Abstract
Monocytes play a pivotal role in various human infectious and inflammatory diseases. To reveal a whole picture of pathophysiologic function of activated human monocytes, this study used the serial analysis of gene expression (SAGE) procedure in lipopolysaccharide (LPS)-stimulated human monocytes. A total of 35 874 tags corresponding to more than 12 000 different transcripts were sequenced. Comparison of gene expression profile with that of resting monocytes revealed the LPS-inducible gene expression profile. Many cytokines and chemokines, including interleukin (IL)-6, IL-1α, IL-1β, tumor necrosis factor (TNF)-α, macrophage inflammatory protein (MIP)-1β, MIP-2β, MIP-2α, liver and activation-regulated chemokine (LARC), MIP-1α, thymus and activation-regulated chemokine (TARC), macrophage-derived chemokine (MDC), regulated on activation, normal T cell expressed and secreted (RANTES), growth-regulated oncogene (GRO) α, and IL-8, were observed in the highest inducible transcripts. Other genes encoding plasminogen activator inhibitor type 2 (PAI-2), Hc-gp39, apolipoproteins, malate dehydrogenase, matrix metalloproteinase-9 (MMP-9), and cyclooxygenase (COX2) were also highly elevated in LPS-stimulated monocytes. Moreover, up-regulation of Naf1β, IL-7 receptor, adenosine receptor A2a, and many novel genes was newly identified. These results suggest that the LPS-inducible gene products may be involved in cell activation and migration, angiogenesis, tissue remodeling, and metabolism, and thus may orchestrate the inflammatory reactions. On the other hand, the expression of numerous sets of novel genes was discovered to be down-regulated on LPS stimulation. This study represents the first comprehensive analysis of LPS-inducible gene expression in human monocytes and provides tremendous novel information for the function of LPS-activated monocytes and targets for diagnosing, monitoring, and treating sepsis and various human infectious and inflammatory diseases.
Introduction
Sepsis is now a contributing factor in more than 100 000 deaths per year in the United States and the annual incidence is nearly 300 000 cases.1,2 In general, the septic response occurs when an invading microbe has circumvented the innate and acquired immune defenses. Lipopolysaccharide (LPS, also called endotoxin), a component of the outer membrane of gram-negative bacteria, is the most potent and well-characterized gram-negative bacterial signal molecule. LPS interacts with CD14 together with Toll-like receptor (Tlr) molecule(s),3-6 a receptor complex on monocytes/macrophages and neutrophils, and activates second messenger and signal transduction pathways. These signals in turn activate transcription factors, mainly nuclear factor (NF)-κB. As a result, LPS triggers a wide variety of cellular responses, including the production of cytokines and chemokines, release of arachidonic acid metabolites, and generation of reactive oxygen and nitrogen intermediates that are responsible for the pathophysiologic reactions.2 7-9 Monocytes play an essential role in infection and inflammation by producing these mediators. Although many LPS-inducible genes have been investigated, no comprehensive study of gene expression in LPS-stimulated monocytes has been reported.
Several methods such as subtractive hybridization10 and differential display11 have been developed to identify changes in expression profiles under different conditions. However, these methods could analyze very limited numbers of transcripts and did not provide the quantitative information. In contrast, the serial analysis of gene expression (SAGE)12-18 is a technique that allows a rapid, detailed analysis of thousands of transcripts simultaneously. This method can be used not only to characterize quantitative information on the abundance of known transcripts but also to identify novel expressed genes. We have recently reported the results of the SAGE technique with human monocytes and macrophages19 and monocyte-derived dendritic cells.20 In this study, we performed SAGE on human blood monocytes and LPS-stimulated monocytes to evaluate the general patterns of gene expression following LPS stimulation.
Materials and methods
Purification of human monocytes
Peripheral blood mononuclear cells (PBMCs) were obtained from the venous blood drawn from 8 healthy volunteers in Tokyo Metropolitan Red Cross Blood Center and were used to prepare human monocytes. PBMCs were isolated by centrifugation on a Ficoll-Metrizoate density gradient (d = 1.077 g/mL, Lymphoprep; Nycomed, Oslo, Norway) and were suspended in RPMI 1640 medium containing 10% FCS (heat-inactivated fetal calf serum, Gibco, Gaithersburg, MD), 100 μg/mL streptomycin, and 100 U/mL penicillin. This medium contained less than 3 pg of LPS per milliliter as assessed by a Limulus amebocyte lysate. PBMCs were then incubated with anti-CD14 monoclonal antibody coated with microbeads, and monocytes were isolated by passing the PBMCs through a magnetic cell separation system (MACS; Miltenyi Biotec, Bergish Gladbach, Germany) with column type VR. The total number of isolated monocytes was 3 × 108. The highly purified monocytes were obtained after the incubation on the plastic tissue culture plate for 30 minutes at 37°C, 5% CO2 in air. More than 99% of the cells were judged to be monocytes by morphology, positive staining for CD14 (LeuM3, Becton Dickinson, San Jose, CA) in a flow cytometric analysis, and nonspecific esterase staining.20To prepare LPS-stimulated monocytes, the freshly isolated monocytes were incubated with 1 μg/mL of LPS (Escherichia coliO55:B5, Difco, Detroit, MI) in RPMI 1640 medium containing 10% FCS in 5% CO2 at 37°C for 3 hours. Monocytes and LPS-stimulated monocytes were prepared from 8 donors.
SAGE protocol
The total RNA of the stimulated cells was isolated by direct lysis using RNAzol B (Cinna/Biotex Laboratories, TEL-TEST, Friendswood, TX). Poly(A)+RNA was further isolated using the FastTrac messenger RNA (mRNA) purification kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. SAGE was performed as described previously.12-15 Poly(A)+RNA (2.5 μg) was converted to double-stranded complementary DNA (cDNA) with a BRL synthesis kit (GIBCO BRL, Rockville, MD) following the manufacturer's protocol with the inclusion of primer biotin-5′-T18-3′. The cDNA was cleaved with NlaIII and the 3′-terminal cDNA fragments were bound to streptoavidin-coated magnetic beads (Dynal, Oslo, Norway). After ligation of oligonucleotides containing recognition sites for BsmFI, the linked cDNAs were released from the beads by digestion with BsmFI. The released tags were ligated to one another, concatenated, and cloned into the SphI site of pZero 1.0 (Invitrogen). Colonies were screened with polymerase chain reaction (PCR) using M13 forward and M13 reverse primers. PCR products containing inserts of more than 400 bp were sequenced with the TaqFS Dyeterminater kit and analyzed using a 377 ABI automated sequencer (Perkin-Elmer, Branchburg, NJ). All electropherograms were reanalyzed by visual inspection to check for ambiguous base and to correct misreads.
The SAGE procedure was performed on mRNA from human monocytes and LPS-stimulated monocytes. Sequence files were analyzed with the SAGE software,12 CGAP SAGE database (http://www.ncbi.nlm.nih.gov/SAGE/), and NCBI's sequence search tool (Advanced BLAST search, http://www.ncbi.nlm.nih.gov/BLAST/). After elimination of linker sequences and the repeated ditags, a total of 93 433 tags representing 57 559 and 35 874 from human monocytes and LPS-stimulated monocytes, respectively, were analyzed. To compare these 2 SAGE libraries, each tag number was normalized using SAGE software12 by calculating each total tag number to 35 800.
Reverse transcriptase-PCR (RT-PCR)
Total RNA (200 ng) was prepared by use of RNAzol B. The RNA was reverse-transcribed in 50 μL of 10 mmol/L Tris-HCl (pH 8.3), 6.5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L dithiothreitol, 1 mmol/L of each dNTP, 2 μmol/L random hexamer, and 2.4 U/μL of Moloney murine leukemia virus reverse transcriptase for 1 hour at 42°C. cDNA, corresponding to 40 ng of total RNA, was boiled for 3 minutes and quenched on ice before amplification by PCR. The conditions for PCR were as follows: in a 50-μL reaction, 0.15 μmol/L of each primer; 125 μmol/L each of dGTP, dATP, dCTP, and dTTP (Toyobo, Osaka, Japan); 50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.3; 1.5 mmol/L MgCl2 and AmplyTaq (Perkin-Elmer). Primers used for RT-PCR are listed in Table 1. Reaction mixtures were incubated in a Perkin-Elmer DNA Thermal Cycler for 25 to 30 cycles (denaturation for 60 seconds at 94°C, annealing for 60 seconds at 58°C, extension for 120 seconds at 72°C).
Statistical analysis
Statistical significance between samples was calculated as described previously.16
Results
Results from LPS-stimulated monocytes
In LPS-stimulated monocytes library, a total of 35 874 tags were sequenced. These tags corresponded to more than 12 000 different transcripts. The expressed genes were searched through the GenBank database to identify individual genes. Table2 shows the top 50 transcripts in LPS-stimulated monocytes. The most expressed genes were identified as macrophage inflammatory protein-1α (MIP-1α), with the expression frequency of 3.34%, followed by ferritin L chain and ferritin H chain. Other highly expressed genes consist of products associated with cytokines, chemokines, protein synthesis, lipid metabolism, and major histocompatibility complex (MHC) class I and class II.
Comparison of gene expression profile between resting and LPS-stimulated monocytes
We previously reported results of SAGE on human monocytes and macrophages.19 To investigate the LPS-stimulation regulated genes in human monocytes, we compared the unstimulated19 and stimulated SAGE tag libraries after normalization of each tag number using SAGE software.12Comparison of expression patterns revealed that most genes (> 20 000 transcripts) were expressed at similar levels between resting monocytes and LPS-stimulated monocytes (Figure 1). However, the expression profiles also revealed 250 transcripts that were expressed at significantly different levels (P < .01). Expression levels of 132 of 250 genes were decreased in LPS-stimulated monocytes as compared with those in unstimulated monocytes. Conversely, 118 transcripts were overexpressed in LPS-stimulated monocytes.
Comparison of gene expression patterns in resting and LPS-stimulated monocytes.
A semilogarithmic plot reveals 40 tags that were decreased more than 10-fold in LPS-stimulated monocytes, whereas 60 tags were increased more than 10-fold. After normalization of the number of each tag, the relative expression of each transcript was determined by dividing the number of tags observed in monocytes or LPS-stimulated monocytes as indicated. To avoid division by 0, we used a tag value 1 for any tag that was not detectable in one of the samples. These ratios are plotted on the abscissa. The number of genes displaying each ratio is plotted on the ordinate. Mono and LPS indicate resting monocytes and LPS-stimulated monocytes, respectively.
Comparison of gene expression patterns in resting and LPS-stimulated monocytes.
A semilogarithmic plot reveals 40 tags that were decreased more than 10-fold in LPS-stimulated monocytes, whereas 60 tags were increased more than 10-fold. After normalization of the number of each tag, the relative expression of each transcript was determined by dividing the number of tags observed in monocytes or LPS-stimulated monocytes as indicated. To avoid division by 0, we used a tag value 1 for any tag that was not detectable in one of the samples. These ratios are plotted on the abscissa. The number of genes displaying each ratio is plotted on the ordinate. Mono and LPS indicate resting monocytes and LPS-stimulated monocytes, respectively.
Table 3 lists the 50 most highly expressed tags in LPS-stimulated monocytes. These LPS-induced transcripts mainly consist of genes encoding proteins associated with cytokines and chemokines including interleukin (IL)-6, IL-1α, IL-1β, tumor necrosis factor (TNF), MIP-1β, MIP-2β, MIP-2α, liver and activation-regulated chemokine (LARC), thymus and activation-regulated chemokine (TARC), macrophage-derived chemokine (MDC), regulated on activation, normal T cell expressed and secreted (RANTES), and growth-regulated oncogene α (GRO α); lipid metabolism such as apolipoprotein C-1, apolipoprotein E, and lysosomal acid lipase; and proteases and inhibitors such as plasminogen activator inhibitor type 2 (PAI-2), matrix metalloproteinase-9 (MMP-9), and cystatin B.
Table 4 shows the top 50 transcripts identified to be decreased in LPS-stimulated monocytes. The mRNAs encoding some ribosomal proteins, DNA-binding proteins such as GOS3, c-fos, jun-D, zing finger protein (tristetraprolin), and C/EBPδ, differentiation-related protein such as MCL1, and cell structure-related proteins such as actin-binding protein p57 and ARC41 were decreased.
RT-PCR of genes represented in the SAGE
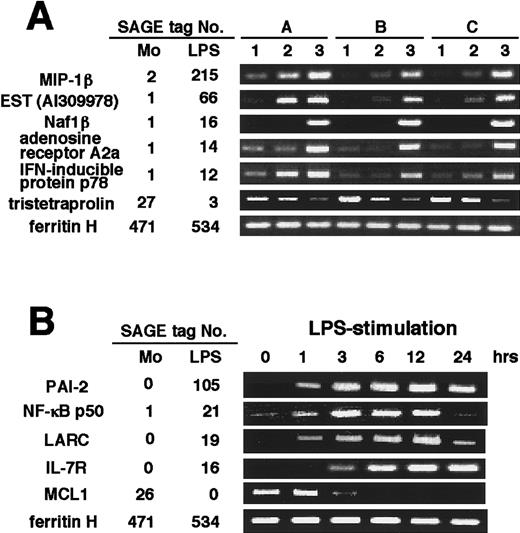
Although we obtained the blood from 8 healthy volunteers to find the average in the gene expression, one question that still arises from these data is if a difference exists in the gene expression among individual donor-derived cells. Another possibility is that the change of gene expression was due merely to cell adhesion to the plastic tissue culture plate.21 22 To address these questions, we arbitrarily selected 6 differentially expressed transcripts and evaluated them in 3 donor-derived samples of unstimulated, incubated for 3 hours without LPS-stimulation, and LPS-stimulated monocytes by RT-PCR (Figure 2A). The expression of each transcript was compared to SAGE data (Tables 3 and 4). MIP-1β (resting monocytes [Mo] 2:LPS-stimulated monocytes [LPS] 215), EST (AI309 978) (Mo 1:LPS 66), Naf1β (Mo 1:LPS 16), adenosine receptor A2a (Mo 1:LPS 14), and interferon (IFN)-inducible protein p78 were highly induced (Mo 1:LPS 12); whereas tristetraprolin (Mo 27:LPS 3) was reduced and ferritin H was expressed almost equally (Mo 471:LPS 534). Although there were some different expression levels of transcripts among the donors, it was confirmed that the induction of the identified genes was due to the specific stimulation of LPS rather than simple adhesion. Moreover, we selected 5 differentially expressed transcripts and examined the time course of the induction of these genes after LPS stimulation (Figure 2B). The expression of the genes encoding PAI-2 (Mo 0:LPS 105), NF-κB p50 (Mo 1:LPS 21), LARC (Mo 0:LPS 19), and IL-7 receptor (IL-7R) (Mo 0:LPS 16) was highly induced up to 12 hours, but that of NF-κB p50 and LARC was barely detected at 24 hours, whereas the expression of MCL1 gene was gradually decreased (Mo 26:LPS 0). These results validate our SAGE data for resting and LPS-stimulated monocytes and establish the general expression profiles of the LPS-inducible genes in human monocytes.
RT-PCR analysis of genes expressed differently in resting and LPS-stimulated monocytes.
(A) RT-PCR was performed on total RNA isolated from human resting monocytes (1), monocytes only incubated for 3 hours without LPS (2), and monocytes cultured for 3 hours with LPS (3), as described in Materials and methods. A, B, and C indicate different donors. Mono and LPS indicate resting monocytes and LPS-stimulated monocytes, respectively. SAGE tag No. is the number of times the tag was identified as shown in Tables 3 and 4. (B) Monocytes were incubated with LPS for 0, 1, 3, 6, 12, 24 hours, respectively, and RT-PCR was performed on these samples from 3 different donor-derived cells. A representative result is shown.
RT-PCR analysis of genes expressed differently in resting and LPS-stimulated monocytes.
(A) RT-PCR was performed on total RNA isolated from human resting monocytes (1), monocytes only incubated for 3 hours without LPS (2), and monocytes cultured for 3 hours with LPS (3), as described in Materials and methods. A, B, and C indicate different donors. Mono and LPS indicate resting monocytes and LPS-stimulated monocytes, respectively. SAGE tag No. is the number of times the tag was identified as shown in Tables 3 and 4. (B) Monocytes were incubated with LPS for 0, 1, 3, 6, 12, 24 hours, respectively, and RT-PCR was performed on these samples from 3 different donor-derived cells. A representative result is shown.
More information on this work is available at our web site (http://www.prevent.m.u-tokyo.ac.jp/SAGE.html).
Discussion
Experimental and clinical studies have provided details about a series of expressions and mechanisms of action of LPS-inducible gene products.7 However, these studies provided only the limited number of the well-known mediators and the global quantitative analysis of gene expression has never been conducted. In this study, we have applied the recently developed SAGE method to allow quantitative analysis of a large number of transcripts in human LPS-stimulated monocytes.
Among thousands of differentially expressed genes, many cytokine and chemokine genes were identified to be highly inducible as expected. The widely studied proinflammatory cytokines, IL-1, IL-6, and TNF, were expressed at higher levels in LPS-stimulated monocytes than resting cells and should play a central role in the initiation of systemic responses (Table 3 and the categorized list in Table5). Chemokines are small proteins with 4 conserved cysteines forming 2 essential disulfide bonds. Four subfamilies, CXC, CC, C, and CX3C chemokines, are distinguished according to the position of the first 2 cysteines. Now chemokine functions extend far beyond leukocyte physiology and include leukocyte trafficking, viral infection, angiogenesis, hematopoiesis, and antitumor effects.23,24 Surprisingly, many chemokines were up-regulated simultaneously on LPS-stimulation. Among them, CC chemokines, including MIP-1β, LARC, MIP-1α, TARC, MDC, RANTES (Table 3 and the categorized list in Table 5), monocyte chemoattractant protein-1 (MCP-1), and MCP-2 (Table 5), were highly induced. These CC chemokines mainly chemoattract monocytes, basophils, eosinophils, and lymphocytes. The other major chemokine subfamily, CXC chemokines, including MIP-2β (GRO γ), MIP-2α (GRO β), GRO α (Tables 3 and5), and IL-8 (Tables 2 and 5), were also increased. Interestingly, all these highly inducible CXC chemokines belong to the subgroup called ELR chemokines, which have a 3-amino acid motif, ELR (glutamate-leucin-arginine) between the N-terminus and the first cysteine. These ELR CXC chemokines have an apparent uniformity of function such as neutrophil chemoattractants and activator, endothelial cell chemoattractants, and angiogenic factors. In contrast, non-ELR CXC chemokines inhibit the angiogenic effects of ELR chemokines and basic fibroblast growth factor (bFGF),23 and the transcripts of this subgroup were not increased significantly in LPS-stimulated monocytes. These data suggest that monocytes are involved in inflammatory angiogenesis.25 It was reported that monocytes adhere to the vascular wall during angiogenesis and monocyte activation plays a major role in angiogenesis and collateral artery growth.26 TNF and bFGF were presumed to be involved in this event.27-29 However, in addition to these factors, our results showed other mediators such as ELR CXC chemokines might contribute to the inflammatory angiogenesis. Moreover, these results indicate that a wide variety of chemokines may orchestrate under pathophysiologic conditions and play a role in inflammatory reactions.
In contrast, the gene expression of receptors of cytokine and chemokine was not significantly changed after LPS stimulation except for the up-regulation of the IL-7R gene (Table 3 and the categorized list in Table 5). Through the IL-7–induced production of inflammatory cytokines by monocytes,30-32 IL-7R may contribute to inflammation and tumor immunity.30
Disseminated intravascular coagulation (DIC) is frequently associated with sepsis and can be a life-threatening bleeding disorder.9 LPS is known to activate several steps in the coagulation cascade. The accelerated coagulation reactions cause small thrombi and emboli throughout the microvasculature followed by a phase of procoagulant consumption and secondary fibrinolysis. In this study, PAI-2, a member of the serine protease inhibitor (Serpin) superfamily,33 was one of the highest induced genes in LPS-stimulated monocytes (Table 3 and the categorized list in Table 5). Suppression of the fibrinolytic system directly contributes to the persistence of intravascular clots. PAI-2 is an inhibitor of urokinase-type plasminogen activator (u-PA) and may be involved in fibrinolytic suppression and modulation of DIC. Furthermore, because u-PA generates plasmin in events involving degradation of extracellular matrix,34 PAI-2 may also contribute to fibrin deposition, tissue remodeling, and cell migration at the site of inflammation.
Other known inflammation-related transcripts such as MMP-9 and inducible cyclooxygenase (COX2) were increased on LPS stimulation (Table 3). These results suggest that the proteinase MMP-9 may be involved in local remodeling and repair,35-37 and the enzyme COX2 may play a critical role in inflammation and modulate the systemic reaction by converting arachidonic acid to prostaglandins and thromboxans.38
Another novel observation is the up-regulation of Naf1β (Nef-associated factor 1β) and adenosine receptor A2a (Table 3). Naf1β is human immunodeficiency virus-1 (HIV-1) Nef-binding protein that increased cell surface CD4 expression.39 The role of Naf1β in LPS-stimulated monocytes remains to be elucidated. Adenosine is a potent endogenous anti-inflammatory agent released by cells under circumstances of metabolic stress, such as ischemia. Currently, at least 4 adenosine receptors, A1, A2a, A2b, and A3, have been identified.40 The increased expression of adenosine receptor A2a is likely to contribute to systemic control of inflammation via local inhibition of the cytokine synthesis.41 42
In this study, many genes were very unexpectedly found to be decreased on LPS stimulation (Table 4). MCL-1 is a member of the Bcl-2 family expressed in early monocyte differentiation43 and has moderate viability-enhancing effects in a spectrum of hematopoietic cells in transgenic mice.44 The down-regulation of “early differentiation gene” MCL-1 may relate to the activated state of the monocytes rather than differentiated state and to the rapid turnover of the mRNA.43 Immediate early genes such as c-fos, jun-D, andtristetraprolin45 and other DNA binding proteins like GOS3 and C/EBPδ were decreased. Our results suggest that these down-regulated transcripts may control the LPS-induced gene expression transiently to the adequate level. Although the biologic significance of the decreased expression of numerous genes mostly remains to be clarified, this discovery may open a new biologic research field of activated monocytes.
In conclusion, we completed SAGE of LPS-stimulated monocytes. Comparison between resting and activated monocytes revealed the comprehensive LPS-inducible gene expression profiles in monocytes. This study confirms the pivotal role of monocytes with a variety of functions in inflammation, tissue injury and repair, angiogenesis, and metabolism. The data revealed in this study should provide basic information necessary for further understanding of the molecular pathogenesis of sepsis and be useful in diagnosing or monitoring human various infectious and inflammatory diseases in combination with a developing DNA microarray system. Furthermore, tag sequences of unknown genes identified here suffice to clone their cDNAs, and some of these gene products may be useful as pharmaceuticals or targets for novel intervention therapy of inflammatory diseases.
Acknowledgments
We are very grateful to Dr V. E. Velculescu, L. Zhang, W. Zhou, B. Vogelstein, and K. W. Kinzler for their help with the SAGE procedures, and also to Dr H. Young for reviewing this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kouji Matsushima, Department of Molecular Preventive Medicine, School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan; e-mail:koujim@m.u-tokyo.ac.jp.