specific PCR using primers SP-4 and AS-5. The invariant G nucleotide at the acceptor splice-site of intron 5 in JK*Aor JK*B alleles is replaced by an A in the Jk(Δ6)null allele. Right, the 221-bp PCR product was amplified from the Jk(Δ6) mutant but not from the wt Jk, Jk(a+b−) or Jk(a−b+), Jk(Δ7), and Jk(S291P) samples demonstrating the specificity of the assay. (B) Left, schematic representation of the Jk(Δ7) DNA genotyping assay by PCR-RFLP using primers SP-6, SP-7, and AS-8. The invariant G nucleotide at the donor splice-site of intron 7 inJK*A or JK*B alleles is replaced by a T in theJk(Δ7) null allele, which creates a Mse I restriction site. Right, the 76-bp PCR product encompassing the splice-site mutation was cleaved by Mse I into 48- and 28-bp fragments in the Jk(Δ7) mutant. As expected the PCR products from wt Jk, Jk(a+b−) or Jk(a−b+), Jk(Δ6), and Jk(S291P) samples were uncut. Single base substitution and intronic sequence are shown by a star and in lower case, respectively. Size of fragments (bp) are given on the right.
All 32 Jknull samples were found homozygous for theJK*B polymorphism using the Mnl I PCR-RFLP previously described20 (data not shown). A 221-bp AS-PCR product characteristic of the Jk(Δ6) mutation was amplified from 17 of the 32 Jknull samples, as well as from variant B.S. used as control, but no product was amplified from Jk(a+b−) and Jk(a−b+) donors (collectively called wt Jk hereafter), from the variant L.P. carrying the Jk(Δ7) mutation, or from the 15 Finnish Jknull, which carry the S291P mutation (Figure 1A, Table2). Nucleotide sequencing further indicated that these 17 Jknull donors, including the 14 Polynesians, were homozygous for the mutation affecting the invariant G residue of the 3′ acceptor splice-site of intron 5 of JK gene, as observed in the variant B.S. (data not shown). PCR-RFLP assays for the Jk(Δ7) mutation showed that the 76-bp PCR product remained uncut in all samples tested, including wt Jk control donors, except for sample L.P. carrying the Jk(Δ7) mutation, in which the PCR product was cleaved into 2 fragments of 48 and 28 bp (Figure 1B, Table 2). These data provided a molecular explanation for 17 of the 32 Jknullsamples and raised the possibility of a new mutation occurring in theJK gene in the Finnish samples.
Analysis of the Jk transcripts in Finnish Jknulldonors
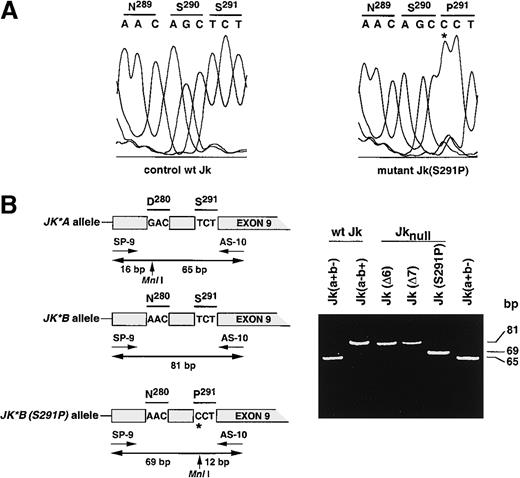
Total reticulocyte RNA isolated from one Finnish Jknull sample was used as template to amplify by hemi-nested PCR the entire Jk cDNA using appropriate primer pairs as described in “Materials and methods.” A 1.2-kb amplified fragment was readily amplified and sequence analysis revealed a T871C nucleotide transition that resulted in a S291P amino acid substitution located in a potential N-glycosylation motif (Figure2A). Because the T871C substitution was correlated with the presence or the absence of an MnlI restriction site like the G838A transition, which caused theJK*A/JK*B polymorphism,20 a hemi-nested PCR-RFLP assay using primers SP-9 and AS-10 (Table 1) to test simultaneously the 2 positions was developed (Figure 2B). As expected, the 81-bp PCR product remained uncut in the Jk(a−b+) control and Jk(Δ6) and Jk(Δ7) mutants, whereas it was cleaved into 2 fragments of 65 and 16 bp in the control Jk(a+b−) sample. In contrast, 2 fragments of 69 and 12 bp were observed with 15 Finnish Jk(S291P) mutants, demonstrating the homozygosity of the mutation in these donors (Figure 2B, Table 2).
Identification of Jk transcript mutation in the Finnish Jknull variant.
(A) Partial sequence analysis of the Jk transcript isolated from a wt Jk donor and from Finnish Jknull variant, Jk(S291P), within exon 9. The 2 Jk transcripts differ by a single base substitution T871C (star) changing serine (S) to proline (P) at position 291 in the Jk polypeptide. The display of the sequence diagram is from the Alf-Express DNA sequencer. (B) Left, schematic representation of the Jk(S291P) DNA genotyping assay by PCR-RFLP using primers SP-9 and SP-10. Exon 9 carries 2 polymorphisms that create 2 Mnl I sites, the first at position 838 corresponds to theJK*A/JK*B polymorphism, and the second at position 871 corresponds to the JK*B (S291P) null allele. Right, the 81-bp PCR product encompassing the 2 polymorphisms was cleaved byMnl I into 69- and 12-bp fragments in the Jk(S291P) mutant. As expected, the products from Jk(a−b+), Jk(Δ6), and Jk(Δ7) samples were uncut, whereas the product from Jk(a+b−) was cut into 16- and 65-bp fragments. Only the upper part of the gel containing the higher fragments is shown. Single base substitution is located by a star and size of fragments (bp) are given on the right.
Identification of Jk transcript mutation in the Finnish Jknull variant.
(A) Partial sequence analysis of the Jk transcript isolated from a wt Jk donor and from Finnish Jknull variant, Jk(S291P), within exon 9. The 2 Jk transcripts differ by a single base substitution T871C (star) changing serine (S) to proline (P) at position 291 in the Jk polypeptide. The display of the sequence diagram is from the Alf-Express DNA sequencer. (B) Left, schematic representation of the Jk(S291P) DNA genotyping assay by PCR-RFLP using primers SP-9 and SP-10. Exon 9 carries 2 polymorphisms that create 2 Mnl I sites, the first at position 838 corresponds to theJK*A/JK*B polymorphism, and the second at position 871 corresponds to the JK*B (S291P) null allele. Right, the 81-bp PCR product encompassing the 2 polymorphisms was cleaved byMnl I into 69- and 12-bp fragments in the Jk(S291P) mutant. As expected, the products from Jk(a−b+), Jk(Δ6), and Jk(Δ7) samples were uncut, whereas the product from Jk(a+b−) was cut into 16- and 65-bp fragments. Only the upper part of the gel containing the higher fragments is shown. Single base substitution is located by a star and size of fragments (bp) are given on the right.
Because the Finnish samples carry a single missense mutation (S291P), we looked for a putative mutation in the promoter region of theJK gene. Amplification and sequencing of the proximal promoter (500 bp) upstream of the erythroid transcription initiation site of the JK gene did not reveal any alteration (data not shown).
N-glycosylation of the Kidd/urea transporter protein
In vitro transcription-translation coupled assays and ex vivo functional studies in Xenopus oocytes were performed to determine the effect of the missense mutation S291P on the expression level and functional properties of the Jk mutant polypeptide.
In the cell-free transcription-translation coupled system, the mutant plasmid pT7TS-Jk(S291P) directed the synthesis of a polypeptide of apparent molecular mass of 36 kd with equal efficiency as the wild-type pT7TS-Jk control plasmid (Figure 3). In the presence of microsomal membranes, a minor band at 36 kd and a major band at 40 kd corresponding to the glycosylated 36-kd product were detected with both plasmids, thus suggesting that the glycosylation pattern of the Jk(S291P) protein mutant was not affected. To substantiate this point, we performed a site-directed mutagenesis of asparagine 211 (N211I) in the consensus N-glycosylation motif predicted to be exposed on the third extracellular loop of the Jk polypeptide and which most likely is glycosylated.24 As expected, the mutant plasmid pT7TS-Jk(N211I) directed only the synthesis of the nonglycosylated 36-kd protein in the presence or absence of microsomal membranes (Figure 3A). No protein material was translated using the pT7TS plasmid alone.
Expression of Jk proteins in transcription-translation coupled reticulocyte lysate system.
(A) Autoradiogram ofl-[35S]-methionine-labeled proteins wt Jk, Jk(S291P), and Jk(N211I) in vitro synthesized from pT7TS-wt Jk, pT7TS-Jk(S291P), and pT7TS-Jk(N211I) vector constructs, respectively, and analyzed by SDS-PAGE. Transcription-translation reactions were done with or without canine pancreatic microsomal membranes as indicated. The pT7TS vector alone was used as negative control. Arrows on the right refer to size of products (kd).
Expression of Jk proteins in transcription-translation coupled reticulocyte lysate system.
(A) Autoradiogram ofl-[35S]-methionine-labeled proteins wt Jk, Jk(S291P), and Jk(N211I) in vitro synthesized from pT7TS-wt Jk, pT7TS-Jk(S291P), and pT7TS-Jk(N211I) vector constructs, respectively, and analyzed by SDS-PAGE. Transcription-translation reactions were done with or without canine pancreatic microsomal membranes as indicated. The pT7TS vector alone was used as negative control. Arrows on the right refer to size of products (kd).
Expression and functional analysis in Xenopusoocytes
To test whether the Jk(S291P) mutant protein was functionally expressed as a urea transporter, the mutant cDNA carrying the T871C mutation was subcloned into the pT7TS vector and the cRNA transcript was synthetized and injected into Xenopus oocytes. The [14C]-urea uptake of Jk(S291P) cRNAs-injected oocytes was significantly increased when compared to water-injected control oocytes (Figure 4A), but was much lower when compared to the [14C]-urea uptake of wt Jk cRNA-injected oocytes (21.71 ± 0.43 versus 10.02 ± 1.66 pmol/oocyte at 1.5 minutes). The corresponding urea permeabilities calculated at 1.5 minutes were 53.0 ± 1.05 and 24.4 ± 4.05 × 10−6cm/s, respectively, versus 0.63 ± 0.14 × 10−6cm/s for water-injected control oocytes.
Expression and functional analysis of Jk proteins inXenopus oocytes.
(A) Time course of [14C]-urea uptake inXenopus oocytes injected with 0.1 ng cRNA encoding the wt Jk and Jk(S291P) polypeptides. As negative control, water-injected oocytes were used. The assay was initiated by suspending individual oocytes in 0.2 mL Barth solution containing 8 μCi/mL [14C]-urea (145 μmol/L) and 5 μCi/mL [3H]-raffinose. The urea uptake was stopped after the indicated time (t = 0, 1.5, 3, and 6 minutes) by addition of 3 mL of ice-cold Barth solution and followed by rapid washes with 2 × 5 mL of the same solution. Then, the oocyte-associated [14C]-radioactivity was determined as previously described.6 Six to 10 individual oocytes were counted for each time. Mean and SE from different experiments are shown. (B) Northern blot analysis of cRNA encoding wt Jk and Jk(S291P) in oocytes. Total oocyte RNAs (15 μg/lane) at the day of injection and 3 days after were separated and hybridized with the [32P]-labeled Jk cDNA probe, as described in “Materials and methods.” Equal loading and absence of degradation were checked by staining with ethidium bromide. Autoradiography was for 1 hour at −80°C. (C) Immunostaining of oocyte sections. Oocytes from the same batches were fixed and sections of injected oocytes were stained with an affinity-purified antibody against the N-terminus of the Kidd/urea transporter protein and visualized with FITC-conjugated anti-rabbit IgG as described in Materials and methods and then imaged using Nikon Eclipse TE300 microscope (Nikon, Paris, France). Images were taken with epifluorescence illumination (magnification, × 40) and treated with a Biocom informatic system of image integration.
Expression and functional analysis of Jk proteins inXenopus oocytes.
(A) Time course of [14C]-urea uptake inXenopus oocytes injected with 0.1 ng cRNA encoding the wt Jk and Jk(S291P) polypeptides. As negative control, water-injected oocytes were used. The assay was initiated by suspending individual oocytes in 0.2 mL Barth solution containing 8 μCi/mL [14C]-urea (145 μmol/L) and 5 μCi/mL [3H]-raffinose. The urea uptake was stopped after the indicated time (t = 0, 1.5, 3, and 6 minutes) by addition of 3 mL of ice-cold Barth solution and followed by rapid washes with 2 × 5 mL of the same solution. Then, the oocyte-associated [14C]-radioactivity was determined as previously described.6 Six to 10 individual oocytes were counted for each time. Mean and SE from different experiments are shown. (B) Northern blot analysis of cRNA encoding wt Jk and Jk(S291P) in oocytes. Total oocyte RNAs (15 μg/lane) at the day of injection and 3 days after were separated and hybridized with the [32P]-labeled Jk cDNA probe, as described in “Materials and methods.” Equal loading and absence of degradation were checked by staining with ethidium bromide. Autoradiography was for 1 hour at −80°C. (C) Immunostaining of oocyte sections. Oocytes from the same batches were fixed and sections of injected oocytes were stained with an affinity-purified antibody against the N-terminus of the Kidd/urea transporter protein and visualized with FITC-conjugated anti-rabbit IgG as described in Materials and methods and then imaged using Nikon Eclipse TE300 microscope (Nikon, Paris, France). Images were taken with epifluorescence illumination (magnification, × 40) and treated with a Biocom informatic system of image integration.
To test for a difference in stability between the wild-type and mutant injected cRNAs, total RNA was isolated from oocytes of the same batches and analyzed by Northern blot, which showed that similar levels of wild-type and mutant cRNAs were detectable 3 days after injection (Figure 4B).
To localize the Jk(S291P) protein in oocytes, immunohistochemistry was performed on oocytes injected with water or cRNAs (Figure 4C). Staining with an affinity-purified antibody directed against theN-terminus of the Kidd/urea transporter protein revealed that the wt Jk protein was clearly expressed at the plasma membrane of oocytes, whereas oocytes expressing the mutant Jk(S291P) protein exhibited a weaker staining of the plasma membrane, with an increased cytoplasm reactivity (Figure 4C). Water-injected controls were negative, showing the specificity of antibody labeling. To provide a quantitative estimation of the plasma membrane staining of wild-type and mutant proteins, densitometric analysis of plasma membrane sections of 6 oocytes (4 fields analyzed per oocyte) for each type of cRNA injected was performed, based on the finding that the fluorescent signal is proportional to the amount of cRNA injected.29Using the Visiolab 2000 program from Biocom, a 70% reduction of membrane insertion of the mutant protein was calculated, as the values for wild-type and mutant proteins were 187.63 ± 7.68 and 78.80 ± 1.58, respectively, versus 31.75 ± 0.64 for water-injected control oocytes (arbitrary units at magnification 20).
Expression of the Jk(S291P) protein in human erythroleukemic K562 cells
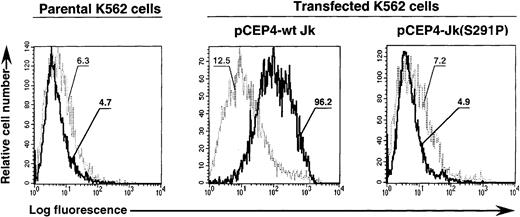
Expression vectors containing the wild-type and mutant cDNAs were transfected in K562 cells as described in “Materials and methods.” After drug selection, flow cytometry analysis indicated that the human monoclonal (and human serum) anti-Jkb strongly reacted with K562 cells transfected with the wild-type but not the mutant cDNA construct (Figure 5). Anti-Jka (polyclonal or monoclonal) did not react with parental or transfected K562 cells (data not shown). The geometric mean ratios were 7.69 and 0.68 for K562 cells transfected with wild-type and mutant constructs (versus 0.74 for parental cells), respectively, indicating that only wild-type construct directed efficient Jk cell surface protein expression in an erythroid context.
Expression of wt Jk and Jk(S291P) polypeptides in K562 transfectants.
K562 cells transfected with pCEP4-wt Jk or Jk(S291P) cDNA constructs were stained with the human monoclonal antibody (MoAb) anti-Jka (1:2 dilution) used as control (light curve) and the human MoAb anti-Jkb (diluted 1:2 in phosphate-buffered saline) (heavy curve), followed by PE-goat antihuman IgG (Fab′2 fragments) and analyzed by flow cytometry. The log of the fluorescence intensity is shown on the abscissa and the cell number on the ordinate. The geometric means were reported for each population analyzed.
Expression of wt Jk and Jk(S291P) polypeptides in K562 transfectants.
K562 cells transfected with pCEP4-wt Jk or Jk(S291P) cDNA constructs were stained with the human monoclonal antibody (MoAb) anti-Jka (1:2 dilution) used as control (light curve) and the human MoAb anti-Jkb (diluted 1:2 in phosphate-buffered saline) (heavy curve), followed by PE-goat antihuman IgG (Fab′2 fragments) and analyzed by flow cytometry. The log of the fluorescence intensity is shown on the abscissa and the cell number on the ordinate. The geometric means were reported for each population analyzed.
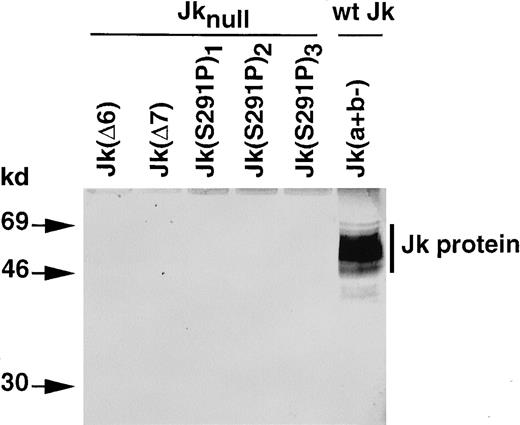
Jk protein from common and Jknull RBCs
Investigation by Western blot analysis indicated that the Jk protein was absent from RBC membrane preparations of all Jknull samples investigated (Figure 6). Indeed, the affinity-purified antibody that recognizes theN-terminus of the Jk protein reacted with a diffuse band of 46 to 69 kd present in membrane proteins from donors with a wt Jk phenotype, Jk(a+b−), but did not react with membrane proteins from the Jknull mutants Jk(Δ6), Jk(Δ7), and Jk(S291P).
Immunoblot analysis of Kidd/urea transporter protein from erythrocyte membranes of individuals with common and rare Jk phenotypes.
The RBC membrane proteins (40 μg) were separated by SDS-PAGE and immunoblotted with an affinity-purified rabbit antibody against theN-terminus of the Kidd/urea transporter protein, as described in Materials and methods. Size of products (kd) on the left.
Immunoblot analysis of Kidd/urea transporter protein from erythrocyte membranes of individuals with common and rare Jk phenotypes.
The RBC membrane proteins (40 μg) were separated by SDS-PAGE and immunoblotted with an affinity-purified rabbit antibody against theN-terminus of the Kidd/urea transporter protein, as described in Materials and methods. Size of products (kd) on the left.
Discussion
Here we report the molecular basis of 32 unrelated Jknull samples, most originating from Polynesia and Finland. All were homozygous for the JK*B polymorphism. These 2 countries have been selected for their genetic homogeneity and a high incidence of the Jknull phenotype. Indeed, the populations within Polynesia are known to be genetically remarkably homogenous given the wide geographic region they inhabit.30 This homogeneity may have been the result of an initial founder effect, and later bottlenecks, coupled with an ongoing network of contact that existed throughout Polynesia during and immediately following settlement.31 Despite the similarities between Polynesians, the incidence of some blood group phenotypes showed differences in frequency between islands. Thus, the ABO and Lewis phenotypes differed significantly32 as do the frequencies of the Jknull phenotypes.33This being so, our investigations showed that the 14 Jknulldonors from Polynesia resulted from the same mutation, Jk(Δ6), previously described in variant B.S of Chinese origin.19The molecular homogeneity found in the ethnic groups, Samoan, Niuean, Tongan, and Hawaiian, for the Jknull phenotype might have been the result of a common ancestor who carried the mutatedJK gene. Moreover, because the mutation has been initially described in the variant B.S. from China, this mutation supports also the scenario that assumes that the common ancestors of the Polynesians as Melanesians and Micronesians are primarily derived from Asia and Southeast Asia.30 Polynesians do show genetic affinities to the populations of Asia. There is a high frequency of the “Polynesian mtDNA motif” in the corridor from the south through the Philippines and east Taiwan.34 Moreover, this mutation is not limited to Southeast Asia and Polynesia, because it has also been found in Australian, American, and European donors (Table 2).
The Jknull phenotype is relatively common in Finland, having a frequency of about 0.03%35 (and unpublished data) and an even distribution over the whole country. All 15 samples examined exhibited a T871C transition of the JK*B allele, resulting in the missense mutation S291P of the Kidd/urea transporter protein. Because this amino acid substitution was located in a NSS motif at position 289 to 291, it was predicted that this potentialN-glycosylation site might be altered. The mere description of this mutation was described when this paper was under revision.36 However, neither functional nor glycosylation studies were carried out by the authors36 to relate the S291P mutation with the Jknull phenotype. Here, we have carried out such an analysis and at first we found that the wild-type and mutant Jk transcript typical of Finnish Jknull cells could be translated with an equal efficiency into major polypeptide species of 36 and 40 kd, as seen by transcription-translation analysis in the absence or in the presence of microsomal membranes, respectively. Further analysis by site-directed mutagenesis demonstrated that the asparagine at position 211 (located in the third predicted extracellular loop of the urea transporter24) of the wt Jk protein was the unique N-glycosylation site used. These findings indicate that the NSS motif at position 289 to 291 is not used as a glycosylation site, which correlates well with its predicted localization in the eighth transmembrane domain of the Jk polypeptide.24 Moreover, the complete absence ofN-glycan on the mutant Jk(N211I) polypeptide indirectly demonstrates that the 3rd potential N-glycosylation site at position 125 to 127, which is located in the 3rd transmembrane domain of the Jk protein, is not used.
Next, we found that the mutant Jk(S291P) protein behaves as a functional urea transporter when expressed in Xenopusoocytes, but the urea permeability of oocytes expressing this protein was 50% lower than that of oocytes expressing the wt Jk protein, although the 2 injected cRNAs were translated in vitro with equal efficiency and had a similar stability for a 3-day period. Immunostaining of paraffin-embedded sections of oocyte membranes expressing the mutant Jk(S291P) and the wt Jk polypeptides, using an affinity-purified antibody to the N-terminus of the Jk protein, showed that there was less of the mutant protein than the wt Jk protein at the cell surface and more intracellularly. Quantitative staining analysis of oocyte membranes revealed that the reduced urea permeability mediated by the mutant Jk(S291P) protein is consistent with a reduced expression of the mutant protein at the oocyte plasma membrane, thus indicating that the specific urea transport activity of the wild-type and mutant Jk protein is similar. The mutant protein appears to be moving to the plasma membrane more slowly, presumably because of the destabilizing S→P mutation.
Oocyte studies clearly demonstrate that the missense mutation S291P does not alter the urea transport properties of the mutant protein, but they do not explain the absence of mutant protein in human RBCs. To clarify this point cell surface expression of wild-type and mutant Jk proteins was investigated in an erythroid context. Using episomal expression vectors in K562 cells it was found that under forced conditions of expression the wild-type but not the mutant Jk(S291P) protein was efficiently expressed at the cell surface. Altogether, these results strongly suggested that the presence of the S→P mutation (located in the eighth predicted transmembrane domain of the Jk protein) prevents or drastically reduces surface expression in erythroid cells and is most likely responsible for the molecular basis of the Finnish Jknull phenotype, because no Jk protein is detected on RBCs from these individuals. Missense mutations presumably associated with the absence or severe reduction of plasma membrane expression have been described for other blood group protein mutants like those occurring in Rhnull (regulator type),37 weak D,38 or Fyxphenotypes.39 40
Our studies suggest that the oocyte expression system is more permissive than the mammalian cell system because a fraction of the mutant Jk protein could reach the cell surface and conferred urea permeability to the injected oocytes. A similar phenomenon has been shown for the common deleted form of the cystic fibrosis transmembrane conductance regulator (CFTR-ΔF508), which is retarded in endoplasmic reticulum and partially expressed at the plasma membrane,41 whereas this mutant protein was not found at the plasma membrane of mammalian cells.42 A difference with the expression machinery of oocytes has also been found for an insect aquaporin mutant AQPcic (AQP-C134S), which could be expressed in yeasts but not in Xenopus oocytes.43
In conclusion, 3 different mutations of the JK gene resulting in the Jknull phenotype have been currently found, which all can be identified by PCR-genotyping assays. The Jk(Δ6) mutation is widely spread among Polynesians and has been found occasionally in donors from the United States, Australia, and Europe. The Jk(Δ7) mutation, however, was found only in the variant L.P. of French origin.19 The missense mutation Jk(S291P) is associated with the Finns. Compared to the high incidence of the Jknull phenotype in Polynesian Islands and Finland, the next challenge will be to understand what pressures of selection and advantage, if any, drive the expansion of this phenotype in these 2 distinct countries.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jean-Pierre Cartron, Unité INSERM U76, INTS, 6 rue Alexandre Cabanel, 75015 Paris, France; e-mail: cartron@infobiogen.fr.

![Fig. 3. Expression of Jk proteins in transcription-translation coupled reticulocyte lysate system. / (A) Autoradiogram ofl-[35S]-methionine-labeled proteins wt Jk, Jk(S291P), and Jk(N211I) in vitro synthesized from pT7TS-wt Jk, pT7TS-Jk(S291P), and pT7TS-Jk(N211I) vector constructs, respectively, and analyzed by SDS-PAGE. Transcription-translation reactions were done with or without canine pancreatic microsomal membranes as indicated. The pT7TS vector alone was used as negative control. Arrows on the right refer to size of products (kd).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/4/10.1182_blood.v96.4.1566/4/m_h81600003003.jpeg?Expires=1766373478&Signature=Ut9MOMzR1esj7ckAVPjXHBc4-uxZBIVaYxNKOaPrq1dYwjGximnnkiqKHyuY1Lm9ZRCHFSk7zZCyrnD2KZEMbvAzLThdhPH6jemFPxlX0lzsNOl-6FAlDYtOrEAxo52HoK3bAA8Naw8sdHcee2cQye8~yi381jBT0Plv~DsndFrk5uXHJBe56lmmNC9pxmQB1HzJKZR8YTQFSL5hov~9vkbttP~ADsTRh2bG9vKjCAnqRPXK9N~wzYXxHSBu7OPecSDRU-bvikgZjugPt74lefdYSAC9yCorEW31m1r0YIj~95rPOE5rcMIPCWHVGjIBq9i~UxubymrKQMNIyX-pCw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Expression and functional analysis of Jk proteins inXenopus oocytes. / (A) Time course of [14C]-urea uptake inXenopus oocytes injected with 0.1 ng cRNA encoding the wt Jk and Jk(S291P) polypeptides. As negative control, water-injected oocytes were used. The assay was initiated by suspending individual oocytes in 0.2 mL Barth solution containing 8 μCi/mL [14C]-urea (145 μmol/L) and 5 μCi/mL [3H]-raffinose. The urea uptake was stopped after the indicated time (t = 0, 1.5, 3, and 6 minutes) by addition of 3 mL of ice-cold Barth solution and followed by rapid washes with 2 × 5 mL of the same solution. Then, the oocyte-associated [14C]-radioactivity was determined as previously described.6 Six to 10 individual oocytes were counted for each time. Mean and SE from different experiments are shown. (B) Northern blot analysis of cRNA encoding wt Jk and Jk(S291P) in oocytes. Total oocyte RNAs (15 μg/lane) at the day of injection and 3 days after were separated and hybridized with the [32P]-labeled Jk cDNA probe, as described in “Materials and methods.” Equal loading and absence of degradation were checked by staining with ethidium bromide. Autoradiography was for 1 hour at −80°C. (C) Immunostaining of oocyte sections. Oocytes from the same batches were fixed and sections of injected oocytes were stained with an affinity-purified antibody against the N-terminus of the Kidd/urea transporter protein and visualized with FITC-conjugated anti-rabbit IgG as described in Materials and methods and then imaged using Nikon Eclipse TE300 microscope (Nikon, Paris, France). Images were taken with epifluorescence illumination (magnification, × 40) and treated with a Biocom informatic system of image integration.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/4/10.1182_blood.v96.4.1566/4/m_h81600003004.jpeg?Expires=1766373478&Signature=rgZi3Y32vx2YIfrOggZ5HD6j2EOXYDmDDn0gaPQ~EsVPvIEgiTZA0difrRRDZXzyQ9zX9syeBNT9CvyWw6kR36Tjll-pydz7j1jE2ARpLOmos~45n9-~VLxnKBt2vlWhZEXUhA2A~Q4Bqb7jfHyTU4Aa6EBGTqtFJ2mHNNYQBPuX552cXNEd2-mbu3NAjyAPook8O1Bj8s~2-2d3c-0rFkK84iKQRiA326ruuW3A7-9b7SF9cYLvcxdKD6dxvUOSMUOTA2JxuoDEIAIUg9GyzCmEWLkdu0bWnN~RZ6JYE9Fh2lMP0AOHuTwU1TIjNvnjA3MHesOPm0L05VPsT5KbCA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)