Abstract
Because there is no known genetic abnormality common to all patients with myeloma, it is important to understand how genetic heterogeneity may lead to differences in signal transduction, cell cycle, and response to therapy. Model cell lines have been used to study the effect that mutations in p53 and rascan have on growth properties and responses of myeloma cells. The U266 cell line has a single mutant p53 allele. Stable expression of wild-type (wt) p53 in U266 cells results in a significant suppression of interleukin (IL)-6 gene expression and in the concomitant suppression of cell growth that could be restored by the addition of exogenous IL-6. Expression of wt p53 also leads to cell cycle arrest and protection from doxorubicin (Dox)- and melphalan (Mel)-induced apoptosis. The addition of IL-6 resulted in cell cycle progression and blocked p53-mediated protection from apoptosis. ANBL6 is an IL-6–dependent cell line that is sensitive to dexamethasone (Dex), Dox, and Mel. IL-6 is able to protect ANBL6 cells from Dex- and Mel- but not Dox-induced apoptosis. To study the effect of an activating mutation in ras, the ANBL6 cell line transfected with either a constitutively activated N- orK-ras gene was used. Both N-ras12 andK-ras12 genes were able to protect ANBL6 cells from apoptosis induced by Dex, Dox, and Mel. These data show that changes inras or p53 can alter the myeloma cell response to IL-6 and demonstrate that the genetic background can alter therapeutic responses.
Introduction
Multiple myeloma is a B-cell cancer characterized by the accumulation of clonal plasma cells in the bone marrow.1 Numerous studies2 have demonstrated that interleukin-6 (IL-6) is a major growth factor for myeloma cells. Typically, IL-6 is produced by the bone marrow stromal cells.3 In addition, the production of IL-6 by normal B cells and a number of myeloma cell lines also suggests an autocrine source of IL-6 in some patients with myeloma.4-7 There are no known genetic abnormalities common to every case of myeloma. Indeed, significant genetic heterogeneity among patient tumor cells should be associated with differences in disease progression and therapeutic response.
Mutations in the p53 gene have been reported in 10% to 20% of tumor samples from patients with myeloma.8,9 Moreover, p53 inactivation is often associated with advanced stages of the disease.9 Transient expression of a wild-type (wt)p53 gene has been shown to suppress IL-6 promoter activity in HeLa cells.10 The functional target for transcription repression by p53 was mapped to the multiple cytokine and second-messenger response element (MRE, −173 to −145) within the IL-6 promoter.10,11 Mutation at the C/EBP β (NFIL-6)-binding site within the MRE blocks the ability of p53 mutants to stimulate IL-6 promoter activity.12 Therefore, dysfunctional p53 may contribute to the deregulation of IL-6 production in myeloma cells.
IL-6 signals through a variety of downstream signaling pathways, including the Ras family of guanosine triphosphate–binding proteins.13 Point mutations at codons 12, 13, or 61 lead to a constitutively active Ras protein. Activating mutations in N- or K-ras are found in multiple myeloma with a frequency of 30% to 50%.8,14-16 It has been shown that activating mutations in either N- or K-ras can provide a proliferation signal independent of any additional IL-6 in the ANBL6 myeloma cell line.17,18 It was also shown that N-ras, but not K-ras, was able to provide protection from apoptosis induced by IL-6 withdrawal or treatment with dexamethasone.18 This demonstrates that mutations in the ras gene may have significant effects on myeloma cell growth and response to therapy.
Given the fact that there is considerable genetic heterogeneity in multiple myeloma, it is important to understand how the genetic background can affect signal transduction, cell cycle, and response to therapy. We used model cell lines to study the effect of p53and ras mutations on the response of myeloma cells to IL-6 and chemotherapeutic agents. The autocrine IL-6–producing U266 cell line carries a single allele of the p53 gene with a inactivating mutation at exon 5.9,19 This cell line provided an excellent model to examine the effect of wt p53 on IL-6 expression. We were also able to use this cell line to study the effect of p53 mutations on myeloma growth and response to therapy. The ANBL6 cell line is an IL-6–dependent line that contains a wtras gene. To study the effect of an activatingras mutation on response to therapy, we used ANBL6 cells that were previously transfected with either a constitutively activatedN-ras12 or K-ras12 gene.18 Our data suggest that changes in the p53 or ras gene can alter the myeloma cell response to IL-6. They also demonstrate that myeloma cells may respond differently to chemotherapeutic agents depending on the genetic background of the cell.
Materials and methods
Cell culture
The autocrine IL-6–producing U266 myeloma cell line was purchased from American Tissue Culture Collection (Rockville, MD), and the ANBL6 cell lines were as previously described.18 All cell lines were maintained in RPMI 1640 supplemented with 10% fetal calf serum, 50 U/mL each of penicillin and streptomycin, 2 mmol/Ll-glutamine (BRL, Gaithersburg, MD), and 0.5 ng/mL IL-6 (R&D Systems, Minneapolis, MN) except as indicated. All cultures were incubated in the absence of IL-6 for 3 days to eliminate the IL-6 effect on the cells and were then centrifuged on Ficoll-Histopaque to eliminate dead cells before each experiment.
Plasmid constructs and transfection
The p53 cDNA was a generous gift from Dr Robert Kratzke (VA Hospital, Minneapolis, MN). The Tet-regulated expression system was purchased from Stratagene (La Jolla, CA). The pTet.TAK.Neo vector was generated by cloning the neomycin gene to pTet.TAK vector at theNotI site. The p53 expression vector (pTet.Splice.p53.Hyg) was constructed by cloning p53 cDNA to the EcoRI site of pTet.Splice, and the hygromycin gene was then cloned to theNotI site of pTet.Splice.
Stable transfection of cells
U266 cells (1 × 107) were collected by centrifugation and resuspended with 400 μL of complete RPMI 1640 containing 10% fetal calf serum and 50 U/mL each of penicillin and streptomycin and 2 mmol/L L-glutamine (BRL). Linearized pTet.TAK.Neo and p53 expression vectors were mixed in a 1:1 molar ratio. pTet.TAK.Neo was linearized with ApalI (NEB, Beverly, MA). pTet.Splice.p53.Hyg was linearized with BstXI (NEB). DNA was added to the cell suspension and transferred to a 0.4-cm gap-width electrocuvette. Cells were electroporated at 300 V and 960 μF in a Gene Pulser (Bio-Rad, Hercules, CA). Cells were then allowed to recover for 20 minutes at room temperature and were transferred to 10 mL complete RPMI 1640 with 0.1 ng/mL IL-6 and incubated for 48 hours at 37°C and 7% CO2 before selection in Geneticin (400 mg/mL; BRL) or Hygromycin (200 μg/mL; CalBiochem). Fresh selective medium was added every 4 hours for 12 days, and then the antibiotic-resistant transfectants were expanded in complete RPMI 1640 with 0.1 ng/mL IL-6.
RNA preparation and reverse transcriptase–polymerase chain reaction
Total RNA was prepared by extraction with TriZol (BRL) according to the manufacturer's instructions. The RNA was quantitated by spectrophotometry, and 1 μg RNA was reversed transcribed with SuperScript II reverse transcriptase (RT) (BRL) at 42°C for 50 minutes with 300 μg oligo(dT) (Pharmacia, Piscataway, NJ) in a 20-μL reaction mixture according to manufacturer's instructions. Five microliters of the reaction mixture from reverse transcription was amplified in a 100-μL reaction with 20 pmol of each primer, 2.5 U Taq polymerase (Amplitaq; PerkinElmer-Cetus, Norwalk, CT), 200 mmol/L dNTP (Pharmacia LKB Biotechnology, Uppsala, Sweden), 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 8.3), and 1.5 mmol/L MgCl2. The reactions were denatured at 95°C for 3 minutes and subjected to 30 cycles of amplification with denaturation at 94°C for 1 minute, annealing at 55°C to 60°C for 30 seconds, and extension at 72°C for 1 minute followed by final extension at 72°C for 15 minutes using a DNA Thermocycler (PerkinElmer-Cetus, Emeryville, CA).
Oligonucleotide primers
All oligonucleotide primers used in this study were synthesized on the Milligen Bioresearch DNA synthesizer (model 8750; Novato, CA). The sequences of the oligonucleotide are listed below:
p53.
CMV: 5′-CGC CAT CCA CGC TGT TTT GAC CTC CAT AG-3′. INIT p53: 5′-GTC ACA GAC TTG GCT GTC CCA-3′
IL-6.
IL-6.5′: 5′-AAC TCC TTC TCC ACA AGC G-3′. IL-6.3′: 5′-TGG ACT GCA GGA ACT CCT T-3′
Porphobilinogen deaminase.
PBGD5′: 5′-TGT CTG GTA ACG GCA ATG CG-3′. PBGD3′: 5′-TCA ATG TTG CCA CCA CAC TG-3′.
[3H] Thymidine incorporation assay
The assay was performed as previously described.20 Briefly, cells were diluted to 2 × 105 cells/mL, and 200 μL of each culture was seeded to 96-well plates in the presence of various concentrations of IL-6 as indicated. The 96-well plates were incubated for 2 days before 1 μCi [3H] thymidine (5 Ci/mmol; Amersham, Arlington Heights, IL) was added to each well. The cells were further incubated for at least 12 hours, but no more than 18 hours, and then harvested onto glass filter paper (Skatron, Sterling, VA). [3H] Thymidine incorporation was measured by liquid scintillation counting (Beckman, Arlington Heights, IL).
Apoptosis and cell cycle analysis
The assay was performed as previously described.20Briefly, cells were washed and diluted to 2 × 105cells/mL with RPMI 1640 with or without dexamethasone (Dex), doxorubicin (Dox), melphalan (Mel), or IL-6 as indicated. Cells were harvested and pelleted at the indicated time points. Cell pellets were suspended with 0.5 mL hypotonic lysis solution (50 μg/mL propidium iodide (Sigma, St. Louis, MO) in 0.1% sodium citrate and 0.1% triton X-100) and incubated at 4°C for at least 3 hours in the dark. Nuclear staining was analyzed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Nuclear extract preparation
Nuclear extracts were prepared as previously described.21 Briefly, 108 cells were washed in ice-cold phosphate-buffered saline, and the cell pellet was lysed in 5 packed cell volumes Buffer A (10 mmol/L HEPES, pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol, 0.1% NP-40) for 2 minutes on ice. Nuclei were pelleted at 9000 rpm for 30 seconds at 4°C and resuspended in 0.5 packed cell volumes Buffer B (20 mmol/L HEPES, pH 7.9, 25% glycerol, 0.42 NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L phenylmethylsulfonyl fluoride (PMSF), 0.5 mmol/L DTT). Tubes were rocked for 30 minutes at 4°C and then centrifuged for 20 minutes to pellet insoluble material. Protein concentration was determined by using the Bio-Rad Protein Assay Kit.
Electrophoretic mobility shift assay
For NF-kB, AP-1, or NFIL-6 mobility shifts, 10 μg nuclear extract was incubated with radiolabeled probe (10 000 cpm) in a binding buffer that contained 5% glycerol, 50 mmol/L NaCl, 10 mmol/L Tris (pH 7.5), 1 mmol/L DTT, 1 mmol/L EDTA, and 0.5 μg poly dI-dC for 30 minutes at room temperature. The samples were electrophoresed on a 4.5% acrylamide gel containing 50 mmol/L Tris (pH 7.8), 0.38 mol/L glycine, and 2 mmol/L EDTA. For the STAT gel shift, 10 μg nuclear extract was incubated with radiolabeled probe (10 000 cpm) in a binding buffer that contained 20 mmol/L HEPES, 0.1 mmol/L EDTA, 1 mmol/L DTT, 0.1 mmol/L PMSF, 1.5% glycerol, and 0.5 μg poly dI-dC for 30 minutes at room temperature. Samples were electrophoresed on a 4.5% acrylamide gel with 5% glycerol and 0.5 × TBE. For competition assays, 100 × molar excess of a specific or a nonspecific probe was incubated with the samples for 5 minutes before the addition of labeled probes. The sequence of the oligonucleotides used were (upper strand shown): NF-kB, TCT CAA CAG AGG GGA CTT TCC GAG AGG CCA TCT GG; AP-1, CTA GAT CCT CTA GAA CTG ACT CAT CGG ATC TAC; NFIL-6, TCG ACT GCA GAT TGC GCA ATC TGC ATC TAC; STAT, AGC TTC ATT TCC CGT AAT CCC TAA AGC T.
Results
Ectopic expression of wt p53 suppresses autocrine IL-6 expression and suppresses cell cycle in U266 cells
To study the effect of p53 mutations on myeloma cell growth and response to therapeutic agents, U266 cells were stably transfected with a wt p53 expression construct under the control of a minus-tetracycline (Tet)-inducible promoter. Reverse transcription–polymerase chain reaction (RT-PCR) shows that the wtp53 gene is expressed in the U266.TAK.p53 stably transfected cell line, but not in the parental cell line transfected with the empty vector alone (Figure 1A). To amplify only the RNA from the transfected gene and not the endogenous gene carrying the mutation, the primers were designed with one recognizing a vector sequence and the other recognizing a sequence in the p53gene. In our hands, the expression of wt p53 is constitutive in the bulk-selected population and not suppressed in the presence of Tet. We chose to examine bulk-transfected populations rather than individual subclones to minimize potential artifacts of integration from limited numbers of subclones. Previous studies10 12have indicated that wt p53 can suppress IL-6 promoter activity and that some mutations in p53 can up-regulate IL-6 expression. RT-PCR shows that IL-6 expression is significantly suppressed in U266 cells by the ectopic expression of wt p53 (Figure 1A). Longer exposure shows some residual expression of IL-6 in the U266.TAK.p53 cell line (data not shown). Additionally, the level of secreted IL-6 measured by enzyme-linked immunosorbent assay is 42.6 pg/mL in U266.TAK-conditioned media compared with only 1.9 pg/mL in U266.TAK.p53-conditioned media.
Wt p53 suppresses autocrine IL-6 expression and suppresses cell cycle in U266 cells.
(A) Wt p53 suppresses autocrine IL-6 expression. Total RNA was isolated from the U266.TAK and the U266.TAK.p53 cell lines. Expression of p53 and IL-6 was detected by RT-PCR. Expression of PBGD was used as a loading control.22 (B) Wt p53 suppresses cell growth. Cells were incubated with or without varying concentrations of IL-6 and then pulsed with [3H] thymidine. [3H] Thymidine incorporation was measured by liquid scintillation counting. Values represent the mean of duplicate samples ± SEM. Data shown are representative of at least 3 independent experiments. (C) Wt p53 reduces the number of cells in the S and G2/M stage of the cell cycle. Cells were treated with or without IL-6 and were harvested for cell cycle analysis. Percentages of cells in each phase of the cell cycle were determined by flow cytometry. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. (D) Expression of wt p53 alters transcription factor activation. Nuclear extracts were prepared, and binding activities were measured by electrophoretic mobility shift assay. ANBL6 extracts were included as a reference for STAT1 and STAT3 dimers.23 SC, 100 × specific competitor; NSC, 100 × nonspecific competitor.
Wt p53 suppresses autocrine IL-6 expression and suppresses cell cycle in U266 cells.
(A) Wt p53 suppresses autocrine IL-6 expression. Total RNA was isolated from the U266.TAK and the U266.TAK.p53 cell lines. Expression of p53 and IL-6 was detected by RT-PCR. Expression of PBGD was used as a loading control.22 (B) Wt p53 suppresses cell growth. Cells were incubated with or without varying concentrations of IL-6 and then pulsed with [3H] thymidine. [3H] Thymidine incorporation was measured by liquid scintillation counting. Values represent the mean of duplicate samples ± SEM. Data shown are representative of at least 3 independent experiments. (C) Wt p53 reduces the number of cells in the S and G2/M stage of the cell cycle. Cells were treated with or without IL-6 and were harvested for cell cycle analysis. Percentages of cells in each phase of the cell cycle were determined by flow cytometry. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. (D) Expression of wt p53 alters transcription factor activation. Nuclear extracts were prepared, and binding activities were measured by electrophoretic mobility shift assay. ANBL6 extracts were included as a reference for STAT1 and STAT3 dimers.23 SC, 100 × specific competitor; NSC, 100 × nonspecific competitor.
A [3H] thymidine incorporation assay was used to examine the effect of wt p53 on the proliferation of U266 cells in the presence of varying concentrations of IL-6 (Figure 1B). The control cell line shows high [3H] thymidine incorporation in the absence of IL-6 and no additional stimulation in response to IL-6. In contrast, the U266.TAK.p53 cell line shows very low [3H] thymidine incorporation in the absence of IL-6, which increases in response to IL-6 in a dose-dependent manner. Consistent with this, the U266.TAK.p53 cell line in the absence of IL-6 shows a loss of cells in the S and G2/M stages of the cell cycle (Figure 1C). The addition of IL-6 results in an increase in the number of cells in cycle from 29% to 44%. There is no evidence of increased apoptosis in wt p53 transfected cells in the absence of IL-6 (Figure 1C). This may be due to a small amount of residual IL-6 expression in the U266.TAK.p53 cell line, serum components that maintain viability, or other factors that we have not examined.
To determine the effect of wt p53 expression on various IL-6–related transcription factors, electrophoretic mobility shift assays were performed in the presence and absence of IL-6 (Figure 1D). NF-κB DNA binding activity remains constant in all the conditions tested and can serve as a control for the integrity of the nuclear extracts. Both AP-1 and STAT1/3 are involved in the IL-6 signal transduction pathway, and their DNA binding activity is down-regulated in the U266.TAK.p53 cell line in the absence of IL-6. Addition of IL-6 restores the binding activity of both AP-1 and STAT1/3. It has been shown that the up-regulation of IL-6 by mutant p53 requires an intact NFIL-6 binding site in the IL-6 promoter12 and that p53 can be directly involved in repressing NFIL-6–dependent gene transcription.24 Consistent with this, there is a suppression of NFIL-6 binding in the U266.TAK.p53 cell line when compared to the control. Addition of IL-6 did not significantly restore NFIL-6 binding. This suggests that wt p53 may suppress IL-6, in part, by down-regulating NFIL-6 activity.
p53-mediated cell cycle arrest protects U266 cells from Dox- and Mel-induced apoptosis
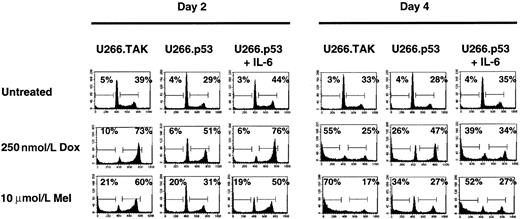
p53 has been shown to play an important role in transducing a signal from damaged DNA to genes that control cell cycle and apoptosis (reviewed in Gotz and Montenarh25). Recent studies7 also demonstrate that cells expressing wt p53 show reduced cytotoxicity in response to DNA damaging agents, such as Dox or alkylating agents such as Mel. Because Dox and Mel are used in the treatment of myeloma,26 27 we wanted to determine whether either would have different effects on apoptosis in U266 cells with mutant or wt p53. U266.TAK or U266.TAK.p53 cells were treated with 0.25 μmol/L Dox or 10 μmol/L Mel for 4 days to examine the effect of p53 on apoptosis. Treatment of U266.TAK cells with Dox or Mel causes an increase in the G2/M stage of the cell cycle by day 2 when compared to untreated cells (Figure 2). Thirty-nine percent of the cells are in the S/G2/M stages compared to 73% and 60% for Dox and Mel, respectively. [3H] Thymidine incorporation assays show that the increase results from G2/M arrest rather than from increased cell cycle because there is little or no [3H] thymidine incorporation in the presence of Dox or Mel (data not shown). The G2/M block in cell cycle is followed by a significant increase in apoptosis by day 4. Expression of wt p53 results in a loss of accumulation in the S/G2/M stages of the cell cycle and significantly suppresses Dox- (26% compared to 55%) or Mel- (34% compared to 70%) induced apoptosis. Notably, the addition of IL-6 stimulates cell cycle progression resulting in an increase in apoptosis (Figure 2).
Expression of p53 protects U266 cells from Dox- and Mel-induced apoptosis.
Cells were incubated in the absence of IL-6 for 3 days before each treatment. Cells were treated with IL-6, Dox, or Mel as indicated and were harvested on days 2 and 4 for cell cycle and apoptosis analyses. Percentages of cells in each stage of the cell cycle were determined by PI staining of nuclei and then by flow cytometry analysis. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. Data shown are representative of at least 3 independent experiments.
Expression of p53 protects U266 cells from Dox- and Mel-induced apoptosis.
Cells were incubated in the absence of IL-6 for 3 days before each treatment. Cells were treated with IL-6, Dox, or Mel as indicated and were harvested on days 2 and 4 for cell cycle and apoptosis analyses. Percentages of cells in each stage of the cell cycle were determined by PI staining of nuclei and then by flow cytometry analysis. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. Data shown are representative of at least 3 independent experiments.
IL-6 protects ANBL6 cells from Dex- and Mel- but not Dox-induced apoptosis
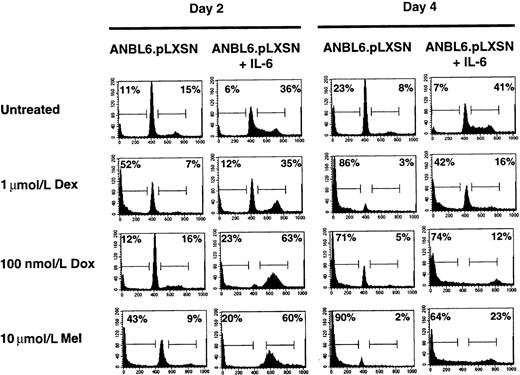
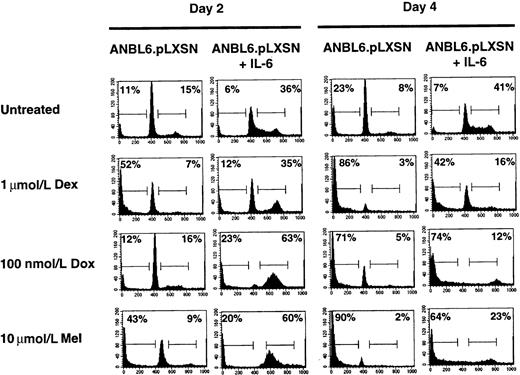
To compare the U266.TAK.p53 cell line with another IL-6–dependent cell line, we used ANBL6 cells. One difference between the 2 cell lines is that ANBL6 expresses a mutant p53 (P.L., unpublished observations). The addition of IL-6 to ANBL6 cells stimulates the cell cycle and provides protection from Dex-induced apoptosis.18 We confirm the IL-6 protection and show that ANBL6 cells treated with Dex in the absence of IL-6 arrest in the G0/G1 stage of the cell cycle before undergoing apoptosis (Figure3). Treatment of the ANBL6 cell line with Dox or Mel in the presence of IL-6 results in an arrest in the S or G2/M stage of the cell cycle, similar to the arrest in the U266.TAK.p53 cell line treated with IL-6 (Figure 3). Unlike the U266.TAK.p53 cell line, however, ANBL6 cells are sensitive to Dox-induced apoptosis, even in the absence of IL-6. Moreover, IL-6 is able to provide some protection from Mel-induced apoptosis in ANBL6 cells. This suggests that differences in the genetic makeup of cells, such as p53 mutations, can affect the response to chemotherapeutic agents.
IL-6 protects ANBL6 cells from Dex- and Mel- but not Dox-induced apoptosis.
Cells were incubated in the absence of IL-6 for 3 days before each treatment. Cells were treated with IL-6, Dex, Dox, or Mel as indicated and were harvested on days 2 and 4 for cell cycle and apoptosis analyses. Percentages of cells in each stage of the cell cycle were determined by PI staining of nuclei and then by flow cytometry analysis. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. Data shown are representative of at least 3 independent experiments.
IL-6 protects ANBL6 cells from Dex- and Mel- but not Dox-induced apoptosis.
Cells were incubated in the absence of IL-6 for 3 days before each treatment. Cells were treated with IL-6, Dex, Dox, or Mel as indicated and were harvested on days 2 and 4 for cell cycle and apoptosis analyses. Percentages of cells in each stage of the cell cycle were determined by PI staining of nuclei and then by flow cytometry analysis. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. Data shown are representative of at least 3 independent experiments.
Activated N-ras or K-ras can protect ANBL6 cells from apoptosis induced by Dex, Dox, or Mel
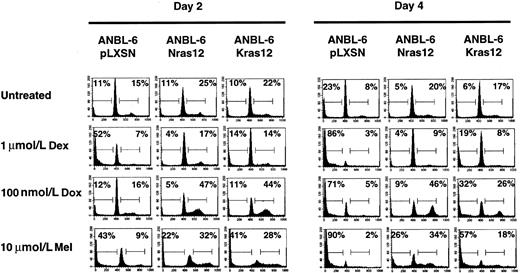
It has been shown18 that activating mutations in N- or K-ras can stimulate proliferation and protect ANBL6 cells from apoptosis induced by IL-6 withdrawal. Based on this and on the results obtained with the U266 cell line, we expected ANBL6 cells transfected with either an activated N-ras12 or K-ras12 gene to be more sensitive than the parental cell line to Dox and Mel because these genes would stimulate cell cycle and DNA synthesis. Similar to the effect of IL-6, mutations in either N-ras orK-ras are able to stimulate the cell cycle and result in S or G2/M phase arrest in cells treated with Dox or Mel (Figure4). Surprisingly, expression of an activating mutation in either N-ras or K-ras is able to provide significant protection from apoptosis induced by Dex, Dox, or Mel. This protection appears to be better in theN-ras12–transfected cell line than theK-ras12–transfected cell line.
Activated N-ras or K-ras can protect ANBL6 cells from apoptosis induced by Dex, Dox, or Mel.
Cells were treated with IL-6, Dex, Dox, or Mel as indicated and were harvested on days 2 and 4 for cell cycle and apoptosis analyses. Percentages of cells in each stage of the cell cycle were determined by PI staining of nuclei and then by flow cytometry analysis. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. Data shown are representative of at least 3 independent experiments.
Activated N-ras or K-ras can protect ANBL6 cells from apoptosis induced by Dex, Dox, or Mel.
Cells were treated with IL-6, Dex, Dox, or Mel as indicated and were harvested on days 2 and 4 for cell cycle and apoptosis analyses. Percentages of cells in each stage of the cell cycle were determined by PI staining of nuclei and then by flow cytometry analysis. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. Data shown are representative of at least 3 independent experiments.
Discussion
In this study, model cell lines were used to examine the effect of specific genetic mutations on cell cycle and apoptosis in myeloma cells. Although the data presented were limited to cell lines and not to fresh patient samples, we feel this approach has some advantages. By making only a single genetic change in a cell line, we are able to examine the specific effects of a change in one particular gene in 2 cell populations that are otherwise genetically identical (Figure5). We used 2 myeloma cell lines that have distinct IL-6 characteristics and introduced a specific genetic change in each line. A wt p53 gene was introduced in the autocrine IL-6–producing U266 cell line to study the effect that changes in p53 have on IL-6 expression and response to therapy. We also used the IL-6–dependent cell line ANBL6, transfected with either an activating mutation in the N-ras or theK-ras gene, to examine the effect of rasmutations on apoptosis induced by various chemotherapeutic agents.
Genetically engineered cell lines can be used to study the effect of a single-gene mutation on cell cycle, apoptosis, and response to therapy.
Genetically engineered cell lines can be used to study the effect of a single-gene mutation on cell cycle, apoptosis, and response to therapy.
Previous studies10 have demonstrated that wt p53 can suppress IL-6 promoter activity. Additionally, some mutants of p53 may actually increase IL-6 expression.12 These results suggest that mutations in p53 may play a role in the aberrant expression of IL-6 in myeloma. Consistent with this, we show that the expression of wt p53 can suppress IL-6 expression in U266 cells. One consequence of IL-6 suppression is the loss of AP-1 and STAT1/3 binding activities, both of which are part of the IL-6 signal transduction pathway. It was recently suggested28 that myeloma cells have elevated levels of constitutive STAT activation. Our data suggest that this may be the result of IL-6 produced either by the myeloma cell or from the environment, because loss of IL-6 expression resulted in a loss of STAT1/3 binding activity.
p53 has been shown to be involved in DNA repair by blocking cells in the G0/G1 stage of the cell cycle, therefore allowing DNA repair before the initiation of DNA synthesis.29 In support of this, it has been demonstrated17,30 that wt p53 can reduce the cytotoxicity of Dox in ovarian cancer and breast cancer cells. Others31 32 have found, however, that a loss of wt p53 gene is associated with resistance to Dox. Our study demonstrates that the expression of wt p53 decreased the cytotoxic effects of Dox and Mel in U266 cells. The protection appears to be dependent on cell cycle arrest at the G0/G1 stage of the cell cycle, and could be a result of a p53-dependent block of the cell cycle, a consequence of loss of IL-6 expression, or both. The ANBL6 cell line also expresses a mutant p53 gene. In the absence of IL-6, these cells are arrested in the G0/G1 stage of the cell cycle but are sensitive to apoptosis induced by Dox or Mel. This suggests that the protection seen in the U266.TAK.p53 cell line may be due to DNA repair facilitated by the presence of wt p53 rather than just a suppressed cell cycle. In the presence of IL-6 there may be a competition between a p53-induced block in the cell cycle and IL-6 driving the cell cycle. Stimulating the cell cycle with IL-6 may not allow for sufficient DNA repair, resulting in the intermediate levels of apoptosis seen in the U266.TAK.p53 cell line in the presence of IL-6 (Figure 2).
Activating mutations in N- or K-ras have previously been shown to stimulate proliferation of ANBL6 cells.18 Based on this and on the apoptosis data from the U266 cells, we expected that theN-ras12– and K-ras12–transfected ANBL6 cells would be more sensitive than the parental ANBL6 line to Dox- or Mel-induced apoptosis. Surprisingly, expression of either an activated N- or K-ras is able to provide protection from apoptosis. This protection appears to be more complete than the protection provided by IL-6. One possible explanation for this may be differences in signal kinetics. IL-6 may provide a transient signal through ras that is quickly down-regulated. The cells with the ras mutation, however, will have a constitutive signal downstream of ras, with a resultant constitutive protection. The mechanism for this is at present unclear.
Previous reports have suggested IL-6 is an antiapoptotic factor. However, as we have shown, IL-6 induction of the cell cycle can have an apoptotic-enhancing effect on DNA-damaging agents. Moreover, we have shown that genetic heterogeneity can have an affect on how myeloma cells respond to IL-6 and chemotherapeutic agents. As new screening methods are applied to determine therapeutic response, genetic reclassification of tumors should provide more improvements in treating patients.
Supported by National Institutes of Health grant PO1 CA62242 and by a grant from the Leukemia Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Brian Van Ness, Cancer Center Research Bldg, Box 806 UMHC, Minneapolis, MN 55455; e-mail: vanne001@tc.umn.edu.

![Fig. 1. Wt p53 suppresses autocrine IL-6 expression and suppresses cell cycle in U266 cells. / (A) Wt p53 suppresses autocrine IL-6 expression. Total RNA was isolated from the U266.TAK and the U266.TAK.p53 cell lines. Expression of p53 and IL-6 was detected by RT-PCR. Expression of PBGD was used as a loading control.22 (B) Wt p53 suppresses cell growth. Cells were incubated with or without varying concentrations of IL-6 and then pulsed with [3H] thymidine. [3H] Thymidine incorporation was measured by liquid scintillation counting. Values represent the mean of duplicate samples ± SEM. Data shown are representative of at least 3 independent experiments. (C) Wt p53 reduces the number of cells in the S and G2/M stage of the cell cycle. Cells were treated with or without IL-6 and were harvested for cell cycle analysis. Percentages of cells in each phase of the cell cycle were determined by flow cytometry. The number in the left corner represents the apoptotic fraction, and the number in the right corner represents the combined S and G2/M stages of the cell cycle. (D) Expression of wt p53 alters transcription factor activation. Nuclear extracts were prepared, and binding activities were measured by electrophoretic mobility shift assay. ANBL6 extracts were included as a reference for STAT1 and STAT3 dimers.23 SC, 100 × specific competitor; NSC, 100 × nonspecific competitor.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/9/10.1182_blood.v96.9.3175/5/m_h82100335001.jpeg?Expires=1766261272&Signature=mkmztmUS0tfVAQD7knsIWvxA7zAyCUu6WPOm9QhaLx5DHkzFITItoHe7-Kfg1XByGShFKfoRTMtpDhM0yp0XVRFFK21W96lqLhxs7Aes8~bgrjLp3zkcfvuAvOioYNJ1Gq4uiehuWW290KgTpfLwbwQ8ACVWh3FGjE~WJg4bsfbaY1EPfHlU1LJpkOdd26YOGF1j0en5cT~kyxE6pmqtFezWQi785vBD7CroWJcMxCh7SpnZusHAZQDil9EVvRfk99HqPAM6XPdv3rRIDbDrZXP8cvDVHrHoXmYbUz06J3OU9Ua6oMlEzMLWKPMcLr1qUvJkls4MXJ4L4wf9lMAj~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)