Abstract
Retinoic acid (RA) signaling is mediated by its nuclear receptors RXR and RAR, which bind to their cognate response elements as a heterodimer, RXR/RAR, and act in concert with coregulatory factors to regulate gene transcription on ligand binding. To identify specific cofactors that interact with the RXR/RAR heterodimer in acute promyelocytic leukemia (APL) cells, a double cistronic construct was used that allowed coexpression of the RXR LBD (ligand binding domain) with the RAR LBD as an affinity matrix to pull down interacting proteins from nuclear extracts prepared from a human APL cell line, NB4. A group of proteins was detected whose interaction with RXR/RAR is ligand inducible. The molecular weight pattern of these proteins is similar to that of a complex of proteins previously identified as DRIP or TRAP, which are ligand-dependent transcription activators of VDR and TR, respectively. The RXR/RAR-interacting proteins from NB4 were confirmed to be identical to the DRIP subunits by comparative electrophoresis, Western blot analysis, and in vitro protein interaction assay. In addition to RXR/RAR, the DRIP component can interact directly with the APL-specific PML-RARα fusion protein. The same DRIP complex is present in RA-resistant APL cells and in a variety of cancer cell lines, supporting its global role in transcriptional regulation.
Introduction
Retinoids are a group of natural and synthetic derivatives of vitamin A that exert a wide variety of effects on biologic processes, such as homeostasis, pattern formation during embryogenesis, and cellular growth and differentiation.1,2Retinoid signaling is mediated by 2 nuclear receptors—retinoic acid receptor (RAR) and retinoic acid X receptor (RXR)—belonging to a large superfamily of transcription factors (nuclear receptors) that respond to hormonal signals, including steroid, vitamin D3, and thyroid hormones.3-6 These nuclear receptors share extensive homologies in their protein structure. The 2 highly conserved domains are the DNA-binding domain (DBD) and the ligand-binding domain (LBD), which also contains a dimerization surface and a ligand-dependent transactivation function, AF-2, at the carboxyl terminus of the receptor. Nuclear receptors interact as homodimers or as heterodimers with the common partner RXR, bind to specific DNA-response elements located in the promoter of the target gene, and regulate transcription.5 6
Although the precise mechanisms by which nuclear receptors regulate gene transcription are unknown, they have been shown to interact directly with components of the basal transcription machinery, perhaps affecting its assembly or function.7-9 Additionally, recent studies have identified a group of nuclear receptor-interacting proteins that are specific coregulatory factors in mediating gene transcription. Coactivators interact with nuclear receptors in a ligand-inducible manner and require the integrity of the AF-2 domain. Identified coactivators include members of the SRC-1 family (SRC-1/NCoA-1/p160,10-12ACTR/pCIP/RAC-3/AIB,13-15 and TIF-2/NCoA-2/GRIP-1)16,17; the cointegrators CBP/p300 and their interacting protein pCAF18,19; other factors including RIP140,20,21 SUG-1/TRIP-1,22,23TIF-1,24 and ARA70.25 Discoveries of histone acetyltransferase activity in CBP/p300, pCAF, and some members of the SRC-1 family suggest that coactivators may function through chromatin structural modifications that allow promoter accessibility for the preinitiation complex, leading to the activation of transcription.26-29 Interestingly, 2 identified receptor corepressors, SMRT and NCo-R,30-32 appear to modify chromatin structure by recruiting histone deacetylases and to repress transcription in the absence of ligand.33 34
A distinct coactivator complex was identified recently by 2 separate groups for its ligand-dependent interaction to the vitamin D3 receptor (DRIP) or to the thyroid hormone receptor (TRAP).35,36 DRIP/TRAP exists as a large macromolecular complex containing at least 15 proteins that comprise a novel set of nuclear receptor coactivators. In vitro transcription assays using either crude cell nuclear extracts or purified components of the general transcription complex suggested that the DRIP/TRAP complex acts as a ligand-dependent positive transcriptional regulator of various nuclear receptors.35 36
Hormonal therapies targeting nuclear receptors are frequently used in the treatment of neoplastic diseases. In acute promyelocytic leukemia (APL), retinoic acid (RA) has been demonstrated to have a dramatic effect on the induction of cytodifferentiation and the maturation of leukemic cells, leading to clinical remission in patients.37-41 APL is characterized by a reciprocal chromosomal translocation, t(15;17), that fuses the PML gene with the retinoic acid receptor α (RARα) gene and generates a chimeric PML-RARα protein.42-45 Expression of the PML-RARα in transgenic mice has confirmed its role in the pathogenesis of APL.46-50 Pharmacologic levels of retinoids can reverse the oncogenic properties of PML-RARα and induce its degradation, restoring normal retinoid signaling and inducing terminal differentiation of APL cells. However, APL cells can develop resistance to RA, and relapse occurs in patients with APL who are treated with RA alone. Proposed explanations for acquired RA resistance include the progressive reduction of RA plasma concentration seen with repeated oral RA dosing and missense mutations in the ligand-binding domain of the fusion PML-RARα gene, reported in in vitro–derived RA-resistant APL subclones and in patients with APL who have had relapses.51-58
We have reported that one such retinoid-resistant subclone of NB4, R4, harbors a dominant-negative PML-RARα mutation, PML-RARα M4, that abolishes ligand binding and inhibits transcription on RA response elements56 by constitutive association with the corepressor, SMRT.59 However, most retinoid-resistant APL cells, both from patient samples and from derived cell lines, do not show mutations in PML-RARα. Although studies on 2 additional RA-resistant subclones, MR2 and MR6, failed to find mutations or deletions in the PML-RARα gene, transient transfection results showed that they have lost their ligand-dependent transcriptional activation on RA response elements, and ligand binding assays demonstrated in each cell line an altered binding of all-trans retinoic acid (t-RA) to nuclear high molecular weight protein complexes.55 This prompted us to compare the interactions with retinoid receptors of RA-dependent coregulatory factors from these cells to those in nuclear extracts of the parental NB4 cell line. To analyze specific nuclear protein interactions in vitro, we used a double cistronic vector that allowed the coexpression of the ligand-binding domains of RXR and RAR,60 which were then immobilized on a glutathione–Sepharose matrix and used as an affinity column to isolate interacting proteins.
We report here the detection of a protein complex in NB4 that interacted with the RXR/RAR heterodimer in a ligand-inducible manner. The complex is virtually identical to a previously identified transcription coactivator complex, known as DRIP or TRAP, for its interaction with VDR and TR, dependent on their respective ligands.35 36 The same complex is also present in 3 RA-resistant NB4 subclones, in U937 subclones that express different levels of PML-RARα, and in various non-APL cell types. These studies suggest a ubiquitous presence of the DRIP complex and support its general role in transcriptional activation.
Materials and methods
Cell culture
The parental NB4 promyelocytic leukemia cell line, 3 RA-resistant subclones, Namalwa B cells, and the myeloid precursor cells KG-1, U937, were grown in RPMI medium (GIBCO BRL, Burlington, Ontario, Canada) supplemented with 10% fetal bovine serum (Upstate Biotechnology, Lake Placid, NY). All cell cultures were incubated at 5% Pco2 at 37°C in humidified air.
Purification of GST fusion proteins
The bicistronic expression construct glutathione-S-transferase (GST)-RXR LBD/H6-RAR LBD and the GST-VDR LBD (amino acid 110-427) were kindly provided by Ronald M. Evans and Leonard P. Freedman, respectively. All GST fusion proteins were overexpressed and purified as described in the GST Gene Fusion System (Pharmacia Biotech, Baie d'Urfe, Quebec, Canada) with the exception that the glutathione–Sepharose pellet was washed once with high-stringency buffer (500 mmol/L NaCl, 50 mmol/L Tris-HCl [pH 8.0], 1 mmol/L EDTA, 0.4% NP40), and 3 times with low-stringency buffer (100 mmol/L NaCl, 50 mmol/L Tris-HCl [pH 8.0], 1 mmol/L EDTA, 0.4% NP40). The amount of purified proteins were measured by Bradford assay (Bio-Rad, Mississauga, Ontario, Canada) and examined by Coomassie blue staining.
Nuclear extract preparation
Nuclear extracts were prepared according to the Dignam method.61 Prepared nuclear extracts were then dialyzed twice against 100-fold excess volume of dialysis buffer (100 mmol/L KCl, 20 mmol/L HEPES-KOH [pH 7.9], 0.2 mmol/L EDTA, 20% glycerol, 1 mmol/L dithiothreitol) for 2 hours each time at 4°C. Extract amounts were measured by Bradford assay (Bio-Rad), and the extracts were stored in aliquots at −80°C.
In vitro GST pull-down assay
Immobilized GST fusion proteins (10-20 μg) were preincubated at 4°C for 1 hour with or without ligand in the binding buffer (20 mmol/L HEPES-KOH [pH 7.9], 180 mmol/L KCl, 0.2 mmol/L EDTA, 0.05% NP-40, 0.5 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L dithiothreitol) containing 1 mg/mL bovine serum albumin. The fusion proteins were then incubated with either 1.2 mg nuclear extracts or 500 000 cpm 35S-labeled in vitro translated protein (TNT Coupled Reticulocyte Lysate System; Promega, Madison, WI) with or without ligand in the binding buffer at 4°C for 4 to 5 hours. Sepharose beads were then washed 3 times with the binding buffer containing 0.1% NP-40. For isolation of the protein complex, the bound proteins were eluted with washing buffer containing 0.2% N-lauroyl sarkosine, separated by SDS-PAGE, and visualized by silver nitrate staining (Bio-Rad). For in vitro translated proteins, the Sepharose beads were boiled in SDS-sample buffer, and proteins were separated by SDS-PAGE and analyzed by autoradiography.
Western analysis
Proteins eluted from the glutathione–Sepharose matrix were analyzed by SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad). Primary antibodies were made 1:1000 dilution in 5% milk in phosphate-buffered saline, and proteins that interacted with the antibodies were detected using the ECL Western blotting detection kit (Amersham, Buckinghamshire, UK).
Results
Proteins interact with retinoid receptors in a ligand-dependent manner in NB4 cells
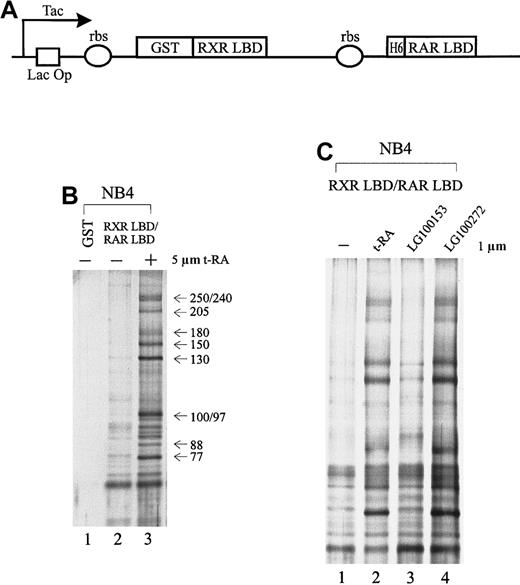
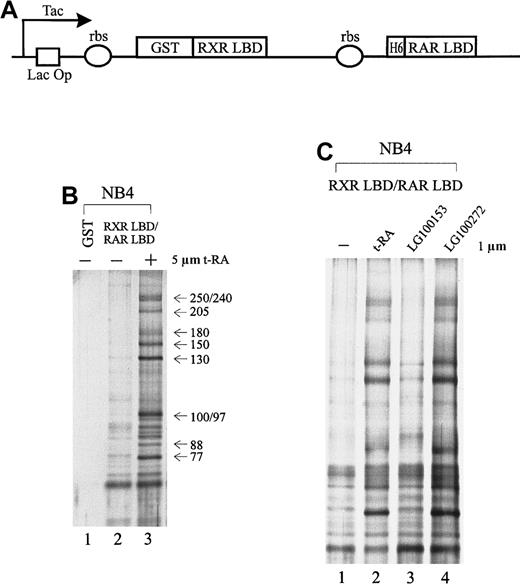
To isolate specific cofactors that interact with the retinoid receptors in APL cells, we overexpressed the retinoid receptor heterodimer ligand-binding domain using a double cistronic expression construct (Figure 1A) in which the RXR LBD was fused to glutathione-S-transferase (GST) and the RAR LBD was tagged with 6 histidine residues.60 The ribosome-binding site preceding each cistron allows the coexpression of both proteins in the form of a heterodimer. The nuclear receptor LBD consists of a transcriptional activation domain AF-2, which is essential for interaction with coactivators. Nuclear extracts prepared from NB4 cells were passed through immobilized GST-RXR LBD/RAR LBD in the absence or presence of 5 μmol/L t-RA. As a result, we isolated a group of proteins consisting of at least 10 subunits, ranging in molecular weight from 77 to 250 kd, that interacted specifically with the retinoid receptor heterodimer only when ligand was present (Figure 1B, lane 3). In contrast, in the absence of ligand or with GST alone, few nonspecific binding proteins were observed (Figure 1B, lanes 1, 2).
Cofactors that interact with the retinoid receptors in APL cells.
(A) Schematic diagram of the double cistronic expression plasmid. The RXR LBD is fused with GST, and the RAR LBD is tagged with 6 histidine residues. Both cDNAs are driven by a Tac promoter under the control of the lac operator. The ribosome-binding site (rbs) preceding each cistron allows the coexpression of both proteins. Proteins can be purified by either glutathione Sepharose or Ni2+ NTA agarose. (B) Ligand-inducible interactions between GST-RXR LBD/RAR LBD and a group of proteins isolated from APL cell line NB4 nuclear extracts. Immobilized GST-RXR LBD/RAR LBD was incubated with 1.2 mg NB4 nuclear extract in the absence (−) or presence (+) of 5 μmol/L all-trans retinoic acid (t-RA) (lanes 2, 3). GST alone was used as a control (lane 1). Bound proteins were eluted, separated by SDS-PAGE, and visualized by nitrate silver staining. Approximate molecular weights of the interacting proteins are indicated at right. (C) Binding of the complex to retinoid receptors in response to selective ligands. Protein interactions were examined in the absence (−) or presence of 1 μmol/L t-RA (lane 2), 1 μmol/L RXR-specific ligand LG100153 (lane 3), and 1 μmol/L RAR-specific ligand LG100272 (lane 4).
Cofactors that interact with the retinoid receptors in APL cells.
(A) Schematic diagram of the double cistronic expression plasmid. The RXR LBD is fused with GST, and the RAR LBD is tagged with 6 histidine residues. Both cDNAs are driven by a Tac promoter under the control of the lac operator. The ribosome-binding site (rbs) preceding each cistron allows the coexpression of both proteins. Proteins can be purified by either glutathione Sepharose or Ni2+ NTA agarose. (B) Ligand-inducible interactions between GST-RXR LBD/RAR LBD and a group of proteins isolated from APL cell line NB4 nuclear extracts. Immobilized GST-RXR LBD/RAR LBD was incubated with 1.2 mg NB4 nuclear extract in the absence (−) or presence (+) of 5 μmol/L all-trans retinoic acid (t-RA) (lanes 2, 3). GST alone was used as a control (lane 1). Bound proteins were eluted, separated by SDS-PAGE, and visualized by nitrate silver staining. Approximate molecular weights of the interacting proteins are indicated at right. (C) Binding of the complex to retinoid receptors in response to selective ligands. Protein interactions were examined in the absence (−) or presence of 1 μmol/L t-RA (lane 2), 1 μmol/L RXR-specific ligand LG100153 (lane 3), and 1 μmol/L RAR-specific ligand LG100272 (lane 4).
As demonstrated above, t-RA appeared to induce the interaction of a specific protein complex to the RXR/RAR heterodimer. Because t-RA in vitro only binds to and activates RAR, we asked whether RXR binding could also be involved in the recruitment of the protein complex by using a synthetic RXR-selective agonist LG100153. Protein interactions were examined with the immobilized RXR LBD/RAR LBD in the presence of LG100153 (Figure 1C, lane 3) and compared with t-RA and a synthetic RAR-specific agonist, LG100272 (TTNPB) (lanes 2, 4); 1 μmol/L of the ligand was used to minimize cross-reactivity. Both selective agonists have been characterized; they have been reported to bind their respective receptors with high affinity and to transactivate selectively the reporter constructs on the respective response elements at this concentration.62-64 Figure 1C shows that t-RA and LG100272 equally induce the binding of the complex to receptor, whereas LG100153 has no effect. This suggests that the ligand-dependent interaction of the protein complex is mediated through the RAR partner of the retinoid receptor heterodimer. Further, we found that binding to RAR of an antagonistic ligand LG100815 did not recruit the protein complex (data not shown), suggesting that conformational changes associated with transcriptional activation are required for binding of the protein complex.
Retinoid receptor heterodimer interacts with the DRIP/TRAP complex
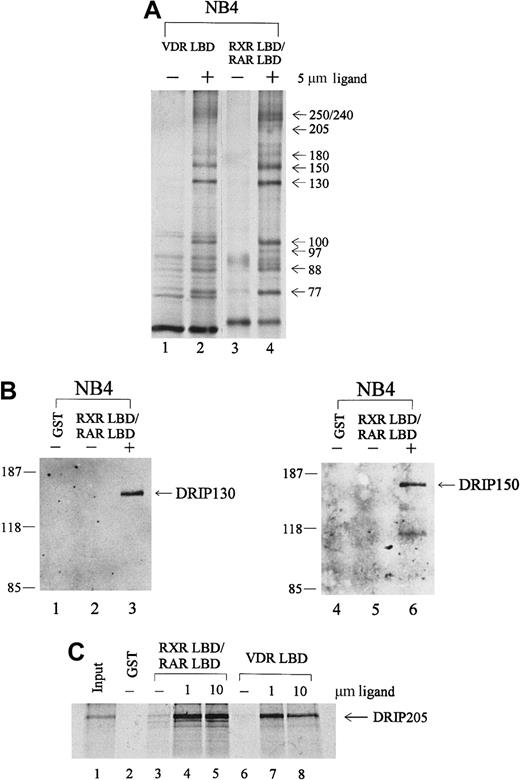
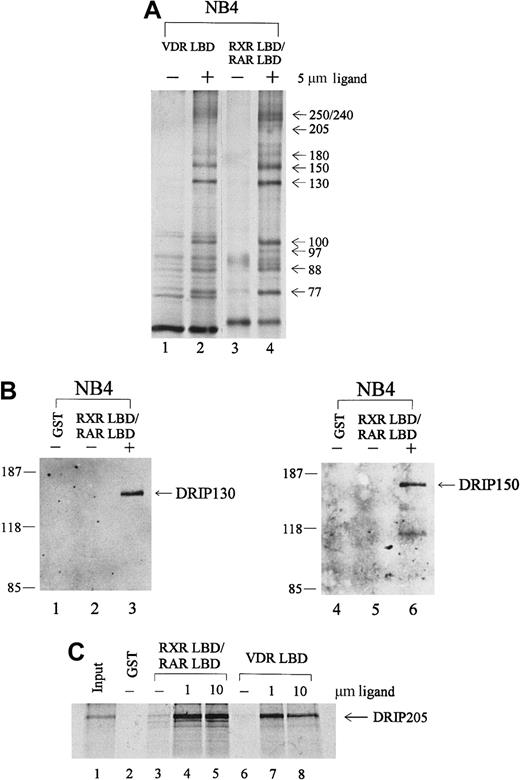
To identify proteins in the RAR/RXR-interacting complex, we performed immunoblot analyses with antibodies to known nuclear receptor coactivators. Anti-CBP, ACTR, and SRC-1 did not react with any of the proteins pulled down from NB4 extracts (data not shown). However, the distribution of these proteins by apparent molecular weight was similar to a recently identified coactivator complex, DRIP or TRAP, that shows ligand-dependent interactions with VDR and TR. To determine whether the protein complex we isolated from APL NB4 cells included proteins within the DRIP complex, we took several approaches. First, a GST pull-down assay was performed with immobilized VDR LBD, compared with the RXR LBD/RAR LBD heterodimer in the absence or presence of their respective ligands, 1,25-dihydroxy vitamin D3 (VD3) and t-RA. As shown in Figure 2A, an essentially identical group of interacting proteins was observed with liganded VDR LBD (lane 2) and RXR LBD/RAR LBD (lane 4). To confirm the identity of individual proteins, we obtained polyclonal antibodies to 2 members of the DRIP complex.65 Immunoblot analyses with antibodies to DRIP130 and DRIP150 confirmed their presence in the retinoid receptor–interacting proteins pulled down from NB4. Both antibodies recognized specific protein bands at their corresponding molecular weights only in the presence of t-RA (Figure 2B, lanes 3, 6).
Retinoid receptors interact with the DRIP/TRAP complex in APL NB4 cells.
(A) Comparison of the protein interactions to retinoid receptor heterodimer with those of the vitamin D3 receptor. Immobilized GST-VDR LBD or GST-RXR LBD/RAR LBD was incubated with 1.2 mg NB4 nuclear extract in the absence (−) or presence (+) of their respective ligands, 1,25-dihydroxy vitamin D3 (lane 2) and t-RA (lane 4). (B) Immunoblot analyses with 2 specific anti-DRIP antibodies. The RXR/RAR-interacting proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to Western blotting using antibodies to DRIP130 and DRIP150, respectively. (C) In vitro interaction of the DRIP205 protein to RXR/RAR heterodimer or VDR. Purified GST or GST-RXR LBD/RAR LBD and GST-VDR LBD were incubated with in vitro translated 35S-labeled DRIP205 in the absence (−) and presence of 1 μmol/L or 10 μmol/L t-RA (lanes 4, 5) and 1,25-dihydroxy vitamin D3 (lanes 7, 8) respectively. Twenty percent of the DRIP205 protein used was shown as input.
Retinoid receptors interact with the DRIP/TRAP complex in APL NB4 cells.
(A) Comparison of the protein interactions to retinoid receptor heterodimer with those of the vitamin D3 receptor. Immobilized GST-VDR LBD or GST-RXR LBD/RAR LBD was incubated with 1.2 mg NB4 nuclear extract in the absence (−) or presence (+) of their respective ligands, 1,25-dihydroxy vitamin D3 (lane 2) and t-RA (lane 4). (B) Immunoblot analyses with 2 specific anti-DRIP antibodies. The RXR/RAR-interacting proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to Western blotting using antibodies to DRIP130 and DRIP150, respectively. (C) In vitro interaction of the DRIP205 protein to RXR/RAR heterodimer or VDR. Purified GST or GST-RXR LBD/RAR LBD and GST-VDR LBD were incubated with in vitro translated 35S-labeled DRIP205 in the absence (−) and presence of 1 μmol/L or 10 μmol/L t-RA (lanes 4, 5) and 1,25-dihydroxy vitamin D3 (lanes 7, 8) respectively. Twenty percent of the DRIP205 protein used was shown as input.
Since previous reports identified DRIP205/TRAP220 as the subunit within the complex that interacts directly with VDR and TR,65-67we assessed whether DRIP205 could interact with the RXR/RAR heterodimer. 35S-labeled DRIP205 protein prepared by in vitro translation was allowed to bind RXR LBD/RAR LBD, and a significant ligand-dependent interaction was observed (Figure 2C, lanes 4, 5). Increasing the t-RA concentration from 1 μmol/L to 10 μmol/L did not further enhance the binding, suggesting that the interaction at 1 μmol/L already reached its maximum. Consistent with the earlier report, DRIP205 showed increased binding to VDR in the presence of its ligand VD3 (lanes 7, 8). We also examined the binding of DRIP205 to APL-specific fusion protein PML-RARα and saw a ligand-dependent interaction (Figure 4C). However, a dominant-negative PML-RARα that has a mutation within AF-2 of the ligand-binding domain56 does not interact with DRIP205 in the presence of ligand. This confirms that the integrity of the AF-2 domain is essential for interacting with DRIP proteins.35
Given the identity by apparent molecular weight of the complex binding to RXR/RAR and to VDR, the identification of 2 DRIP subunits for which specific antibodies were available, and the observation that DRIP205 could bind directly with the liganded retinoid receptor, we concluded that the RXR/RAR-interacting proteins isolated from NB4 cells were identical to those in the DRIP/TRAP complex.
Identical DRIP complex is present in RA-resistant APL cell lines
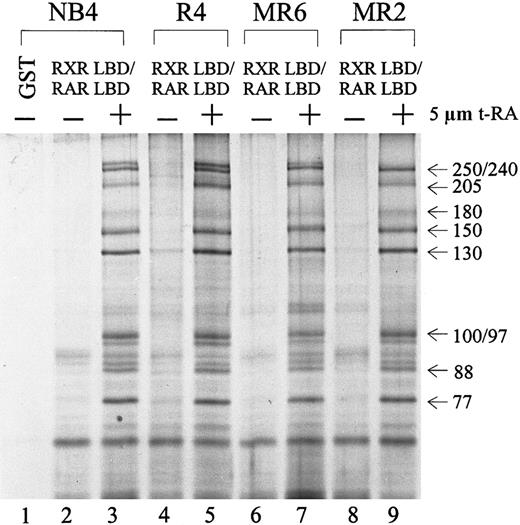
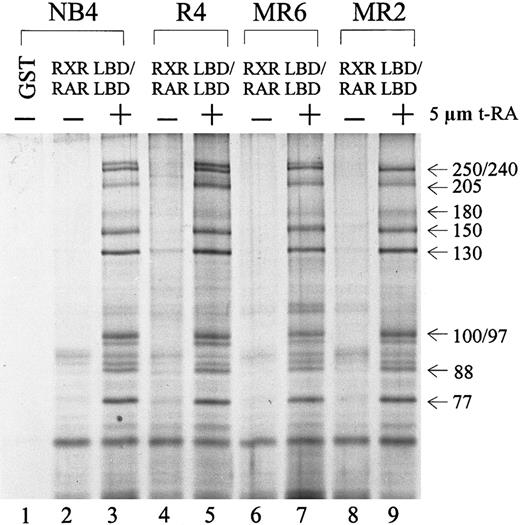
Because several RA-resistant cell lines display altered interactions between their retinoid receptors and cofactors in the presence of t-RA,55 we asked whether the particular DRIP complex isolated from NB4 cells was also present in its RA-resistant subclones. Protein complex interactions with the RXR/RAR in 3 resistant subclones—R4, MR6, and MR2—were compared to those in NB4. Figure3 shows an identical pattern of t-RA–dependent interacting proteins in all 4 cell lines examined (lanes 3, 5, 7, 9). To examine whether the DRIP complexes isolated from the resistant clones have altered affinity to retinoid receptors in comparison with NB4 cells, we reduced the concentration of t-RA in the binding assay to 100 nmol/L and still obtained an identical pattern of interaction in all cell lines (data not shown). This finding suggests that these RA-resistant cell lines maintain normal DRIP complexes that interact with retinoid receptors in a ligand-dependent manner. Thus, abnormalities in either expression or function of the protein components within the DRIP complex do not appear to account for the RA unresponsiveness in these resistant APL cells.
Ligand-inducible interaction of the DRIP/TRAP complex to RXR/RAR heterodimer in NB4 and 3 RA-resistant NB4 subclones.
Nuclear extracts (1.2 mg) prepared from different cell lines were incubated with 10 to 20 μg of the immobilized GST-RXR LBD/RAR LBD in the absence (−) or presence (+) of 5 μmol/L t-RA. GST alone was used as a negative control (lane 1). The approximate molecular weights of the interacting proteins are indicated at right.
Ligand-inducible interaction of the DRIP/TRAP complex to RXR/RAR heterodimer in NB4 and 3 RA-resistant NB4 subclones.
Nuclear extracts (1.2 mg) prepared from different cell lines were incubated with 10 to 20 μg of the immobilized GST-RXR LBD/RAR LBD in the absence (−) or presence (+) of 5 μmol/L t-RA. GST alone was used as a negative control (lane 1). The approximate molecular weights of the interacting proteins are indicated at right.
Expression of the PML-RARα oncoprotein does not affect function of the DRIP complex
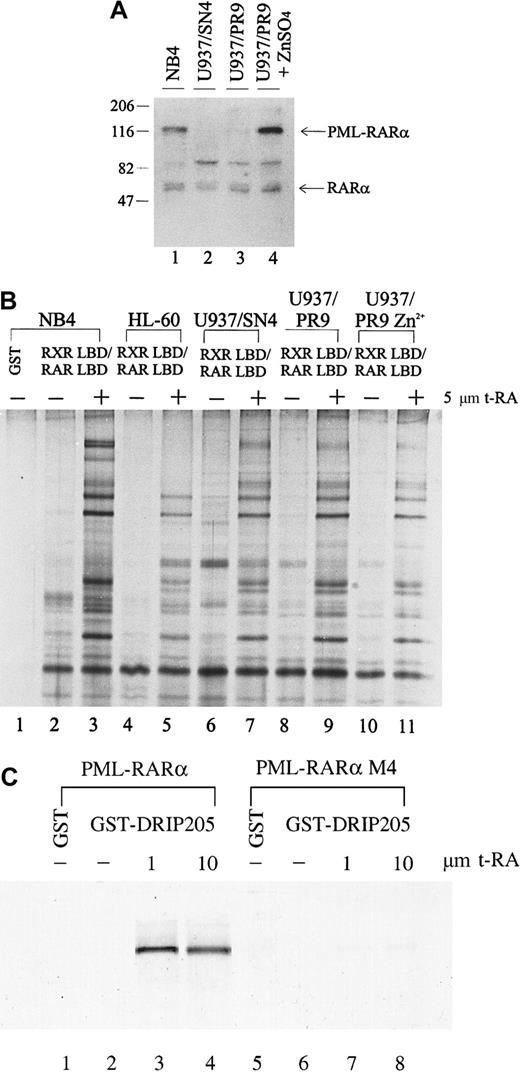
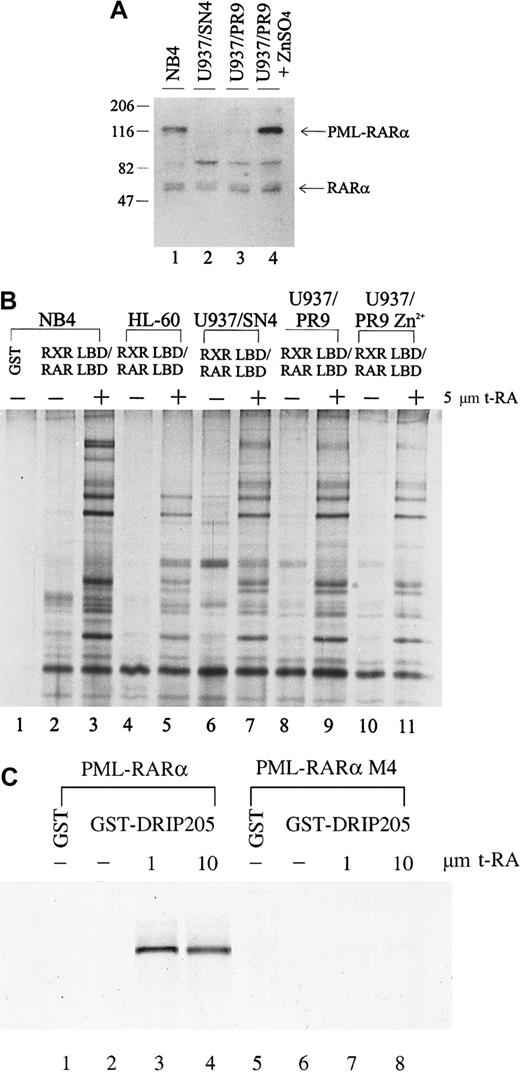
The chimeric PML-RARα gene plays a dual role in the phenotype of APL: it blocks cytodifferentiation at physiologic levels of RA but responds to pharmacologic concentrations of RA. The myeloid precursor U937 cell line differentiates poorly in response to RA; however, when stably expressing PML-RARα, these cells show increased sensitivity to RA-induced differentiation.68 To determine whether expression of the PML-RARα fusion protein affects either the expression of the DRIP components or their interaction with retinoid receptors, we compared the interaction between the DRIP complex and the retinoid receptors in U937 clones that differ in their PML-RARα expression. U937/PR9 is a subclone stably transfected with PML-RARα under the control of a Zn2+-inducible promoter, so that high levels of PML-RARα could be induced in these cells by treatment with ZnSO4. We measured the PML-RARα protein levels in U937/PR9 cells with or without the induction by 100 μmol/L ZnSO4 for 24 hours and compared them with those of natural PML-RARα–expressing NB4 and a U937/SN4 subclone transfected with vector alone (Figure 4A). NB4 expressed PML-RARα, whereas U937/SN4 did not (lanes 1, 2). U937/PR9 cells without Zn2+ induction had a low level of PML-RARα expression, which could be greatly induced by the treatment of 100 μmol/L ZnSO4 (lanes 3, 4). All cell lines examined expressed equal levels of RARα. When the binding of the DRIP complex to RXR/RAR in these cells was examined, we found that all cells displayed identical ligand-dependent DRIP interaction to the receptors regardless of their PML-RARα expression levels (Figure 4B, lanes 7, 9, 11). These observations suggest that induced expression of the oncoprotein PML-RARα does not alter the expression or repress the function of the DRIP components. Furthermore, consistent with our findings in the RA-resistant APL cell lines, U937 clones with different sensitivity to RA have identical patterns of ligand-dependent DRIP binding.
Interaction between the DRIP complex and the retinoid receptors in U937 clones that differ in their PML-RARα expression.
(A) Expression of RARα and fusion PML-RARα proteins in NB4 and U937 subclones by Western blot analysis. Twenty micrograms of the nuclear extracts were run on SDS-PAGE, transferred, and probed with an anti-RARα antibody recognizing its F domain. Positions of RARα and the fusion PML-RARα proteins are indicated. (B) Interaction of the DRIP/TRAP complex with retinoid receptors in U937 subclones (lanes 6-11), HL-60 cells (lanes 4, 5). (C) Direct interaction between the PML-RARα protein and the DRIP205. In vitro translated35S-labeled PML-RARα and mutant PML-RARα M4 were incubated with purified GST-DRIP205 in the absence (−) and presence (+) of 1 μmol/L or 10 μmol/L t-RA (lanes 3, 4, 7, 8). GST alone was included as the negative control (lanes 1, 5).
Interaction between the DRIP complex and the retinoid receptors in U937 clones that differ in their PML-RARα expression.
(A) Expression of RARα and fusion PML-RARα proteins in NB4 and U937 subclones by Western blot analysis. Twenty micrograms of the nuclear extracts were run on SDS-PAGE, transferred, and probed with an anti-RARα antibody recognizing its F domain. Positions of RARα and the fusion PML-RARα proteins are indicated. (B) Interaction of the DRIP/TRAP complex with retinoid receptors in U937 subclones (lanes 6-11), HL-60 cells (lanes 4, 5). (C) Direct interaction between the PML-RARα protein and the DRIP205. In vitro translated35S-labeled PML-RARα and mutant PML-RARα M4 were incubated with purified GST-DRIP205 in the absence (−) and presence (+) of 1 μmol/L or 10 μmol/L t-RA (lanes 3, 4, 7, 8). GST alone was included as the negative control (lanes 1, 5).
The PML-RARα protein retains most functional domains of RARα and functions as an RA-dependent transcriptional activator in a cell type- and a promoter-specific manner.43-45 To assess whether PML-RARα itself interacts with the DRIP complex directly, we performed in vitro protein interaction assays. In vitro translated35S-labeled PML-RARα protein was allowed to interact with the purified GST-DRIP205 in the absence or presence of increasing concentrations of t-RA (Figure 4C). As a result, PML-RARα showed a clear ligand-dependent interaction with the DRIP205 protein. On the contrary, PML-RARα M4, a dominant-negative mutant PML-RARα identified in an RA-resistant cell line that has lost its ligand-binding ability,56 failed to bind DRIP205. We also tested for an in vitro interaction between the PML protein and DRIP205 but did not observe any direct binding (data not shown), confirming that the interaction of PML-RARα to the DRIP complex is mediated through its RARα portion.
We extended our analysis to other non-APL cancer cell lines, including myelocytic leukemia cell lines HL-60 (Figure 4B, lanes 4, 5) and KG-1, the Namalwa B-cell line, and the breast cancer cell line MDA231. We found that KG-1, Namalwa B cells, and MDA231 all exhibited the same RXR/RAR-interacting complex as observed in NB4 cells (data not shown). Initially, HL-60 cells did not show the receptor interaction of the 3 high-molecular-weight protein components at 205, 240, and 250 kd, respectively (Figure 4B). However, when higher concentrations of protease inhibitors were used in the preparation of HL-60 nuclear extract, a separate GST pull-down experiment detected the DRIP205 subunit within the complex (data not shown). Northern analysis also revealed that HL-60 expressed all 3 genes at levels similar to those of NB4 (data not shown).
Discussion
Great efforts have been made in recent years to understand the diverse functions of nuclear receptors. Crystallography studies of several nuclear receptors' LBD structures and discoveries of various receptor coregulatory factors have brought insights to these complex hormonal signaling pathways.69-73 We report here the isolation of a group of distinct proteins from APL NB4 cells that interact with the LBD of the retinoid receptor heterodimer in a ligand-dependent manner. As shown by receptor-selective ligands, the interaction of this complex is mediated through RAR, not RXR (Figure1). The proteins in the complex are identified as virtually identical to those in the coactivator complex DRIP/TRAP on the basis of comparative electrophoresis, immunoblot analyses, and a direct protein–protein interaction assay (Figure 2). Expression of the APL-specific oncoprotein PML-RARα does not repress expression of the DRIP proteins or their interactions with retinoid receptors (Figure 4). Rather, the PML-RARα protein can interact directly with the DRIP component in an RA-induced manner, similar to that of wild-type RARα. To our knowledge, this is the first demonstration that the DRIP complex interacts with RXR/RAR and PML-RARα in a retinoid-dependent fashion. The DRIP complex was initially purified from Namalwa B cells by a GST pull-down strategy similar to that presented here,35whereas the TRAP complex was isolated by affinity purification of a FLAG epitope-tagged TR from HeLa cells grown in the presence of thyroid hormone.36 The DRIP/TRAP complex was demonstrated by in vitro transcription assays to be a ligand-dependent transcription activator.
Our immunoblot analyses with antibodies to known coactivators, including SRC-1, ACTR, and CBP, did not identify them among the DRIP proteins. This is consistent with the sequencing data on the DRIP subunits, whose coding sequences share little homology with the SRC-1 family of the coactivators and thus belong to a distinct family of coactivators. Indeed, a recent report by Treuter and coworkers67 showed that the DRIP205/TRAP220 subunit competes with the SRC-1 family of coactivators and with CBP for binding to the nuclear receptors and that their binding to the receptor LBD AF-2 domain is mutually exclusive.
We found the ubiquitous presence of the DRIP complex in all tested resistant APL clones and in several non-APL cancer cell lines, varying from leukemia cells to breast cancer cells. Our results show that 3 APL clones that have lost transcription and differentiation responses to RA retain the same DRIP complex as NB4 and exhibit functional ligand-dependent association of the complex to the receptor at concentrations of t-RA as low as 100 nmol/L (Figure 3; data not shown). Because these cells are resistant to more than 100-fold higher concentrations of t-RA, these data strongly suggest that reduced affinity of DRIP/TRAP to retinoid receptors is not the cause of RA resistance in these cells. In accordance with this finding, when U937 cells that differentiate poorly in response to RA are induced to respond by the expression of PML-RARα in U937/PR9 cells, there is no change in expression and ligand-dependent binding of the DRIP complex to RXR/RAR (Figure 4A,B). These data suggest that the DRIP complex may play a broader role in gene regulation, whose function is not limited to any specific nuclear receptor–induced cell differentiation pathway. This hypothesis is further supported by the identification of the DRIP205 subunit as a p53-regulatory protein (RB18A).74 In addition, several new reports have found essentially the same complex DRIP/TRAP as a coactivator for other families of transcription factors. Ito et al75,76 showed that the DRIP/TRAP proteins are virtually identical to an SRB- and a Med-containing cofactor complex, SMCC. The SMCC contains homologs of a subset of yeast mediator–holoenzyme components that interact with RNA polymerase II and that have been implicated as global and specific activators for genes such as p53 and VP16. Furthermore, a similar complex, ARC (activator-recruited cofactor) was isolated by Naar et al77,78 to bind to and to activate sterol-responsive, element-binding protein SREBP-1a and other transcriptional activators, including VP16 and NF-κB.77 78 In conclusion, others and we have found that the identical complex interacts with several classes of transcriptional activators and that loss or gain of nuclear receptor–mediated transcriptional activation and differentiation does not influence DRIP expression and ligand-dependent binding to receptors. These results reflect a convergence of transcriptional regulation, suggesting that the DRIP/TRAP complex functions as a global transcription cofactor for a spectrum of pathways.
Acknowledgments
We thank Dr Ronald M. Evans for providing the bicistronic RXR/RAR expression vector, Dr Richard L. Momparler for 1,25-dihydroxy vitamin D3 ligand, Dr Reid Bissonnette (Ligand Pharmaceuticals) for RXR- and RAR-selective ligands, Dr Pierre Chambon for the anti-RARα antibody, and Dr Pier Giuseppe Pelicci for the U937/SN4 and U937/PR9 cell lines.
Supported by a grant from the Medical Research Council of Canada (MRC). W.S. was supported by an MRC Studentship Award, and W.H.M. is a Scholar of the MRC.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wilson H. Miller Jr, 3755 Chemin de la Cote-Ste-Catherine, Montreal, Quebec, Canada H3T 1E2.