Abstract
Identification of the 25 known human blood group molecules is of fundamental importance for the fields of erythroid cell biology and transfusion medicine. Here we provide the first molecular description of the “Dombrock” blood group system. A candidate gene was identified by in silico analyses of approximately 5000 expressed sequence tags (ESTs) from terminally differentiating human erythroid cells. Transfection experiments demonstrated specific binding of anti-Dombrock and confirmed glycosylphosphatidylinositol membrane attachment. Dombrock expression is developmentally regulated during erythroid differentiation and occurs at highest levels in the fetal liver. Homology studies suggest that the Dombrock molecule is a member of the adenosine 5′-diphosphate (ADP)–ribosyltransferase ectoenzyme gene family. Genotypic comparisons suggest Doa versus Dob antigenicity results from a single amino acid substitution within an encoded arginine-glycine-aspartic acid (RGD) motif of the molecule.
Introduction
Based upon their clinical importance for transfusion medicine, enormous effort has been directed toward the study of 25 genetically distinct molecules on human erythrocytes collectively known as the human blood groups.1 Over 200 antigenic variations of these molecules have been defined by serology and genetic linkage studies. Most of the molecules have been cloned and identified as a functionally diverse group of membrane transporters, complement regulatory molecules, adhesion molecules, and ectoenzymes.2 Molecular genetic studies have strongly linked blood group polymorphisms to the severity of malarial disease.3,4 In addition, polymorphic loci associated with blood antigenicity may be useful for linkage studies for a variety of other diseases.5 Hence, molecular and functional aspects of human blood groups provide a rich source of information relevant for studies of erythroid cell biology, human polymorphisms, and transfusion medicine.
The Dombrock (Do) blood group was discovered with serological tests of blood and named after the original Doa serum donor in 1965.6 Nearly a decade passed before the discovery of the antithetical antigen (Dob).7 These antigens are quite informative as genetic markers withDoa gene frequencies of 0.42, 0.33, 0.12, and 0.07 in Northern European, Black American, Japanese, and Thai populations, respectively.8 Three additional antigens (Gya, Hy, and Joa) that are carried on the Dombrock blood group molecule are “high incidence” antigens, with gene frequencies predicted at greater than 99% in all populations studied.9 Clinically, the blood group has not been associated with hemolytic disease of the newborn, but severe hemolytic transfusion reactions due to the presence of anti-Dombrock antibodies have been reported among adults.10,11 Detection of Dombrock-mediated hemolysis is difficult, although the clinical relevance of detecting anti-Dombrock has recently been emphasized for sickle cell disease patients receiving multiple blood transfusions.12
Despite more than 30 years of antigen-based studies of the Dombrock blood group system, the identity of the Dombrock carrier molecule itself has not been determined. Dombrock polymorphisms were originally linked to chromosome 1,13 but a more recent linkage study suggests that the gene resides on the short arm of chromosome 12 (12p).14 Studies of erythrocytes from patients with paroxysmal nocturnal hemoglobinuria suggest that DO antigens reside on a glycosylphosphatidylinositol (GPI)-anchored protein.15By using these 2 criteria, the 12p linkage and GPI membrane attachment, we performed in silico analyses of 4926 expressed sequence tags (ESTs) from terminally differentiating human erythroid cells to identify a candidate Dombrock molecule gene. The identity of the gene,DOK1, and its characterization as the Dombrock carrier molecule are reported here.
Materials and methods
Cells and cell culture
This study used parental cell lines (American Type Culture Collection, Rockville, MD); GPI− cells (gift from Dr M. Edward Medof, Institute of Pathology, Case Western Reserve University, Cleveland, OH); and primary cells, which were obtained from healthy blood donors. Culture methods and flow cytometric sorting of the erythroid precursor cells have been described elsewhere.16
Library construction and sequencing
A complementary DNA (cDNA) library from human erythroid precursor cells was constructed using SMART PCR (polymerase chain reaction) cDNA Library Construction Kit (Clontech, Palo Alto, CA) according to the manufacturer's directions, but with slight modifications. Briefly, reverse transcription was performed in the presence of 1 μmol/L peptide nucleic acid (PNA) oligos (N-terminal)-biotin-GTC-CAC-CCG-AAG-CTT-G-(C-terminal) and (N-terminal)-biotin-C(T/C)T-GAA-GTT-CTC-AGG-A-(C-terminal) designed to specifically suppress reverse transcription of globin transcripts. Synthesized cDNA was digested with SfiI and size-selected on a 1% agarose gel. cDNA fragments smaller than 800 base pair (bp) were discarded. Large-scale sequencing of the library was done by the National Institutes of Health (NIH) Intramural Sequencing Center, Bethesda, MD. Sequencing analyses revealed that less than 2% of the ESTs shared homology with globin gene transcripts. More extensive sequencing of the DOK transcripts was performed in the laboratory using the dRhodamine Terminator Cycle Sequencing Kit (Perkin Elmer Applied Biosystems, Foster City, CA). Sequence assemblies and analyses were performed using Sequencher 3.1 software (Gene Codes Corporation, Ann Arbor, MI). Single nucleotide polymorphic (SNP) sites were analyzed using the Gene Inspector (Textco, West Lebanon, NH) software package.
Fluorescence in situ hybridization
Metaphase spreads derived from 5-bromo-deoxyuridine synchronized peripheral lymphocytes of a normal male were used as a template. The DNA probe containing DOK1 was labeled with digoxigenin 11-dUTP (uridine 5′-triphosphate) by nick-translation, and hybridization signals were detected with rhodamine-conjugated antidigoxigenin antibodies (Roche Molecular Biochemicals, Indianapolis, IN). The condition of hybridization, detection of hybridization signals, digital-image acquisition, processing, and analysis were performed as previously described.17 Chromosomes were identified by converting DAPI-banding into G-simulated banding using the IP Lab Image Software (Scanalytics, Fairfax, VA). We analyzed 15 metaphases.
Reverse transcriptase–polymerase chain reaction
Total RNA was purified from cells using the TRIzol reagent (Life Technologies, Rockville, MD) according to the supplied protocol. Reverse transcriptase (RT) was performed using the oligo-(dT) primer and Superscript Reverse Transcriptase (Life Technologies) at 42°C for 50 minutes, as suggested by the manufacturer. We used 1 μL RT reaction in 50 μL PCR amplification with the appropriate primers as follows: initial denaturation at 95°C for 1 minute, denaturing at 95°C for 15 seconds, annealing at 68°C for 15 seconds, and extension for 20 cycles at 72°C for 20 seconds. The PCR products were resolved in 1.2% agarose gel and stained with ethidium bromide. For DOK1 amplification, 5′-CCATTCCTGCTGCTCCTCTCT-3′ and 5′-TCTGGGGTAGAACTTTTCCTTGGT-T-3′ primers were used (expected product size, 247 bp). For human housekeeping porphobilinogen deaminase (PBGD) amplification, 5′-GGCTCTGCGGAGACCAGGAGTCAG-3′ and 5′-TTACCAGACATGGCTCCGCTTGGA A-3′ primers were used (expected product size, 153 bp). For CD34 amplification, 5′-GACCGCGCTTTGCTTGCTGAGTTTG-3′ and 5′-GACCGCGCTTTGCTTGCTGAGTTTG-3′ primers were used (expected product size, 324 bp).
Northern blot analysis
RNA (10 μg) was electrophoresed in 1% agarose denaturing gel and transferred to the Hybond (Amersham Pharmacia Biotech, Piscataway, NJ) nylon membrane using a downward transfer method18 in transfer buffer (Ambion, Austin, TX). RNA was cross-linked to the membrane using UV Stratalinker 1800 instrument (Stratagene, La Jolla, CA). The membrane was prehybridized in ULTRAhyb hybridization buffer (Ambion) at 42°C for 30 minutes. The buffer was replaced with the fresh ULTRAhyb containing a 32P-labeled probe and hybridized at 42°C for 16 hours. The membrane was washed in low- and high-stringency buffers (Ambion) according to the manufacturer's protocol and subjected to autoradiography. Probes for Northern blotting were generated by PCR, purified by QIAquick PCR Purification Kit (Qiagen, Valencia, CA), and labeled using DNA Labeling Beads, −dCTP (cytidine 5′-triphosphate) (Amersham Pharmacia Biotech). Human Immune System Multiple Tissue Northern (MTN) Blot II (Clontech) was processed as recommended by the manufacturer.
Plasmids and probes
The plasmid pDOK1IRES2-EGFP (pDI2E) was constructed by transferring the EcoR1/SalI fragment containing the entire ORF of DOK1 into a similarly digested pIRES2-EGFP (pI2E) vector (Clontech). The EcoR1/SalI fragment originated from the candidate DOK1 clone selected by in silico screening of the cDNA library constructed from human erythroid precursor cells using the SMART PCR cDNA Library Construction Kit. The library was converted into a plasmid form as suggested by the manufacturer, and all inserts were unidirectionally cloned into SfiI sites of the pTriplEx2 vector. The following primers were used to generate PCR products used as probes: 5′-ATGGGTCCATTGATCAACAGATGCAAGA-3′ and 5′-TTATACTCTGCTTTTGGAAAAGATGATGA-3′ for DOK1 (945-bp product; amplified from reverse-transcribed HEL total RNA) and 5′-CCATCTTCTTCAAGGACGACGGCAACTA-3′ and 5′-GGGCGGACTGGGTGCTCAGGTAG-3′ for EGFP (330-bp product; pEGFP-N1 vector was a template). Primers 5′-GCTCGTCGTCGACAACGGCTC-3′ and 5′-CAAACATGATCTGGGTCATCTTCTC-3′ were used to generate the β-actin probe (probe length, 353 bp). The bacterial artificial chromosome (BAC) clone (GenBank accession no.AC007655) was obtained from the Roswell Park Cancer Institute, Buffalo, NY. The presence of the DOK1 in BAC AC007655 DNA was confirmed by PCR.
Flow cytometric analysis
K562 cells were transfected either with pI2E or pDI2E and grown for 21 days in G418-supplemented media (Biofluids, Gaithersburg, MD). Green fluorescent protein (GFP)–expressing cells were sorted and expanded for 4 weeks in G418 media. Untransfected K562 cells and cells transfected with pI2E or pDI2E were prepared for flow cytometry using a 2-step protocol with the clinical sera samples. Prior to staining, each serum was diluted 1:10 with cold phosphate-buffered saline (PBS) and filtered through a 0.45-μm cellulose acetate filter. We used 50 μL diluted serum to stain 105 cells transfected with pI2E or pDI2E. The cells were incubated at 4°C for 45 minutes, washed with cold PBS, and stained with goat antihuman immunoglobulin G (IgG) phycoerythrin (PE) F(ab′)2 fragment (Sigma, St Loius, MO) at a final dilution of 1:50. After a 30-minute incubation at 4°C, the cells were washed twice in PBS. Appropriate negative controls with the second-step F(ab′)2 fragment or serum alone were completed in both untransfected cells and transfected pools.
Analyses were performed with Coulter Epics Elite flow cytometer (Beckman-Coulter, Hileah, FL) using a 488-nm argon laser. GFP was detected at 520-530 nm and PE at 555-595 nm. Gated live cells expressing GFP were analyzed for the DOK1 expression. Positive DOK1 cell populations were defined as those having a mean PE fluorescence at levels greater than 2 SDs above the negative controls. At least 2000 gated events were collected for each analysis.
PI-PLC treatment
The release of GPI-anchored proteins from the surfaces of intact cells was attained by using phosphatidylinositol phospholipase C (PI-PLC) from Bacillus thuringiensis (Oxford Glycosciences, Wakefield, MA) according to the manufacturer's directions. The cells were incubated in 200 μL tris[hydroxymethyl] aminomethane (Tris)–buffered saline (TBS) (10 mmol/L Tris [pH 7.5] and 150 mmol/L sodium chloride [NaCl]) containing 2 U/mL enzyme for 1 hour at 37°C and washed before analysis.
Results
Identification of a candidate gene
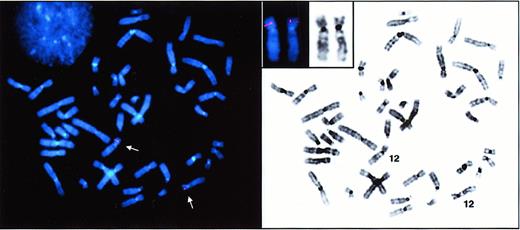
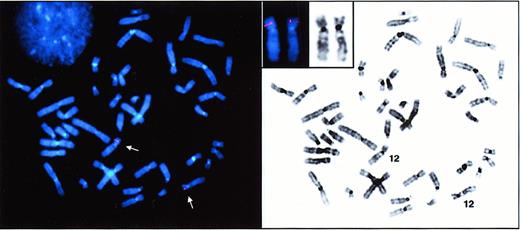
We previously demonstrated the usefulness of genomic-based studies for the discovery and investigation of genes expressed in proliferating erythroid cells.19 To search for genes that encode molecules expressed on circulating erythrocytes such as the Dombrock molecule, we rationalized that a genomic analysis of more differentiated erythroid cells may be required. For this purpose, peripheral blood mononuclear cells from blood donors were cultured in the presence of erythropoietin (EPO), and flow cytometry was used to isolate highly purified populations of erythroid cells undergoing terminal differentiation after 12 days in culture.16 A unidirectional cDNA library was constructed from sorted cells, and a database comprising about 5000 ESTs was generated. We applied 2 criteria to screen the database for Dombrock candidates in silico: (1) The candidate gene should localize to 12p in the region previously linked to Dombrock polymorphism, and (2) a signal peptide required for surface localization of GPI-anchored proteins should be encoded in the 5′-sequence. Based upon the examination of individual Basic Logical Alignment Search Tool (BLAST) homology comparisons for each sequence, one candidate EST from our library conformed to these 2 criteria. The clone shared significant homology with a recently sequenced BAC clone (GenBank accession no. AC007655) derived from 12p. Notably, that BAC sequence also carries a marker (D12S932) in the genomic region that was previously linked to the Dombrock blood group.14 Fluorescence in situ hybridization (FISH) analysis was performed to confirm the location of the BAC clone in this region of chromosome 12 (Figure 1).
FISH detection of DOK1.
Metaphase spreads from synchronized human peripheral lymphocytes were hybridized with a digoxigenin-11-dUTP–labeled DOK1 probe. The hybridization signals were detected with rhodamine-conjugated antidigoxigenin antibodies. Normal metaphase spread after FISH (left panel) shows localization of DOK1 on chromosome 12 (arrows). Inverted DAPI banding (right panel) simulates G-banding for chromosomal identification. Inset displays subchromosomal localization of the gene on 12p12.3-13.1.
FISH detection of DOK1.
Metaphase spreads from synchronized human peripheral lymphocytes were hybridized with a digoxigenin-11-dUTP–labeled DOK1 probe. The hybridization signals were detected with rhodamine-conjugated antidigoxigenin antibodies. Normal metaphase spread after FISH (left panel) shows localization of DOK1 on chromosome 12 (arrows). Inverted DAPI banding (right panel) simulates G-banding for chromosomal identification. Inset displays subchromosomal localization of the gene on 12p12.3-13.1.
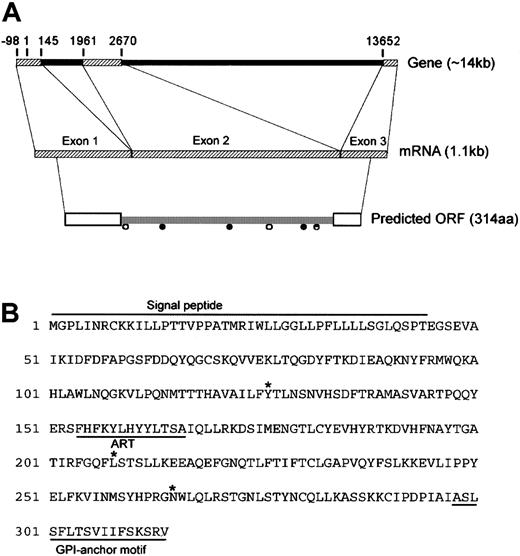
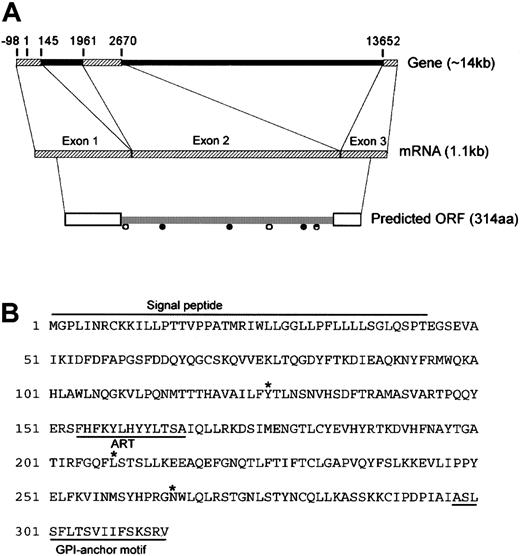
Full-length sequencing of the candidate cDNA clone (referred to here as DOK1) revealed a 1.1-kb cDNA with an open reading frame (ORF) of 945 bp (Figure 2). The gene comprises 3 exons spanning 14 kb. The DOK1 ORF encodes a protein of 314 amino acid residues, with the characteristic N-terminal signal peptide and C-terminal GPI-anchor attachment motif. Several possible sites for N-linked glycosylation and N-myristoylation are present within the coding region. An ADP-ribosyltransferase (ART) motif that is highly homologous to the first exon of the ART gene family member ART4 was predicted within exon 2.20 Alignment studies suggest that DOK1 and ART4 share 2 exons transcribed from 12p.
Molecular organization of DOK1.
(A) Schematic representation of the DOK1 gene. Exons and introns are depicted as hatched bars and filled bars, respectively. The numbers show exon/intron boundaries relative to the position of theDOK1 start codon at position 1. Open boxes show signal peptides, ● indicates putative N-linked glycosylation sites, and ○ indicates putative N-myristoylation sites. (B) Predicted amino acid sequence of DOK1.Signal peptide depicts the predicted signal peptide; GPI-anchor motif, the predicted GPI anchor; ART, the ART motif; and *, the amino acid position corresponding to identified SNP sites.
Molecular organization of DOK1.
(A) Schematic representation of the DOK1 gene. Exons and introns are depicted as hatched bars and filled bars, respectively. The numbers show exon/intron boundaries relative to the position of theDOK1 start codon at position 1. Open boxes show signal peptides, ● indicates putative N-linked glycosylation sites, and ○ indicates putative N-myristoylation sites. (B) Predicted amino acid sequence of DOK1.Signal peptide depicts the predicted signal peptide; GPI-anchor motif, the predicted GPI anchor; ART, the ART motif; and *, the amino acid position corresponding to identified SNP sites.
DOK1 expression pattern
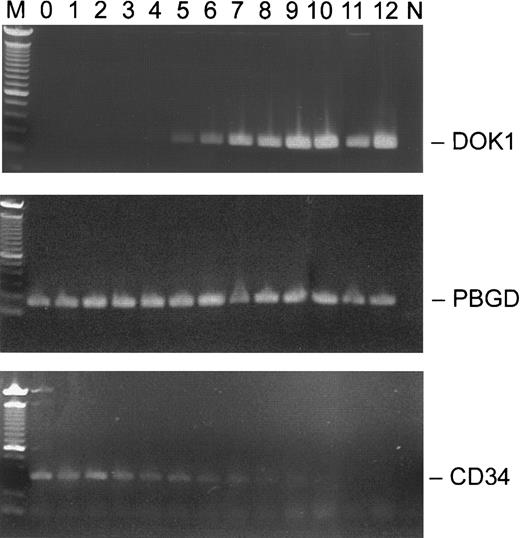
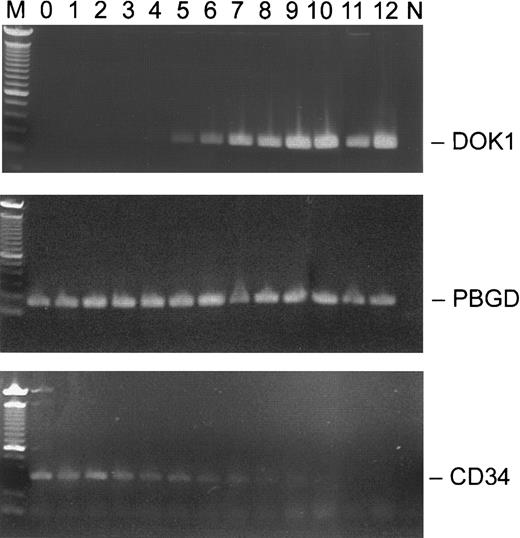
To examine the expression of DOK1 in erythroid cells, the RT-PCR assay was performed using RNA from mobilized CD34+ peripheral blood cells cultured for 2 weeks in the presence of EPO. During that period, the CD34+ cells differentiate into erythroid precursors expressing glycophorin A on their plasma membranes. Intron-spanning PCR primers were chosen to detect expression of the DOK1, CD34, andPBGD genes. As shown in Figure3, the DOK1 expression was not detected before day 4 in culture. The relative signal became stronger over the next week and correlated with the loss of the CD34expression. By day 12, the DOK1 signal was comparable to that of PBGD, while the CD34 expression was no longer seen.
Expression of
DOK1 in CD34+ EPO-stimulated cells.CD34+ cells were grown in the presence of 1 U/mL EPO, and total RNA was purified from the cells collected on respective days 0-12. The RT-PCR products (top panel, DOK1; middle panel, PBGD; and lower panel, CD34) from each day were separated in 1.2% agarose gel. M indicates a 100-bp DNA ladder, and N indicates no DNA control.
Expression of
DOK1 in CD34+ EPO-stimulated cells.CD34+ cells were grown in the presence of 1 U/mL EPO, and total RNA was purified from the cells collected on respective days 0-12. The RT-PCR products (top panel, DOK1; middle panel, PBGD; and lower panel, CD34) from each day were separated in 1.2% agarose gel. M indicates a 100-bp DNA ladder, and N indicates no DNA control.
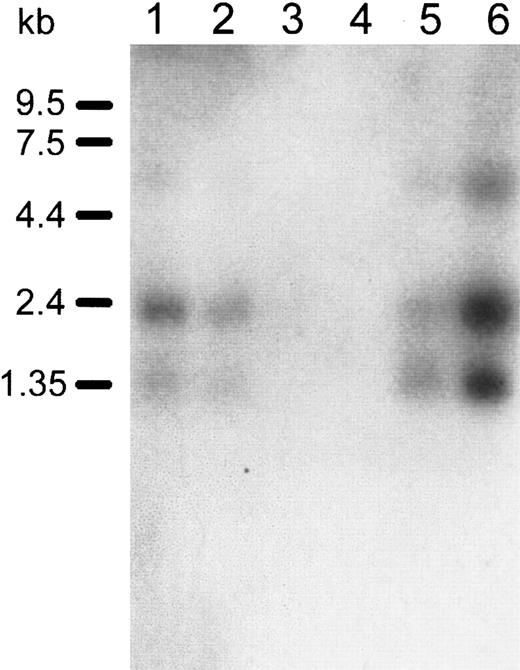
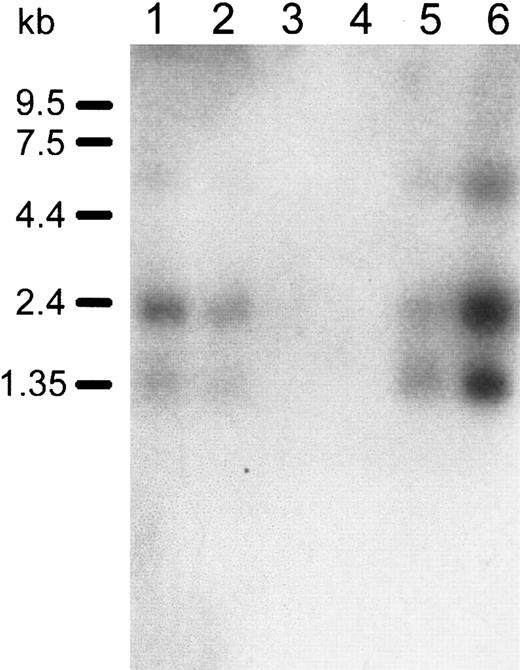
Human Immune System MTN Blot II was used to assess the expression ofDOK1 in hematologic tissues. The DOK1 probe hybridized to 3 bands having sizes of about 1, 2, and 7 kb. The bands were apparent in the spleen, lymph node, bone marrow, and fetal liver (Figure4). There was no hybridization observed in thymus or peripheral blood leukocytes. The strongest signal was present in the lane containing RNA from the fetal liver, which is consistent with erythroid cell production in that organ. A homology search of DOK1 against EST databases also suggests the DOK1gene is primarily expressed in the fetal liver and spleen (data not shown).
Northern blot analysis of
DOK1 expression in human hematologic tissues.Lanes 1 to 6 contain the following, respectively: 2 μg polyA+RNA from the human spleen, lymph node, thymus, peripheral blood leukocytes, bone marrow, and fetal liver. A32P-labeled DOK1 probe was used for hybridization. RNA size marker bands are indicated in the left margin of the blot.
Northern blot analysis of
DOK1 expression in human hematologic tissues.Lanes 1 to 6 contain the following, respectively: 2 μg polyA+RNA from the human spleen, lymph node, thymus, peripheral blood leukocytes, bone marrow, and fetal liver. A32P-labeled DOK1 probe was used for hybridization. RNA size marker bands are indicated in the left margin of the blot.
Serologic testing of DOK1 as molecular carrier of Dombrock antigens
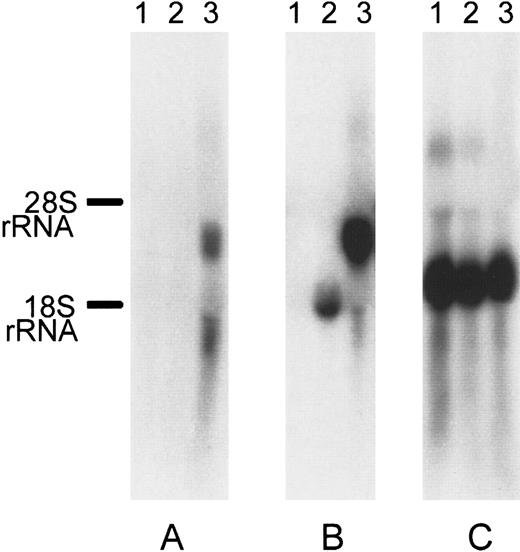
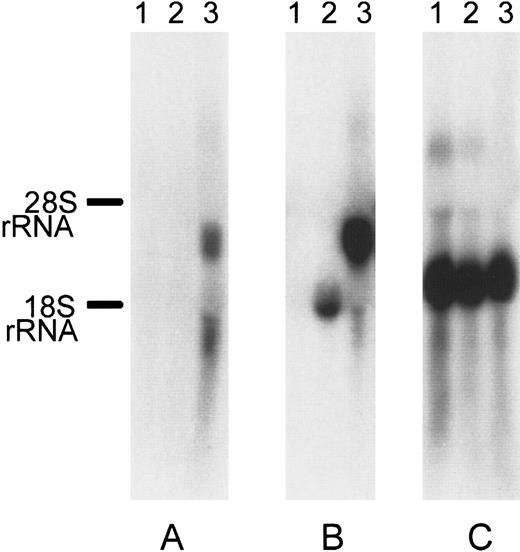
We further screened several cell types (K562, peripheral blood leukocytes, MEG, Jurkat, and HEL) for the expression of DOK1using RT-PCR (data not shown). Only HEL erythroleukemic cells demonstrated expression of DOK1. Because the K562 erythroleukemic cell line had no detectable expression ofDOK1, those cells were chosen for serologic testing. To create stable transfectants, we subcloned the DOK1 open reading frame into a commercially available vector, pIRES2EGFP, to create the plasmid pDI2E. Identification of stable K562 transfection pools was achieved by G418 selection for at least 2 weeks and by GFP expression. Northern blotting of untransfected K562 cells and K562 cells transfected either with pI2E or pDI2E confirmed expression of DOK1 only in pDI2E-transfected cells (Figure 5).
Northern blot of pI2E- versus pDI2E-transfected K562.
Each lane contains 10 μg total RNA from untransfected K562 cells (lane 1), cells transfected with the pI2E vector (lane 2), and cells transfected with the pDI2E vector (lane 3). The 32P-labeled probes were used for hybridizations. (A) Membrane hybridized with a DOK1 probe. (B) Membrane hybridized with an EGFP probe. (C) Membrane hybridized with a β-actin probe.
Northern blot of pI2E- versus pDI2E-transfected K562.
Each lane contains 10 μg total RNA from untransfected K562 cells (lane 1), cells transfected with the pI2E vector (lane 2), and cells transfected with the pDI2E vector (lane 3). The 32P-labeled probes were used for hybridizations. (A) Membrane hybridized with a DOK1 probe. (B) Membrane hybridized with an EGFP probe. (C) Membrane hybridized with a β-actin probe.
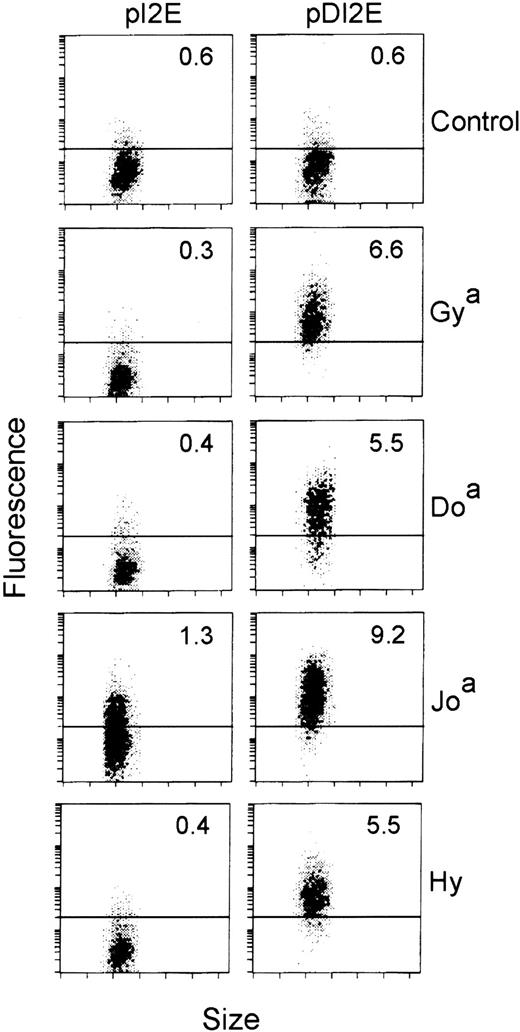
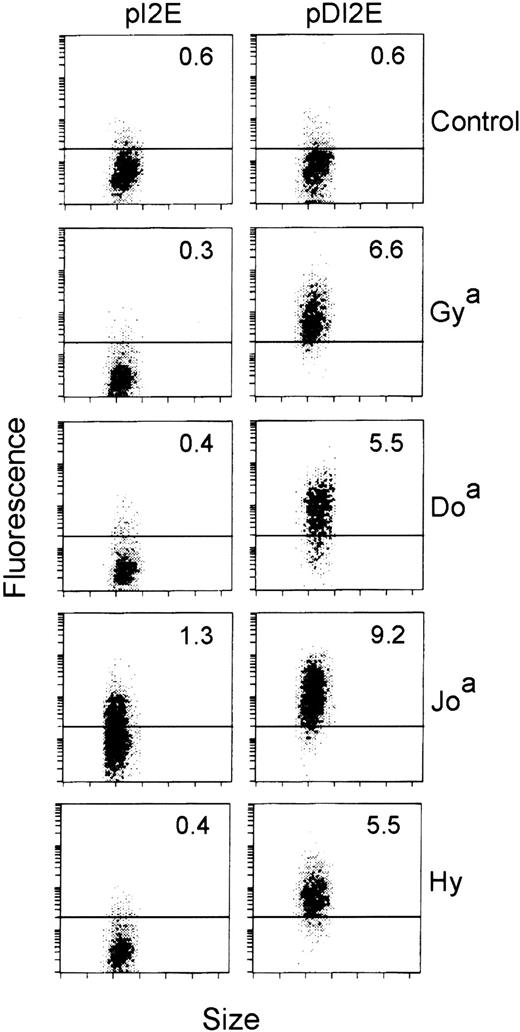
We then assayed the cells expressing GFP among the pDI2E- and pI2E-transfected populations for Dombrock serum antibody binding. Sera negative for Dombrock agglutinating antibodies were used as controls. Dombrock serum antibody binding to the DOK1-transfected cells (pDI2E) resulted in a shift in mean fluorescence to levels greater than 2 SDs above the control cells (pI2E-transfected). As shown in Figure6, Dombrock-specific serum binding was demonstrated for all of the antigens tested. Overall, 14 sera representing 4 of 5 antigens were assayed, although Dobantisera was not available. Of the 14 sera, 9 demonstrated specific, high-level binding to the DOK1-expressing cells (Table). Only 1 of 5 anti-Hy sera demonstrated specific binding to the DOK1-expressing population.
Flow cytometric analyses of pI2E- versus pDI2E-transfected K562 cells.
K562 cells were transfected with pI2E (DOK1−) or pDI2E (DOK1+) DNA and grown in G418-containing media. Stable transfectants were stained with serum containing antibodies to Dombrock antigens (Gya, Doa, Joa, and Hy), counterstained with PE-labeled F(ab′)2 fragments, and analyzed by flow cytometry. The numbers shown at the upper right of each panel are the mean fluorescence values (PE channel) of the transfected cells. The control serum contained no Dombrock antibodies, as measured by indirect hemagglutination assays.
Flow cytometric analyses of pI2E- versus pDI2E-transfected K562 cells.
K562 cells were transfected with pI2E (DOK1−) or pDI2E (DOK1+) DNA and grown in G418-containing media. Stable transfectants were stained with serum containing antibodies to Dombrock antigens (Gya, Doa, Joa, and Hy), counterstained with PE-labeled F(ab′)2 fragments, and analyzed by flow cytometry. The numbers shown at the upper right of each panel are the mean fluorescence values (PE channel) of the transfected cells. The control serum contained no Dombrock antibodies, as measured by indirect hemagglutination assays.
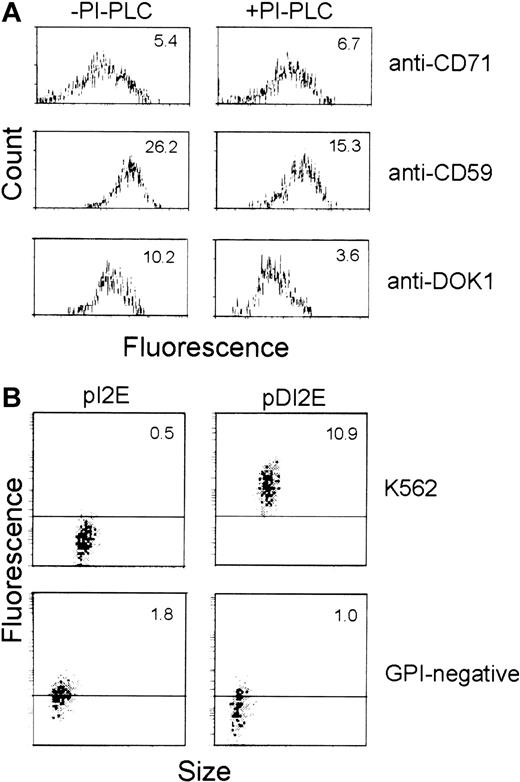
DOK1 encodes a GPI protein
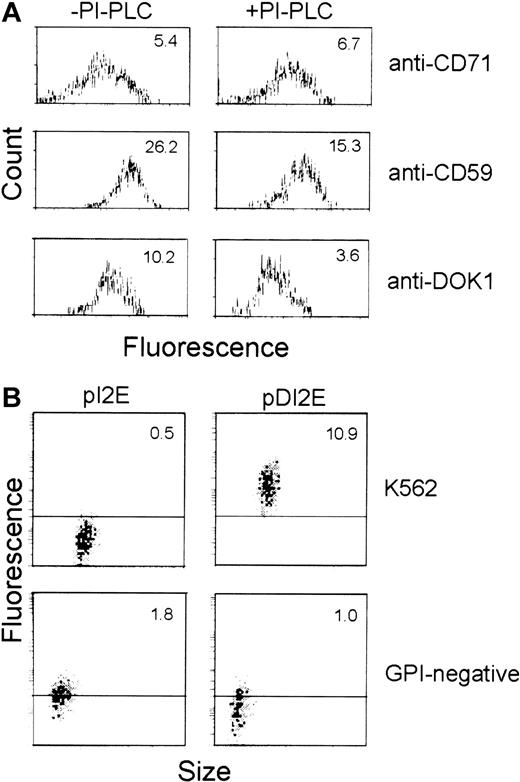
Because the Dombrock molecule is reportedly GPI-anchored, we performed tests to determine whether DOK1 encodes a GPI protein. Flow cytometry was used to compare the level of anti-Dombrock sera binding to pDI2E-expressing cells before and after treatment with PI-PLC. Assays of CD71 (transmembrane protein) and CD59 (GPI protein) cleavage were used as negative and positive controls, respectively. PI-PLC–mediated cleavage of GPI proteins was measured after staining with anti-CD71, anti-CD59, or anti-Dombrock antibodies (Figure7A). PI-PLC action produced no significant changes in mean fluorescence in cells stained with anti-CD71 antibodies (mean fluorescence, 5.4 vs 6.7). In contrast, the level of CD59 on the surface membrane was significantly reduced (mean fluorescence, 26.2 vs 15.3). Staining with Dombrock-specific antisera revealed a pattern similar to that of anti-CD59 stained cells (mean fluorescence, 10.2 vs 3.6). This 3-fold reduction of mean fluorescence is consistent with findings that DOK1 is anchored to the plasma membrane via GPI.
GPI anchoring of DOK1.
(A) K562 cells were transfected with pDI2E construct, and stable transfectants were selected in G418-containing media. The cells were then incubated in PI-PLC (+) or buffer (−) prior to analysis. Flow cytometry was performed using FITC-conjugated anti-CD71, anti-CD59, and Dombrock antisera for comparison. (B) K562 cells and the GPI− cell line transfected either with pI2E or pDI2E DNA and selected in G418-containing media were stained with Dombrock antisera and analyzed by flow cytometry. The mean fluorescence of each population is shown at the top of each panel.
GPI anchoring of DOK1.
(A) K562 cells were transfected with pDI2E construct, and stable transfectants were selected in G418-containing media. The cells were then incubated in PI-PLC (+) or buffer (−) prior to analysis. Flow cytometry was performed using FITC-conjugated anti-CD71, anti-CD59, and Dombrock antisera for comparison. (B) K562 cells and the GPI− cell line transfected either with pI2E or pDI2E DNA and selected in G418-containing media were stained with Dombrock antisera and analyzed by flow cytometry. The mean fluorescence of each population is shown at the top of each panel.
pDI2E was also introduced into K562 cells unable to express GPI-anchored proteins on their plasma membranes (GPI−cells) to independently confirm that DOK1 is a GPI-anchored protein.20 Parental K562 and GPI− K562 cells were transfected with pI2E and pDI2E. The expression of DOK1on the surface of the cells was analyzed by flow cytometry following the staining with anti-Dombrock serum (Figure 7B). The parental K562 cells transfected with pDI2E exhibited strong binding to anti-DOK1 antibodies, while pI2E-transfected K562 cells did not. The GPI− cells transfected either with pDI2E or pI2E failed to show any specific binding of the anti-Dombrock serum, while GFP expression from the internal ribosome entry site (IRES) of pI2E or pDI2E was observed in both cases. Hence, DOK1 demonstrated PI-PLC sensitivity on K562 cells and a lack of surface expression among GPI− cells. Both properties are consistent with the in silico prediction that DOK1 is a GPI protein.
Single nucleotide DOK1 polymorphisms
To determine whether genetic polymorphisms correlate with blood group antigenicity, we isolated genomic DNA or reticulocyte RNA from 8 blood donors of defined serology, ie, 4 Do(a+b−) vs 4 Do(a−b+), and looked for donor-specific differences in the DOK1. The DOK1 coding regions from the samples were sequenced and aligned. The alignments revealed 3 SNP sites within the coding region of DOK1 (Figure 2). While 2 SNP sites did not alter the predicted amino acid primary structure (Y126 and L208), the third site predicted a mutation in the protein sequence (N265D). Notably, the N265D mutation falls within an RGD adhesion motif of the molecule. All 3 SNP sites were consistent among the 8 donors, with N265 found in the 4 Do(a+b−) donor samples and D265 in the 4 Do(a−b+) donor samples.
Discussion
Molecular genetic analyses of the 25 known human blood group molecules are imperative for erythroid cell biology and transfusion medicine.1,22 23 Prior to this study, the Dombrock system was 1 of 4 blood group systems (Dombrock, Scianna, P, and Raph) that had not been cloned and sequenced. While genetic-based studies of enucleated red cells are inherently difficult, recent advances in erythroid cell biology and genomics will likely facilitate the integration of molecular biology with the vast amounts of transfusion-related information gathered over the last century. As demonstrated here, the combination of in silico genetic analyses, basic studies of GPI-anchored protein expression, and clinical blood group serotyping led to the identification of the Dombrock blood group molecule as a member of the ADP-ribosyltransferase gene family. The demonstrated success of this approach attests to the value of bench, bedside, and Internet browser for translational research.
Several lines of evidence reported here support our conclusion that theDOK1 gene encodes the Dombrock blood group glycoprotein. The candidate cDNA clone nearly perfectly aligned with a single copy genomic clone mapped to 12p by FISH analysis. The messenger RNA (mRNA) expression pattern appears to be developmentally linked to erythropoiesis and parallels that of other erythroid markers (Figure 3and data not shown). Notably, the highest level of expression was detected in fetal liver, which suggests possible differential erythroid expression levels during human ontogeny. GPI anchoring to the membrane was shown by PI-PLC sensitivity and confirmed by a lack of surface expression in GPI-deficient cells. Most importantly, the use of clinically derived serum samples allowed us to specifically demonstrate the binding of Dombrock antibodies to the DOK1 gene product. While the majority of sera tested bind specifically to the DOK1 gene product, the lack of Hy antisera binding among 4 of 5 donors was not predicted. This may reflect differences in the Hy epitope structure on the K562 versus mature erythroid membranes. Variability in serum antibody binding has been similarly reported for other blood group antigens.24
The identification of an ART motif within exon 2 strongly suggests that this blood group molecule may be functional as an erythrocyte ADP-ribosyltransferase. Such activity has been identified in avian, bovine, and human erythrocytes.25 However, expression of the ART gene family member (ART4) that aligns with exons 2 and 3 of DOK1 has not been previously reported among erythroid cells. The multiple bands observed in Northern blot analysis (Figure 4) also suggest the existence of other splice variants from the 12p locus. The possibility of alternate splicing prompted us to analyze the EST database for other expressed sequences aligning with the DOK1 region. One EST (GenBank accession No. AA905180) aligns with the 5′-region of DOK1, but the first exon containing the signal sequence appears to be spliced out. This splicing variant predicts an intracellular expression pattern. Two more ESTs (GenBank accession Nos. AI149009 and AW194834) align well with a region in the second intron of DOK1, which implies alternative splicing of the C-terminal portion of the mature DOK1 protein. Interestingly, other cytoplasmic, surface, or secretedART gene family members have been described elsewhere.26
This study was limited to the examination of the DOK1 gene structure and membrane expression in the context of transfusion medicine. Investigations of enzymatic activity will be essential for determining the biological role of erythroid DOK1 expression. ART on red cells may in part be responsible for NAD clearance and the resulting low concentrations of NAD measured in serum. Alternatively, erythrocyte ART may have a more active role in the post-translational modification of protein substrates in contact with erythroid surfaces. Other members of the ART enzyme family are known to be involved in susceptibility to immune-mediated diabetes mellitus.27 The loss of Dombrock molecules due to defective GPI anchoring may also be relevant for understanding the pathogenesis of paroxysmal nocturnal hemoglobinuria.28 Erythrocyte expression of Dombrock and other red cell ectoenzymes (Yt and Kell) provides steady-state levels of these enzymes, and tissue distribution is confined to the vascular space. Thus, in addition to hemoglobin, erythrocytes may be viewed as transporters of several biologically active molecules including membrane-associated ectoenzymes.
While nonpolymorphic regions of the protein or sugar moieties on this heavily glycosylated protein may contribute to Dombrock antigenicity,29 we hypothesize that the N265D polymorphism provides the molecular basis for the Doa and Dob phenotypes. The location of this polymorphism within an RGD motif of the DOK1 molecule has several important implications. RGD motifs within adhesive ligands are commonly involved in cell-to-cell interactions involving integrin binding.30 Interestingly, integrin binding and modification via ART enzymes have been demonstrated during skeletal muscle development.31 RGD amino acid substitution involving the aspartic acid moiety has also been correlated with overall antigenicity of RGD-containing proteins.32 In addition, the polymorphic nature of this motif within the Dombrock carrier molecule suggests that this blood group may have arisen due to some evolutionary advantage gained secondary to a mutation of the RGD motif. Notably, Plasmodium falciparum may influence the adhesive properties of erythrocytes through the expression of RGD-containing proteins during the erythrocytic stage of infection.33 Further investigation of this Dombrock polymorphism in the context of malaria should be pursued because population-based studies have demonstrated a significant increase in the RGD-encoded carrier molecule, Dob, among African and Asian populations.
In addition to improving our understanding of functional aspects of the molecule, the cloning of the gene encoding the Dombrock protein and the determination of the molecular basis of the Doa/Dob polymorphism pave the way to improve important aspects of clinical care. While antibodies in the Dombrock blood group system are not associated with hemolytic disease of the newborn, these antibodies have caused hemolytic transfusion reactions.10,11 Dombrock antibodies are notoriously difficult to identify in the transfusion setting,12 as they are usually present in serum containing several other antibodies. In addition, commercially available reagent red cells are rarely Dombrock-typed. For this reason, reliable typing reagents are not available, and blood for transfusion is mainly selected by testing with the patient's serum. This practice is unsatisfactory because the antibody may not be present in demonstrable amounts in vitro, while causing hemolysis in vivo. Our work provides, for the first time, the bases by which genotyping methods can be used to type the patient, and more importantly, to screen for antigen-negative donor blood.
Acknowledgments
We thank Dr Alan N. Schechter for critical reading of the manuscript and Dr Susan Leitman in the NIH Department of Transfusion Medicine, Bethesda, MD, for providing donor buffy coats. Lindsay Mitchell, Terry Lee, and Aryan Kay provided excellent technical assistance. Drs Gerard Bouffard and Jeff Touchman from the NIH Intramural Sequencing Center, Bethesda, MD, are recognized for the exemplary sequencing services. The accession number for the human DOK1 sequence in GenBank is AF290204.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jeffrey L. Miller, Laboratory of Chemical Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bldg 10, Rm 9B17, Bethesda, MD 20892; e-mail: jm7f@nih.gov.