Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome with eight complementation groups. Four of the FA genes have been cloned, and at least three of the encoded proteins, FANCA, FANCC, and FANCG/XRCC9, interact in a nuclear complex, required for the maintenance of normal chromosome stability. In the current study, mutant forms of the FANCA and FANCG proteins have been generated and analyzed with respect to protein complex formation, nuclear translocation, and functional activity. The results demonstrate that the amino terminal two-thirds of FANCG (FANCG amino acids 1-428) binds to the amino terminal nuclear localization signal (NLS) of the FANCA protein. On the basis of 2-hybrid analysis, the FANCA/FANCG binding is a direct protein-protein interaction. Interestingly, a truncated mutant form of the FANCG protein, lacking the carboxy terminus, binds in a complex with FANCA and translocates to the nucleus; however, this mutant protein fails to bind to FANCC and fails to correct the mitomycin C sensitivity of an FA-G cell line. Taken together, these results demonstrate that binding of FANCG to the amino terminal FANCA NLS sequence is necessary but not sufficient for the functional activity of FANCG. Additional amino acid sequences at the carboxy terminus of FANCG are required for the binding of FANCC in the complex.
Introduction
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome characterized by multiple congenital anomalies, progressive bone marrow failure, and cellular sensitivity to DNA cross-linking agents (reviewed in Auerbach et al1 and Garcia-Higuera et al2). According to somatic cell fusion studies, FA is comprised of 8 distinct complementation groups.3 Four of the FA genes, including the FANCA, FANCG, FANCC, and FANCF genes, have been cloned.4-8 The FANCG gene is identical to the XRCC9 gene.9 Two additional genes, for FANCD10 and FANCE,11 have been mapped. The 4 cloned FA proteins have little or no homology to each other or to other proteins in the database, and little is known regarding their cellular function. On the basis of the similar clinical and cellular phenotypes observed among the 8 FA complementation groups, the FA proteins appear to cooperate in a common cellular pathway.
Cells derived from patients with FA display a broad range of abnormalities (reviewed in Liu et al12 and D'Andrea and Grompe13). FA cells are sensitive to DNA cross-linking agents and oxygen radicals14,15 and have spontaneous chromosome breakage.16 FA cells also have cell cycle abnormalities and exhibit a prolongation in the G2 phase of the cell cycle.17,18 FA cells have increased cellular sensitivity to the apoptotic effects of tumor necrosis factor α and interferon γ.19,20 Most recently, FA cells have been demonstrated to have a defect in the fidelity of DNA double-strand break repair, suggesting a primary defect in a pathway regulating DNA repair.21 22 It remains unclear whether these cellular abnormalities correspond to all FA complementation groups or to only a subset. Also, many of the abnormalities described for FA cells are epiphenomena and may not relate directly to the primary cellular defect.
Increasing evidence demonstrates that the FA proteins cooperate in a novel cellular pathway.23 Three of the cloned proteins, FANCA, FANCG, and FANCC, bind and interact in a protein complex.24-26 Interestingly, this protein complex is not observed in FA cells derived from other FA complementation groups, including groups B, E, F, and H.27 These results suggest that the products of other FA genes, such as the recently cloned FANCF protein,8 regulate the formation of the complex, perhaps by serving as other adaptor proteins or enzymes regulating protein complex assembly. The FA-D complementation group is distinct from the other groups. The FA protein complex forms normally in FA-D cells, suggesting that the product of the FANCD gene functions downstream of complex formation.
Patient-derived mutant forms of FA proteins have been instructive in delineating functional domains of these proteins and in defining the biochemical features of the FA pathway. Point mutations in the carboxy-terminus of FANCC, such as the patient-derived FANCC-L554P mutation,5 block complex formation.28 Also, point mutations in the carboxy terminus of FANCA, especially in the region of the partial leucine zipper, disrupt the complex.29 Point mutations, such as the FANCA(H1110P), are defective in FANCC binding, FANCA phosphorylation, and nuclear localization, thereby defining many of the normal biochemical events in the FA pathway.27
In the current study, we performed a detailed structure/function analysis of FANCA and FANCG to determine the regions of the proteins required for assembly and for nuclear accumulation. On the basis of this analysis, the amino terminal two-thirds of FANCG binds directly to an amino-terminal domain of FANCA containing the nuclear localization signal (NLS) region. Interestingly, an FA patient-derived truncated mutant form of FANCG, lacking the carboxy terminal 39 amino acids, binds to FANCA but fails to recruit FANCC to the complex and fails to complement an FA-G cell line. This result suggests that the 39 amino acids at the carboxyl terminus of FANCG are required for both functional activity and FANCC binding in the complex.
Materials and methods
Cell culture
Epstein-Barr virus-transformed lymphoblasts were maintained in RPMI media supplemented with 15% heat-inactivated fetal calf serum and were grown in a humidified 5% CO2-containing atmosphere at 37°C. COS cells were grown as previously described.30The FA-G lymphoblast line EUFA316 was provided by Dr Hans Joenje.
Construction of expression vectors encoding FA proteins
For construction of HA-tagged FA proteins, the complementary DNA (cDNA) encoding the 9 amino acid hemagglutinin (HA) epitope tag was ligated into the pcDNA3 vector (Invitrogen, San Diego, CA). FANCA and FANCG deletion mutants were generated by polymerase chain reaction (PCR) from the full-length FANCA7 and FANCG cDNA4 9 templates and were subcloned into the HA-pcDNA3 vector. The cDNA inserts were confirmed by DNA sequencing.
For construction of FANCG pMMP-puro mutants, the cDNAs encoding various truncation mutants of FANCG were amplified by PCR and subcloned into the retroviral expression vector, pMMP-puro.31 The FANCG(Q26R), FANCGΔexon13, and FANCG1749ΔA mutations were introduced by PCR with the use of the pfu polymerase (Stratagene, La Jolla, CA). The cDNA inserts were verified by DNA sequencing.
For construction of FANCG and FANCA fusion proteins in yeast 2-hybrid interaction studies, the cDNAs encoding full-length FANCG or FANCG (amino acid 134-278) were subcloned into the EcoRI and BamHI sites of the vector, pAS2-1 (Clontech, Palo Alto, CA). The cDNA encoding FANCA (amino acid 1-230) was subcloned into the BamHI and Xho sites of pACT2 vector (Clontech). The cDNA inserts were verified by DNA sequencing.
Cotransfection of FANCA and FANCG cDNAs in COS cells
For protein expression screening, COS cells were transfected as previously described.32 Forty-eight hours after transfection, cells were directly solubilized in SDS sample buffer, boiled, and loaded to SDS/PAGE. Western blots were performed by using a monoclonal antibody against HA (monoclonal #11) (Bibco Inc, Berkeley, CA).
For immunoprecipitation experiments, COS cells were plated onto 10-cm plates to achieve 40% confluence on the day before transfection. Transfection was performed by using maxiprep plasmid DNAs (5 μg/plate) and 40 μL of lipofectamine (Life Technologies, Gaithersburg, MD) per plate. Two days after transfection, cells were scraped in cold phosphate-buffered saline and pelleted. Immunoprecipitation was performed as previously described.24
35S-labeled in vitro translation
FANCG and FANCA mutant cDNAs, subcloned into the pcDNA vector, were translated with amino acid mix together with35S-methionine (1000 Ci/mmol; Amersham Pharmarcia, Piscataway, NJ) according to manufacturer's specifications (TNT Lysate Coupled Transcription/Translation kit, Promega, Madison, WI). For FANCG and FANCA binding experiments, the in vitro-translated protein products were mixed for 30 minutes at 30°C in reaction buffer, 50 mmol/L Tris, 150 mmol/L NaCl, 5 mmol/L EDTA, 0.5% NP-40, 1% Triton, 3% glycerol, pH 7.4, followed by 30 minutes on ice. The reactions were diluted with the same reaction buffer, and immunoprecipitations were performed. The samples were subjected to SDS-PAGE, and the dried gel was exposed to x-ray films (Kodak, Rochester, NY).
FANCA-NLS peptide competition assay
Two peptides were synthesized for the assay. The sequence of FANCA-NLS-wt was GRRRAWAELLAGRVKREKYN, and the sequence of FANCA-NLS-mut2 was GNNNAWAELLAGNVIDEQYN.30 In vitro-translated FANCG protein was preincubated with either NLS-wt or NLS-mut2 peptides for 30 minutes at 30°C in the reaction buffer described in the previous experiment, before mixing with in vitro-translated HA-FANCA (amino acid 1-240) protein. The concentrations of the peptides are indicated in the figure legends. Immunoprecipitation and SDS-PAGE were performed as previously described.
Interaction of FANCG and FANCA in yeast
FANCG/pAS2-1 and FANCA-NLS/pACT2 were cotransformed into yeast strain Y190 and grown on selective media (-Trp, -Leu). Yeast cells were lysed, according to standard protocol (Clontech). The cell lysate samples were subject to SDS-PAGE. The expression of both FANCG and FANCA proteins were confirmed by Western blots with antibodies against the GAL4 DNA-binding domain and activation domain (Santa Cruz, Santa Cruz, CA), respectively. Clones that expressed both proteins were tested for their ability to activate GAL4 transcription by colony-lift filter assay with proper controls.
Retroviral infection of FA cell lines
The indicated pMMP constructs were transfected by lipofection into 293 producer cells (human embryonic kidney cells) expressing the VSV-G envelope protein.33 Retroviral supernatants were collected on day 5 following lipofection and contained 4.6 × 106 infectious units/mL, as estimated by Southern blot analysis of infected NIH-3T3 cells (data not shown).
FA lymphoblasts were infected with the various pMMP supernatants by a 6-hour incubation in the presence of 8 μg/mL polybrene. Infected cells were washed free of viral supernatant and resuspended in growth media. After 48 hours, cells were transferred to media containing puromycin (1 μg/mL). Dead cells were removed over a Ficoll cushion after 5 days, and surviving cells were grown under continuous selection in puromycin. FA fibroblast lines were retrovirally transduced as previously described.30
Mitomycin C assay
Mitomycin C (MMC) sensitivity assays for lymphoblasts were performed as previously described.30
Immunofluorescence microscopy
Immunofluorescence microscopy of human fibroblasts was performed as previously described.30
Results
The amino terminal region of FANCA binds to FANCG. Recent studies30 34 have shown that the amino terminal region of FANCA is required for FANCC coimmunoprecipitation, nuclear localization, and functional complementation of an FA-A cell line. This amino terminal region of FANCA contains a functional bipartite NLS. To further assess the importance of the amino terminal region of FANCA in FANCG binding, we generated various FANCA mutants (Figure1A). Initially, we cotransfected COS cells with the cDNAs encoding these HA-epitope tagged FANCA mutant proteins along with the wild-type FANCG cDNA (Figure 1B). We used an anti-FANCG antiserum to immunoprecipitate FANCG and FANCG binding proteins. As previously described, the full-length FANCA protein coimmunoprecipitated with FANCG (data not shown). A truncated mutant form of FANCA (FANCA 1-240), containing the amino terminal 240 amino acids of FANCA, was also coimmunoprecipitated with FANCG (upper blot, lane 5). In contrast, a truncated mutant form of FANCA (FANCA 722-1455), containing the carboxy terminal 723 amino acids of FANCA, failed to coimmunoprecipitate with FANCG (upper blot, lane 6), even though the FANCA 722-1455 truncated mutant was well expressed (lower blot, lane 6). These differential binding data (summarized in Figure1A) demonstrate that the amino terminal 240 amino acids of FANCA are necessary and sufficient for FANCG binding.
The amino terminal region of FANCA is required for FANCG binding.
(A) Schematic representation of wild-type (wt) and truncated mutant forms of FANCA. The two indicated mutant FANCA polypeptides contain an HA-epitope tag at their amino terminus. Results of binding studies are summarized as (+) and (−). NLS, nuclear localization signal; Lz, leucine zipper. (B) COS cells were cotransfected with the indicated FANCA cDNA mutants and the full-length FANCG cDNA. Immunoprecipitation (lanes 1-6) was performed with an anti-FANCG antiserum (upper blot) and an anti-HA antibody (middle blot). Immunoblotting was done with an anti-HA monoclonal antibody or with an anti-FANCG antiserum, as indicated. WCE denotes whole cell extract.
The amino terminal region of FANCA is required for FANCG binding.
(A) Schematic representation of wild-type (wt) and truncated mutant forms of FANCA. The two indicated mutant FANCA polypeptides contain an HA-epitope tag at their amino terminus. Results of binding studies are summarized as (+) and (−). NLS, nuclear localization signal; Lz, leucine zipper. (B) COS cells were cotransfected with the indicated FANCA cDNA mutants and the full-length FANCG cDNA. Immunoprecipitation (lanes 1-6) was performed with an anti-FANCG antiserum (upper blot) and an anti-HA antibody (middle blot). Immunoblotting was done with an anti-HA monoclonal antibody or with an anti-FANCG antiserum, as indicated. WCE denotes whole cell extract.
Amino terminal two-thirds of FANCG binds to FANCA
To determine the region of FANCG required for FANCA binding, we next generated a large series of cDNAs encoding mutant FANCG proteins (Figure 2A). One of these FANCG mutant proteins (FANCG1749ΔA) was originally identified by mutational screening of a known FA-G patient. Another variant FANCG protein (FANCGΔexon13) is modeled from a splice variant of the FANCG gene found in normal human lymphoblasts, resulting from skipping of exon 13 (Y. Kuang, unpublished observation). We cotransfected COS cells with a subset of cDNAs encoding HA-epitope tagged FANCG mutant proteins along with the wild-type FANCA cDNA (Figure 2B). We used an anti-FANCA antiserum to immunoprecipitate FANCA and FANCG proteins. The full-length FANCG protein coimmunoprecipitated with FANCA (Figure 2B, lanes 3, 12, and 18). Interestingly, a truncated mutant form of FANCG, containing the amino terminal 428 amino acids of FANCG, coimmunoprecipitated with FANCA (Figure 2B, lane 19). Further truncations of the amino terminal region of FANCG resulted in loss of FANCA binding (lanes 4, 5, and 13). These binding data (summarized in Figure 2A) demonstrate that the amino terminal 428 amino acids of FANCG are necessary and sufficient for FANCA binding.
The amino terminal two-thirds of FANCG (amino acids 1-428) is required for FANCA binding.
(A) Schematic representation of wild-type and mutant forms of FANCG. Seven FANCG polypeptides contain an HA-epitope tag at their amino terminus. FANCGΔexon13 (modeled from a splice variant observed by RT-PCR from normal human cells that results from exon 13 skipping) has 551 amino acids plus 5 altered amino acids. FANCG1749ΔA (derived from a known FA-G patient) has 583 amino acids plus 9 altered amino acids resulting from a frame shift. Results of binding studies are summarized as (+) and (−). Lz, leucine zipper. (B) COS cells were cotransfected with the indicated HA-FANCG mutant cDNAs and the full-length (wild-type) FANCA cDNA. Immunoprecipitation (lanes 1-5, 11-13, and 17-19) was performed with an anti-FANCA antiserum. Whole cell extracts (WCE) were analyzed in lanes 6-10, 14-16, and 20-22. Immunoblotting was done with an anti-HA monoclonal antibody. In lane 19, HA-FANCG 1-428 comigrated with the immunoglobulin heavy chain at 50 kD. * Data not shown.
The amino terminal two-thirds of FANCG (amino acids 1-428) is required for FANCA binding.
(A) Schematic representation of wild-type and mutant forms of FANCG. Seven FANCG polypeptides contain an HA-epitope tag at their amino terminus. FANCGΔexon13 (modeled from a splice variant observed by RT-PCR from normal human cells that results from exon 13 skipping) has 551 amino acids plus 5 altered amino acids. FANCG1749ΔA (derived from a known FA-G patient) has 583 amino acids plus 9 altered amino acids resulting from a frame shift. Results of binding studies are summarized as (+) and (−). Lz, leucine zipper. (B) COS cells were cotransfected with the indicated HA-FANCG mutant cDNAs and the full-length (wild-type) FANCA cDNA. Immunoprecipitation (lanes 1-5, 11-13, and 17-19) was performed with an anti-FANCA antiserum. Whole cell extracts (WCE) were analyzed in lanes 6-10, 14-16, and 20-22. Immunoblotting was done with an anti-HA monoclonal antibody. In lane 19, HA-FANCG 1-428 comigrated with the immunoglobulin heavy chain at 50 kD. * Data not shown.
FANCG binds directly to the NLS of FANCA
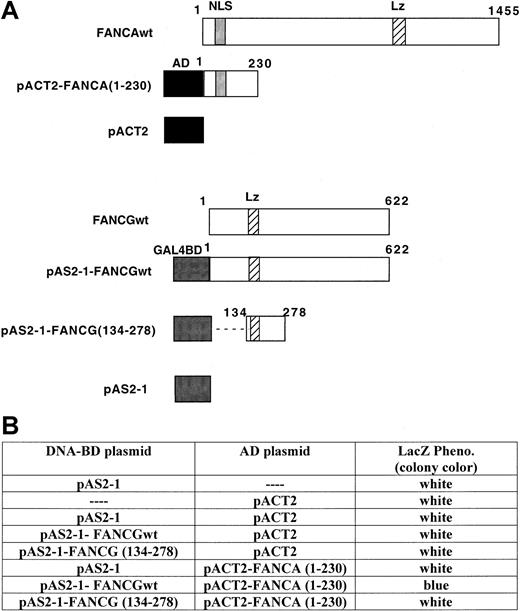
Several groups,24,35 including our own, have used in vitro-translated proteins to demonstrate that FANCA binds to FANCG. To further investigate whether the interaction of FANCA and FANCG is truly a direct protein-protein interaction, we used the yeast 2-hybrid method (Figure 3).36-39 The amino terminal 230 amino acids of FANCA were translationally fused to the activation domain of GAL4 in the pACT2 expression vector (Figure 3A). In addition, variable regions of the FANCG protein were translationally fused to the DNA-binding domain in the pAS2-1 expression vector. The various FANCA and FANCG proteins were coexpressed in Y190 cells. A strong interaction between the amino terminus of FANCA and FANCG was observed, based on the generation of blue colonies in the colony-lift filter assay (Figure 3B). These data further demonstrate that the amino terminus of FANCA interacts with the amino terminus of FANCG and that the interaction is direct.
Analysis of FANCA/FANCG binding interaction by yeast 2-hybrid method.
(A) Diagrams of the constructs used in the yeast 2-hybrid assays. FANCA 1-230 was subcloned in-frame, downstream of the GAL4 activation domain (AD) (amino acids 768-881) in the pACT2 vector. FANCGwt and FANCG (amino acids 134-278) were subcloned in-frame, downstream of the GAL4 DNA-binding domain (GAL4BD) (amino acids 1-147) in the pAS2-1 vector. (B) Colony-lift filter assay to assess the binding between FANCA and FANCG proteins. Yeast strain Y190 was cotransformed with the plasmids indicated. The expression of proteins was confirmed by immunoblotting of whole yeast extract with antibodies against GAL4-AD or GAL4 DNA-BD (data not shown).
Analysis of FANCA/FANCG binding interaction by yeast 2-hybrid method.
(A) Diagrams of the constructs used in the yeast 2-hybrid assays. FANCA 1-230 was subcloned in-frame, downstream of the GAL4 activation domain (AD) (amino acids 768-881) in the pACT2 vector. FANCGwt and FANCG (amino acids 134-278) were subcloned in-frame, downstream of the GAL4 DNA-binding domain (GAL4BD) (amino acids 1-147) in the pAS2-1 vector. (B) Colony-lift filter assay to assess the binding between FANCA and FANCG proteins. Yeast strain Y190 was cotransformed with the plasmids indicated. The expression of proteins was confirmed by immunoblotting of whole yeast extract with antibodies against GAL4-AD or GAL4 DNA-BD (data not shown).
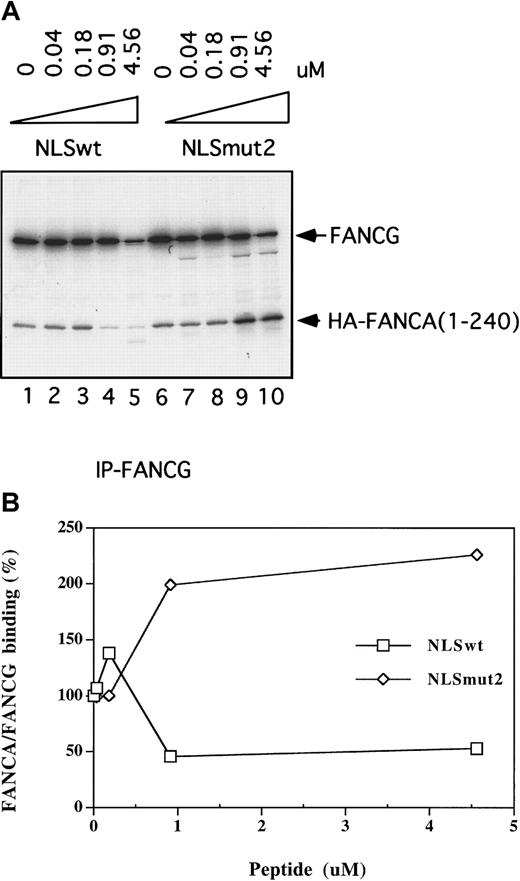
To further implicate the NLS region of FANCA as the direct binding site of FANCG, we next performed the coimmunoprecipitation of in vitro-translated FANCA 1-240 and FANCG in the presence of a specific peptide inhibitor (Figure 4). Interestingly, a 20 amino acid peptide containing the NLS region of FANCA30 specifically competed the coimmunoprecipitation of FANCA 1-240 and FANCG. A mutant peptide, in which all basic amino acids of the NLS have been mutated, failed to compete the binding of FANCA and FANCG. Our previous studies30 had shown that these amino acid changes also disrupt the functional activity of the NLS motif. Densitometric scanning of the autoradiograph in Figure 4A demonstrated that the inhibition was dependent on the concentration of free peptide (Figure 4B). One-half maximal inhibition was detected at a wild-type peptide concentration of approximately 0.9 μmol/L. These data further confirm that FANCG binds directly to the NLS region of FANCA.
Competition of FANCA/FANCG binding by the 20 amino acid peptide from the NLS region of FANCA.
(A) 35S-labeled, in vitro-translated FANCG(wt) protein was preincubated with the indicated concentrations of FANCA-NLS-wt peptide or FANCA-NLS-mut2 peptide for 30 minutes. 35S in vitro-translated, FANCA 1-240 was subsequently added, and an immunoprecipitation was performed with an anti-FANCG antiserum. (B) The autoradiograph in (A) was subjected to densitometric scanning and plotted. FANCA/FANCG binding was normalized as the ratio of HA-FANCA 1-240 over FANCG. The binding in the absence of peptide was set as 100%.
Competition of FANCA/FANCG binding by the 20 amino acid peptide from the NLS region of FANCA.
(A) 35S-labeled, in vitro-translated FANCG(wt) protein was preincubated with the indicated concentrations of FANCA-NLS-wt peptide or FANCA-NLS-mut2 peptide for 30 minutes. 35S in vitro-translated, FANCA 1-240 was subsequently added, and an immunoprecipitation was performed with an anti-FANCG antiserum. (B) The autoradiograph in (A) was subjected to densitometric scanning and plotted. FANCA/FANCG binding was normalized as the ratio of HA-FANCA 1-240 over FANCG. The binding in the absence of peptide was set as 100%.
Amino terminal region of FANCG is necessary but not sufficient for functional complementation of an FA-G cell line
We next tested mutant forms of the FANCG protein for their ability to functionally complement the MMC sensitivity of an FA-G lymphoblast line, EUFA316. The cDNAs encoding various forms of FANCG were retrovirally transduced into EUFA316 cells, and stably transduced cells were analyzed for MMC sensitivity (Figure5). As previously shown,4the wild-type full-length FANCG corrected the MMC sensitivity of FA-G cells (closed circles). Three truncated forms of FANCG (FANCG 1-428, FANCGΔexon13, and FANCG1749ΔA), failed to complement the FA-G cells. The failure of the patient-derived FANCG1749ΔA protein to complement the FA-G cells confirms that this is a bona fide FANCG mutation.
The amino terminal region of FANCG is necessary but not sufficient for functional complementation of an FA-G cell line.
The FA-G lymphoblast line, EUFA316, was transfected with cDNAs encoding various forms of the FANCG protein. G418-selected cells were analyzed for the function of the various FANCG mutant proteins, using the MMC assay. Samples shown are parental EUFA316 cells (○), EUFA316 cells transduced with FANCG(wt) (●), with FANCG 1-428 (▵), with FANCGΔexon13 (■), or with FANCG1749ΔA (♦). Similar results were obtained when the FANCG cDNAs were transfected into the FA-G fibroblast line, FAG326SV (data not shown).
The amino terminal region of FANCG is necessary but not sufficient for functional complementation of an FA-G cell line.
The FA-G lymphoblast line, EUFA316, was transfected with cDNAs encoding various forms of the FANCG protein. G418-selected cells were analyzed for the function of the various FANCG mutant proteins, using the MMC assay. Samples shown are parental EUFA316 cells (○), EUFA316 cells transduced with FANCG(wt) (●), with FANCG 1-428 (▵), with FANCGΔexon13 (■), or with FANCG1749ΔA (♦). Similar results were obtained when the FANCG cDNAs were transfected into the FA-G fibroblast line, FAG326SV (data not shown).
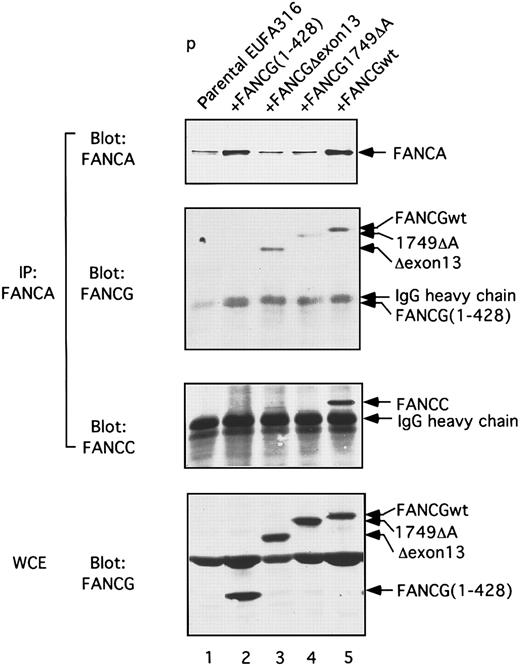
We next analyzed the ability of the truncated forms of the FANCG protein to form a complex with the endogenous FANCA and FANCC proteins in these transfected FA-G lymphoblasts (Figure6). Whole cell extract Western blotting (Figure 6, bottom blot) indicated that the three FANCG-truncated proteins (lanes 2, 3, and 4) were expressed at a similar level, compared to FANCG(wt) (lane 5). This control experiment suggests that the failure to complement the FA-G cell line by these mutant FANCG proteins (Figure 5) did not result from the low level of expression of these proteins. As previously shown,24immunoprecipitation of the FANCA protein with an anti-FANCA antiserum resulted in the coimmunoprecipitation of wild-type FANCC (lane 5, anti-FANCC immunoblot) and wild-type FANCG (lane 5, anti-FANCG immunoblot). The mutant FANCG proteins, FANCG 1-428, FANCGΔexon13, and FANCG1749ΔA, were also found to bind in a complex with FANCA. Interestingly, FANCC was not detected in the complex (lanes 2, 3, and 4). Taken together, these data demonstrate that the binding of FANCC in the FANCA/FANCG/FANCC protein complex and the functional complementation of FA-G cells requires the carboxy terminal 39 amino acids of FANCG.
Truncated mutant forms of the FANCG protein bind to FANCA but not to FANCC.
The FA-G lymphoblast line, EUFA316, expressing the various indicated forms of the FANCG protein, was analyzed for FANCA/FANCC/FANCG binding. Proteins extracted from the indicated EUFA316 transfectants were immunoprecipitated with an anti-FANCA antiserum, and the immune complexes were analyzed by immunoblotting with either anti-FANCA, anti-FANCG (N-terminal), or anti-FANCC antisera. In lane 2, IP-FANCA/Blot-FANCG, FANCG 1-428 migrated slightly faster than immunoglobulin heavy chain. Alternatively, whole cell extracts (WCE) were immunoblotted directly.
Truncated mutant forms of the FANCG protein bind to FANCA but not to FANCC.
The FA-G lymphoblast line, EUFA316, expressing the various indicated forms of the FANCG protein, was analyzed for FANCA/FANCC/FANCG binding. Proteins extracted from the indicated EUFA316 transfectants were immunoprecipitated with an anti-FANCA antiserum, and the immune complexes were analyzed by immunoblotting with either anti-FANCA, anti-FANCG (N-terminal), or anti-FANCC antisera. In lane 2, IP-FANCA/Blot-FANCG, FANCG 1-428 migrated slightly faster than immunoglobulin heavy chain. Alternatively, whole cell extracts (WCE) were immunoblotted directly.
Carboxy terminal truncated mutant forms of FANCG localize to the nucleus
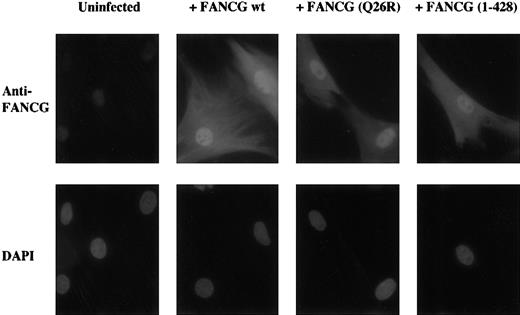
As a control, we next examined the cellular localization of the various mutant forms of FANCG by immunofluorescence (Figure7). Various forms of FANCG were expressed in the FA-G fibroblast line, DF3. These fibroblasts are derived from an FA-G patient, have a homozygous mutation in the FANCG gene (FANCG 1665 G > C), and fail to express endogenous FANCG protein (Y. Kuang, unpublished observation). DF3 fibroblasts were functionally complemented by retroviral transduction with the wild-type FANCG cDNA (data not shown). The wild-type FANCG protein and the mutant FANCG 1-428 protein localized to the nucleus of the transduced DF3 cells similarly (Figure 7). The failure of the FANCG 1-428 protein to complement the cells was, therefore, not due to its failure to localize to the nucleus. In addition, we tested a variant form of FANCG with a Q to R mutation at amino acid 26. This variant allele was found in cells from several FA-G patients and normal control subjects, suggesting that the Q26R change is a polymorphism and is not a pathogenic mutation. Consistent with this hypothesis, FANCG(Q26R) protein binds to FANCA and FANCC and corrects the MMC sensitivity of the transduced FA-G cells (data not shown). Moreover, the FANCG(Q26R) protein accumulates normally in the nucleus of transduced cells (Figure 7).
A truncated mutant form of the FANCG protein translocates to the cell nucleus but fails to function.
The primary FA-G fibroblast line, DF3, was transduced with retroviral constructs encoding the indicated wild-type or mutant forms of the FANCG protein. Following retroviral transduction, the cells were analyzed by immunofluorescence with anti-FANCG antisera or by staining with the DNA-specific dye, DAPI (4,6-diamidino-2-phenylindole). In parallel, the cells were analyzed for correction of MMC sensitivity (data not shown). The wild-type FANCG and FANCG(Q26R) protein corrected the MMC sensitivity of the DF3 cells, but the FANCG 1-428 mutant failed to complement the cells.
A truncated mutant form of the FANCG protein translocates to the cell nucleus but fails to function.
The primary FA-G fibroblast line, DF3, was transduced with retroviral constructs encoding the indicated wild-type or mutant forms of the FANCG protein. Following retroviral transduction, the cells were analyzed by immunofluorescence with anti-FANCG antisera or by staining with the DNA-specific dye, DAPI (4,6-diamidino-2-phenylindole). In parallel, the cells were analyzed for correction of MMC sensitivity (data not shown). The wild-type FANCG and FANCG(Q26R) protein corrected the MMC sensitivity of the DF3 cells, but the FANCG 1-428 mutant failed to complement the cells.
Discussion
Several lines of evidence demonstrate that the FA proteins, including FANCA, FANCG, and FANCC, interact in a common cellular pathway, leading to the accumulation of the FA protein complex in the nucleus. Absence of this nuclear protein complex, resulting from biallelic germline mutations of any one of these FA genes, correlates with chromosome instability and a broad array of cellular and clinical abnormalities. The protein products of additional (uncloned) FA genes appear to be required for the assembly, stability, and nuclear transport of the FA complex.27 In the current study, we identified the regions of FANCG and FANCA required for their binding interaction and identified a carboxy terminal domain of FANCG required for its function and for the recruitment of FANCC to the complex. We also identified patient-derived truncated mutant forms of FANCG that coimmunoprecipitated with FANCA but failed to bind FANCC and failed to functionally complement FA-G cells.
Several structural features of the interaction between the FANCA and FANCG proteins were identified. First, we have identified the regions of the FANCA and FANCG proteins required for their interaction. The amino terminal two-thirds of FANCG binds to an amino terminal region of FANCA. Second, we used a 2-hybrid analysis to determine that the amino terminus of FANCA binds directly to FANCG. Consistent with these studies, FANCA binds to FANCG even in cells derived from other FA complementation groups.24 These results confirm that the products of other FA genes are not required for the FANCA/FANCG protein interaction. Direct binding between FANCA and FANCG has also been recently reported by another group.40
Third, we have demonstrated that FANCG binds directly to the NLS region of FANCA. This interaction is competed by a 20 amino acid peptide corresponding to the amino terminal region of FANCA. Mutations in the NLS region that disrupt nuclear localization of FANCA30also disrupt the interaction between FANCA and FANCG. Disruption of the FA protein complex with this NLS peptide may provide an experimental strategy for promoting MMC sensitivity in otherwise normal cells. Given the direct interaction of FANCG with the NLS of FANCA, alternative models may account for their possible functional interaction in the process of nuclear uptake. FANCG may cooperate with importin-α or act as an importin protein itself, thereby functioning to enhance or regulate nuclear import.41 42 Whether or not the FANCG protein interacts with other proteins involved in nuclear translocation, such as importin-α, importin-β, or nucleoporins, remains to be determined.
Fourth, the interaction of FANCA and FANCG is distinct from the interaction of FANCA and FANCC. Although the FANCA/FANCG binding interaction is direct, the FANCA/FANCC and FANCG/FANCC interactions appear to be indirect. FANCA and FANCC interactions appear to be weak, based on binding of in vitro-translated proteins24 or 2-hybrid analysis.40,43 The binding of FANCA and FANCC may, therefore, require additional adaptor proteins or posttranslational modifications. Interestingly, the FANCA/FANCC interaction is not observed in several other FA complementation groups, including groups B, E, F, and H, suggesting that their interaction is regulated by other FA gene products.27 For instance, the recently cloned FANCF protein8 may also be a component of the FA protein complex required for FANCC binding. Accordingly, the purification of the FA protein complex may allow the identification of other FA gene products.
Fifth, our data elucidate various structural requirements for the interaction of FANCC with the FANCA/FANCG complex. Our previous studies30 demonstrated that the amino terminal NLS region of FANCA is required for FANCC binding. Because the NLS region of FANCA binds directly to FANCG, FANCA/FANCG binding appears to be required for FANCC binding in the complex. Also, point mutations in the leucine zipper region of FANCA disrupt the FANCC interaction, suggesting that this FANCA region also contributes to FANCC binding. Finally, in the current study, the three FANCG mutant proteins, FANCG 1-428, FANCGΔexon13, and FANCG 1749ΔA, bind FANCA but fail to recruit FANCC to the complex. These results demonstrate that the carboxy terminal region of FANCG is required for FANCC interaction with the complex.
The FANCG1749ΔA mutant protein appears to be representative of a class of patient-derived mutant FANCG proteins. This patient-derived mutant protein binds in a complex with FANCA but fails to bind to FANCC and fails to function. Identification of other FANCG truncation or missense mutations in FA-G patients by mutational screening may help to delineate the precise C-terminal functional domain of FANCG.
It is interesting that mutant forms of FA proteins bind and form complexes with other endogenous wild-type FA proteins. For instance, previous studies24,44 have shown that the FANCA mutant protein, FANCA(H1110P), forms a complex with wild-type FANCG. In the current study, we have shown that the carboxy terminal truncated forms of FANCG bind wild-type FANCA. Whether or not these mutant proteins have dominant negative activity in vitro or account for the phenotypic variation among FA patients remains to be determined. Our observations demonstrate that formation of the FANCA/FANCG protein complex is necessary but not sufficient for functional activity of the FA pathway. Similarly, in FA-D cells the FA protein complex forms normally, yet these cells remain sensitive to MMC.27
Finally, our results confirm and extend the results of Kruyt et al.45 These investigators determined that the amino terminal 36 amino acids of FANCA form a novel interaction with the FANCG protein and identified two different carboxy terminal regions of FANCG (encompassing amino acids 400-475 and 585-622) required for FANCA binding. However, these investigators failed to detect FANCC binding in the FANCA/FANCG complex, although the FANCC interaction has been verified by other independent investigators.25 26 Our results confirm the contribution of FANCG amino acids 400-428 to FANCA binding and demonstrate that the carboxy terminus of FANCG (amino acid 585-622) is required for FANCC binding and optimum FANCA binding in the complex. Indeed, our data demonstrate that FANCC binding stabilizes the FANCA/FANCG interaction, because the FANCG1749ΔA mutant (lacking amino acids 583-622) binds less well to wild-type FANCA (Figure 6). Our results demonstrate by 2-hybrid analysis that the FANCA/FANCG binding is direct and support the presence of a partial binding site within the amino terminal two-thirds of FANCG. This FANCG binding site is sufficient for FANCA binding and nuclear accumulation of the complex, but it is insufficient for FANCC binding, stabilization of the FA complex in the nucleus, or biological function.
Acknowledgment
We thank members of the D'Andrea laboratory for helpful discussions.
Supported by grants RO1HL52725-04 and PO1HL54785-04 from the National Institutes of Health. A.D.D. is a Scholar of the Leukemia Society of America. I.G.-H. is supported by the Cancer Research Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan D. D'Andrea, Dana-Farber Cancer Institute, Division of Pediatric Oncology, Harvard Medical School, 44 Binney St, Boston, MA 02115; e-mail: alan_dandrea@dfci.harvard.edu.