Abstract
We have previously reported that expression of the erythropoietin (Epo) gene in mouse embryonal cells was not induced by hypoxia, although hypoxia induced other hypoxia-inducible genes. This study identifies retinoic acid (RA) as an inducer for Epo production in the embryonal carcinoma cell lines P19 and F9. RA induced Epo production through the transcriptional activation of the Epo gene in an oxygen-independent manner. With the use of reporter assays in P19 cells, it is shown that a direct repeat of the nuclear hormone receptor-binding motif separated by a 2-bp spacer (DR-2) in the hypoxia-response enhancer was responsible for the transcriptional activation by RA. Electrophoretic mobility shift assays show that nuclear extracts from P19 cells contained RA receptor complexes that bound to DR-2. In human hepatoma Hep3B cells, an orphan receptor, hepatocyte nuclear factor-4, strongly augmented hypoxic induction of the Epo gene in cooperation with hypoxia-inducible factor-1 (HIF-1) by binding to DR-2, whereas in P19 cells, the interaction of RA receptors with DR-2 was sufficient for RA-induced transcriptional activation of the Epo gene without the requirement of the HIF-1 site. These results suggest that DR-2 regulates expression of the Epo gene by acting as the binding site for different transcription factors in different types of cells.
Introduction
Erythropoietin (Epo) is the major physiologic stimulator of red blood cell formation in mammals.1-4 The main Epo production sites are the kidney in adults and the liver in fetuses. In addition, at least 3 other sites (the brain,5-10 uterus,11 and oviduct12) have been shown to produce Epo, although the levels are low compared with those in the kidney and liver. Brain Epo acts on neurons,13-16 preventing their death from ischemic insult.10,17,18 Uterine Epo is implicated in estrous cycle–dependent angiogenesis,11 and the oviductal function of Epo is not known.
Hypoxia is a signal that can stimulate Epo production intensely.4 A number of genes have been shown to be induced by hypoxia19; these include Epo,20,21some glycolytic enzymes such as lactate dehydrogenase A (LDHA),22,23 glucose transporter 1,24vascular endothelial growth factor,25 nitric oxide (NO) synthase,26 heme oxygenase 1,27transferrin,28 and its receptor.29 Hypoxic induction of these proteins is mostly due to transcriptional activation. The Epo gene possesses a 50-bp hypoxia-response enhancer (3′ enhancer) at 120 bp downstream of the polyA signal.30-33 This 3′ enhancer consists of 3 segments.34,35 The highly conserved sequence (5′RCGTG3′) near the 5′ end of the 3′ enhancer is a binding site for hypoxia-inducible factor 1 (HIF-1), which is a transcriptional factor essential for activation of the hypoxia-inducible genes.35-38 Although the middle segment is not well conserved in the mouse (5′CATGG3′) and human (5′CACAG3′) genes, mutation of this region abolishes the responsiveness to hypoxia.34-36 Thus far, the specific protein that binds to this region has not been demonstrated. The third segment at the 3′ end of the 3′ enhancer is a direct repeat (DR) of a hexanucleotide consensus nuclear receptor–binding half-site (5′TGACCTCTTGACCC3′) separated by a 2-bp spacer (DR-2).34,35,39,40 Binding of hepatocyte nuclear factor-4 (HNF-4), a member of the nuclear hormone receptor superfamily, to this element dramatically increases the hypoxic inducibility of the Epo gene in Hep3B cells.41
In addition to hypoxia, various substances have been shown to stimulate Epo production. Insulin and insulin-like growth factors stimulate Epo production by astrocytes.42 Nuclear receptor ligands such as thyroid hormone and 17β-estradiol enhance Epo production in the perfused kidney and female reproductive organs (uterus and oviduct), respectively.11,12,43 Retinoic acid (RA) increases the serum Epo level in vitamin A–depleted rats.44 We previously showed that embryonic stem (ES) cells and embryonal carcinoma (EC) cells, P19 cells, produce Epo.45 Expression of the Epo gene in P19 cells was not hypoxia inducible, for an unknown reason, although that of the LDHA gene was inducible. No mutations have been found in the promoter and 3′ enhancer of the Epo gene cloned from P19 cells.45 In the present study, we examined the effects of nuclear receptor ligands to understand how Epo production in the embryonic cells is regulated and found that only RA stimulates Epo production.
There are 2 families of RA receptors, retinoic acid receptors (RARs) and retinoid X receptors (RXRs).46,47 Ligand-induced homodimers or heterodimers of RAR and RXR control gene transcription by binding to retinoic acid response elements (RAREs), DRs of nuclear hormone receptor—binding motif (5′YGACCY3′) with various numbers of nucleotide spaces (DR-n).47,48 A survey of the 5′- and 3′-flanking regions conserved in mouse and human Epo genes revealed the potential RAREs in the 5′-flanking region (DR-4 and DR-10) and the 3′-flanking region (DR-2), which agree with the elements pointed out by Blanchard et al.39 Here we report that RA-induced stimulation of Epo production in P19 cells is mediated through DR-2 in the 3′ enhancer.
Materials and methods
Cell culture and Epo production
P19 cells were maintained in minimal essential medium α medium (αMEM; GIBCO BRL, Grand Island, NY) supplemented with 10% fetal calf serum (FCS; Bio Whittaker, Walkersville, MD) in a gelatin-coated dish, and Hep3B and F9 cells were maintained in Dulbecco modified Eagle medium (DMEM; GIBCO BRL) supplemented with 10% FCS. When the effects of nuclear receptor ligands on cultured cells are examined, the cells are usually cultured in media containing charcoal-treated serum to remove the effects of serum-derived ligands. Because proliferation of P19 and F9 cells, however, is largely retarded in the charcoal-treated medium, they were cultured in the medium supplemented with untreated serum. Similarly, Hep3B cells were cultured in the medium containing untreated serum. To examine Epo production, we plated P19 and F9 cells (5 × 104/well) and Hep3B cells (2 × 105/well) in 2 6-well plates (Nunc A/S, Roskilde, Denmark) and cultured them for 20 hours. Then the media were replaced with fresh media containing the indicated substances, and the cells were cultured for the indicated periods in 21% or 2% oxygen. To form aggregates, P19 cells (1 × 105/mL per dish) were grown in Petri dishes 10 cm in diameter (Sekisui Chemical Co, Ltd, Osaka, Japan) for 48 hours. Under these conditions, P19 cells form aggregates spontaneously. Epo in the spent media and Epo mRNA content were measured as described previously.12 45 The hypoxic (2% or 5% oxygen) condition was generated in an oxygen-regulated incubator. Actinomycin D (Sigma, St. Louis, MO) was added at a final concentration of 0.025 μg/mL.
Preparation of Luc reporter plasmids under the control of mouse Epo promoter and 3′ enhancer
A 0.12-kb mouse Epo promoter (mP) was synthesized and subcloned into the SacI and BglII site upstream of the Luc reporter gene in pPGV-B2 (Toyo Ink Mfg. Co, Ltd, Tokyo, Japan) to generate pWT (Table 1). A mouse Epo 3′ enhancer (mE; lower sequence in Figure 3A) was synthesized and subcloned into the SalI and BamHI sites of pWT to produce pWT-WT. The mutant plasmids were constructed by the same procedures.
Transient assay of reporter gene
Plasmids were transfected into P19 cells or Hep3B cells as described previously.45 After transfection, the medium was replaced with fresh growth medium containing all-trans RA. The plate was incubated for another 20 hours in 21% or 2% oxygen. Assays of Luc and β-galactosidase were performed as described previously.45 Expression of the Luc gene was represented as the ratio of Luc to β-galactosidase.
Preparation of β-galactosidase reporter plasmids under the control of human Epo promoter and 3′ enhancer
A β-galactosidase reporter plasmid (p4En0.2Z), in which the β-galactosidase gene was under the control of the human Epo promoter and the 3′ enhancer (Figure 3B, bottom), was constructed as described previously.49 Briefly, 4 tandemly repeatedApaI–PvuII fragments of human Epo 3′ enhancer, an ApaI–Eco52I fragment of human Epo promoter, and β-galactosidase gene from pCH110 were ligated and subcloned into the SacI and PstI sites of pUC18.
Preparation of permanent clones harboring p4En0.2Z
P19 cells were cotransfected with p4En0.2Z and pKSV10neo. Drug-resistant clones were screened in the presence of 400 μg/mL of G418. P19/Z cells were established with 2 rounds of cloning by limiting dilution. Hep3B/Z cells (F6D2 in the previous report) were established as described previously.49
Staining of β-galactosidase with X-gal
Hep3B cells and aggregates of P19 cells were washed with 10 mmol/L phosphate-buffered saline, pH 7.4 (PBS), and fixed with a mixture of 2% formaldehyde and 0.2% glutaraldehyde. After staining for 24 hours in the X-gal solution, the cells were placed in PBS containing 1 mmol/L EDTA. The cells were photographed under a phase-contrast microscope using the Axiovert 135 (Zeiss, Germany) as a magnifier.
Protein extractions
Nuclear extracts were prepared according to the method described by Andrews and Faller.50 Briefly, P19 cells (approximately 1 × 107/well) were scraped into 1.5 mL of cold PBS. The cells were pelleted and resuspended in 400 μL of cold buffer A (10 mmol/L HEPES-KOH, pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol [DTT], and 0.2 mmol/L phenylmethylsulfonyl fluoride [PMSF]). The cells were allowed to swell on ice for 10 minutes and then were vortexed for 10 seconds. The samples were centrifuged and the supernatant was discarded. The pellet was resuspended in 200 μL of cold buffer C (20 mmol/L HEPES-KOH, pH 7.9, 25% glycerol, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, and 0.2 mmol/L PMSF) and was incubated on ice for 20 minutes. Cellular debris was removed by centrifugation for 2 minutes at 4°C. The supernatant was used as a nuclear extract. For preparation of whole-cell extracts, pelleted cells were resuspended in whole-cell extract buffer (50 mmol/L Tris-HCl, pH 8.0, 450 mmol/L NaCl, 1% Triton X-100, and 0.2 mmol/L PMSF) and centrifuged. The supernatant was used as a whole-cell extract.
Electrophoretic mobility shift assay
Oligonucleotides containing the DR-2 element were used as probes and competitors. The sequence for wild-type DR-2 was 5′CCGGCTGACCTCTTGACCCCTCTGGGCTTG3′ (DR-2 WT, where DR-2 is underlined) and that for mutant DR-2 was 5′CCGGCTGCAATCTTACCTCCTCTGGGCTTG3′ (DR-2 MT, where mutations are underlined). These single-strand oligonucleotides were annealed with the complementary oligonucleotides, and the resulting double-strand fragments were end-labeled by filling in 5′ overhangs with α-[32P]dCTP (3000 Ci/mmol) using Klenow fragment. DNA-protein binding reactions were carried out for 10 minutes at room temperature in a total volume of 10 μL containing 4 μg nuclear extract, 2.5 μg poly(dI-dC), and 1 × 105cpm of radiolabeled probe in binding buffer (20 mmol/L HEPES-KOH, pH 7.6, 4% type-400 Ficoll, 40 mmol/L NaCl, 1 mmol/L MgCl2, and 0.5 mmol/L DTT). The products were analyzed by electrophoresis in 4% nondenaturing polyacrylamide gels. Electrophoresis was performed at 80 V in 0.25 × TBE (1 × TBE is 89 mmol/L Tris, 89 mmol/L boric acid, and 2.5 mmol/L EDTA) buffer at 4°C, and the dried gels were autoradiographed. For competition experiments, we added a 100-fold molar excess of an unlabeled probe to the binding reaction before addition of a labeled probe. In supershift analysis, 4 μg of antibody was incubated with a nuclear extract in PBS for 1 hour at 4°C before the binding reaction was done. Antibodies to RARα (sc-551X) and RXRβ (sc-831X) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). In the experiments to examine the binding of HIF-1 with HIF-1 probes, electrophoretic mobility shift assay (EMSA) was performed in the same way except that nuclear extracts were prepared from P19 cells cultured for 5 hours in 2% or 21% oxygen. The oligonucleotide sequence for the wild-type HIF-1 site was 5′gatcGCCCTACGTGCTGCCTCGCATG3′ (HIF-1 WT, where the HIF-1 site is underlined) and that for the mutant HIF-1 site was 5′gatcGCCCTAAAAGCTGCCTCGCATG3′ (HIF-1 MT, where mutations are underlined).
Immunoblot analysis
Protein extracts (15 μg) were subjected to electrophoresis through 6% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to PVDF membrane (Pall Corporation, NY). The membrane was blocked with Block Ace (Dainippon Pharmaceutical Co, Ltd, Osaka, Japan) and incubated with an anti–HIF-1α antibody (Novus Biologicals, Littleton, CO) at a 1:750 dilution in PBS-T (PBS and 0.5% Tween-20) containing 10% Block Ace. Horseradish peroxidase–conjugated anti–mouse IgG (Amersham Pharmacia Biotech, Piscataway, NJ) was used as a secondary antibody at a 1:3000 dilution, and Super Signal Chemiluminescent Substrate (Pierce, Rockford, IL) was used for detection.
Results
Stimulation of Epo production in the EC cells by RA
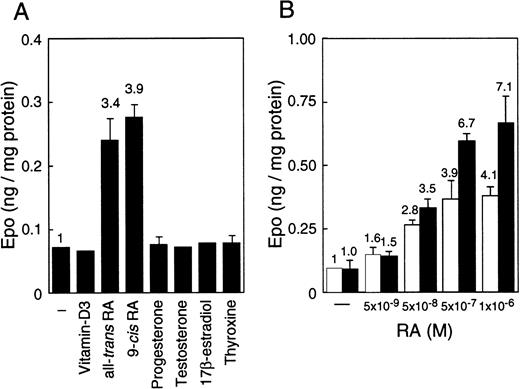
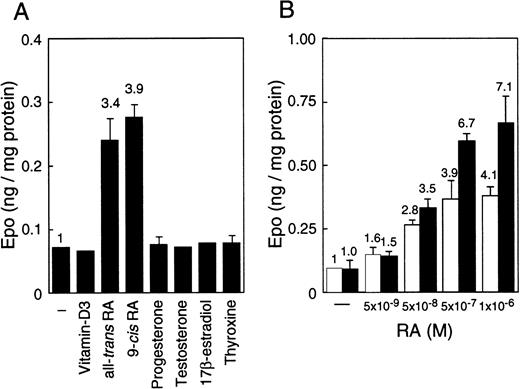
EC cell lines (P19 and F9) cultured under normoxia produce more Epo than Hep3B cells, which is the hepatoma cell line that produces Epo by responding to hypoxia.45 The production in these EC cell lines, however, is not induced by hypoxia.45 To find an inducer of the expression of the Epo gene in EC cells, we examined the effects of a number of nuclear receptor ligands on Epo production in these cells. Epo production was induced up to 4-fold in P19 cells only when cultured with all-trans RA or 9-cis RA (Figure 1A). Both RAs also induced Epo production in F9 cells, and the stimulation degree was similar to that of P19 cells (data not shown). In contrast, only a slight stimulation of Epo production (approximately 1.1- to 1.3-fold) by RAs was observed in Hep3B cells (data not shown). No other ligands tested (10−6 to 10−8 mol/L) stimulated Epo production either in P19 cells (Figure 1A) or in F9 cells or Hep3B cells (data not shown). Aggregate culture of P19 cells has been used as a model system for studying early mammalian development; the P19 cells differentiate into neurons and astrocytes in addition to relatively small numbers of fibroblast-like cells.51 As shown in Figure 1B, all-trans RA stimulated Epo production in P19 cells cultured as a monolayer and as aggregates in a dose-dependent manner, but the degree of stimulation was significantly higher in aggregates than in monolayer cells.
Stimulation of Epo production in P19 cells by RA.
Epo concentration in the spent medium and the total cellular proteins were determined. Epo production is given as nanograms of Epo per milligram cellular protein. (A) P19 cells were cultured with or without the indicated substances at 5 × 10−7 mol/L for 36 hours. (B) P19 cells were cultured with the indicated concentrations of all-trans RA for 48 hours in monolayers (■) and aggregate cultures (▪). Each value is the mean ± SD of triplicate experiments.
Stimulation of Epo production in P19 cells by RA.
Epo concentration in the spent medium and the total cellular proteins were determined. Epo production is given as nanograms of Epo per milligram cellular protein. (A) P19 cells were cultured with or without the indicated substances at 5 × 10−7 mol/L for 36 hours. (B) P19 cells were cultured with the indicated concentrations of all-trans RA for 48 hours in monolayers (■) and aggregate cultures (▪). Each value is the mean ± SD of triplicate experiments.
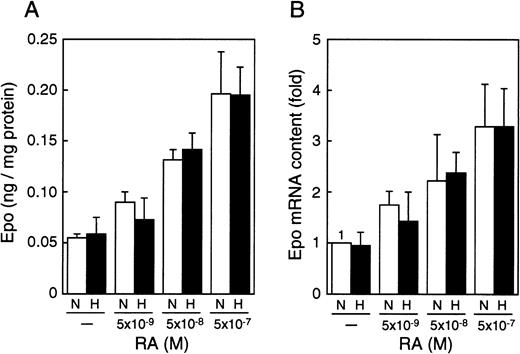
Correlation between Epo production and Epo mRNA, and the effect of oxygen
To determine whether the RA-induced stimulation of Epo production operates at the level of its mRNA, we extracted the total RNA from P19 cells cultured with different concentrations of all-trans RA under normoxia (21% oxygen) or hypoxia (2% oxygen) and measured Epo mRNA in the total RNA by using real-time polymerase chain reaction (PCR). Both the production of Epo and the level of its mRNA were increased by RA in a dose-dependent manner, and the RA-induced increase in Epo production closely paralleled that in mRNA (compare Figure2A and 2B). Moreover, the RA-induced increase in Epo mRNA was inhibited by actinomycin D (data not shown). These results suggest that the RA-induced production of Epo is caused by transcriptional activation of the Epo gene. In contrast, the oxygen concentration altered neither the induction of Epo production nor the level of Epo mRNA in the presence of RA, indicating that the hypoxia inducibility of the Epo gene in P19 cells is not restored by RA treatment.
RA increases Epo production and accumulation of Epo mRNA in an oxygen-independent manner.
P19 cells were cultured with the indicated concentrations of all-trans RA for 20 hours in 21% oxygen (N) or 2% oxygen (H). (A) Epo protein (in nanograms) per milligram cellular protein is shown. (B) The ordinate shows fold induction of Epo mRNA over the control value. The Epo mRNA content when cultured without RA in 21% oxygen (N) was defined as 1. Each value is the mean ± SD of triplicate experiments.
RA increases Epo production and accumulation of Epo mRNA in an oxygen-independent manner.
P19 cells were cultured with the indicated concentrations of all-trans RA for 20 hours in 21% oxygen (N) or 2% oxygen (H). (A) Epo protein (in nanograms) per milligram cellular protein is shown. (B) The ordinate shows fold induction of Epo mRNA over the control value. The Epo mRNA content when cultured without RA in 21% oxygen (N) was defined as 1. Each value is the mean ± SD of triplicate experiments.
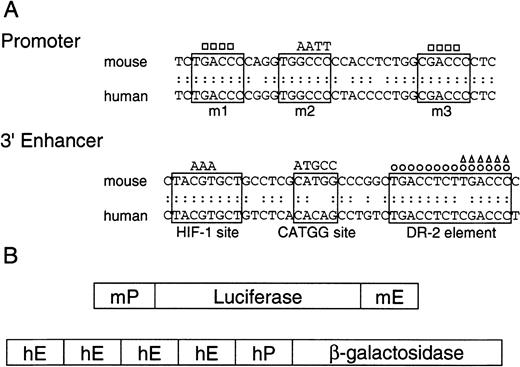
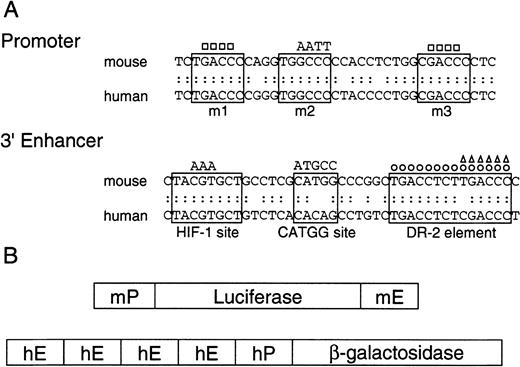
DR-2 in the 3′ enhancer is responsible for the Epo gene to respond to RA
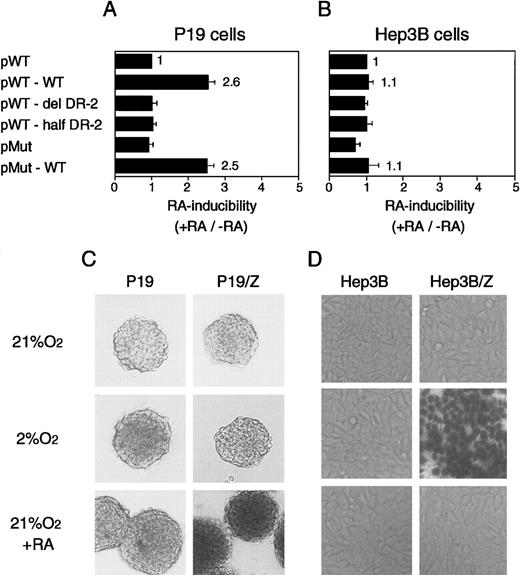
RAREs consist of the direct repetition of 2 core motifs or closely related degenerate motifs with a variable length of nucleotide spaces. A survey of the regions conserved between the mouse and human Epo genes showed that 2 potential RAREs (DR-4 and DR-10) exist in the promoter region (a 32-bp segment located 82 bp upstream of the transcription start site; the upper sequence in Figure3A). The other potential site found is DR-2 (HNF-4 binding site) in the 3′ enhancer (the lower sequence in Figure 3A). To test which site is responsible for the Epo induction by RA, we constructed the plasmids harboring the Luc reporter gene interposed between the mouse 120-bp promoter (mP) and the mouse Epo 3′ enhancer (mE) with the wild-type sequences or its mutated sequences (Figure 3B, top). The mutated position either in potential RAREs in the promoter region (m1, m2, and m3) or in the 3′ enhancer is shown in Figure 3A as a circle or a triangle (Table 1). These plasmids were transfected into P19 cells as a monolayer culture with pβactβgal to normalize transfection efficiency. After the transfected cells were incubated under normoxia with or without all-trans RA, the activities of Luc and β-galactosidase in cell lysates were assayed. As shown in Figure4, P19 cells transfected with the plasmid containing the wild-type 3′ enhancer yielded 2.5-fold RA induction, whereas no RA induction was seen in the cells transfected with the plasmid containing the DR-2–deleted 3′ enhancer (Figure 4A). Deletion of the half-site in DR-2 abolished the RA inducibility in P19 cells. Mutations in the promoter did not change its activity (pMut) or the RA inducibility (pMut-WT). Thus, in P19 cells, DR-2 in the 3′ enhancer is indispensable for the transcriptional activation of the Epo gene in response to RA, whereas the potential RAREs in the promoter region are not required. When Hep3B cells were used, however, no RA inducibility was observed with any reporters (Figure 4B), which is in agreement with the results reported previously.39
Promoters and 3′ enhancers of human and mouse Epo genes, and schematic representation of the constructs.
(A) Nucleotide sequences of promoters and 3′ enhancers in mouse and human Epo genes, and the mutant fragments used in the present experiments. The upper sequences show the promoter regions that contain nuclear hormone receptor response element half-sites enclosed in solid lines. The mutations are shown either as base substitution (AATT) or deletion (■). The lower sequences show 3′ enhancers, where 3 important segments are enclosed in solid lines. The DR-2 element is an HNF-4 binding site in Hep3B cells. The mutations are shown either as base substitution (AAA and ATGCC) or deletion (○, ▵). (B) Schematic representation of the reporter gene constructs used. The upper construct shows the Luc reporter plasmids consisting of the mouse Epo promoter (mP) and 3′ enhancer (mE), and the lower construct shows the β-galactosidase reporter plasmid consisting of the human Epo promoter (hP) and 3′ enhancer (hE).
Promoters and 3′ enhancers of human and mouse Epo genes, and schematic representation of the constructs.
(A) Nucleotide sequences of promoters and 3′ enhancers in mouse and human Epo genes, and the mutant fragments used in the present experiments. The upper sequences show the promoter regions that contain nuclear hormone receptor response element half-sites enclosed in solid lines. The mutations are shown either as base substitution (AATT) or deletion (■). The lower sequences show 3′ enhancers, where 3 important segments are enclosed in solid lines. The DR-2 element is an HNF-4 binding site in Hep3B cells. The mutations are shown either as base substitution (AAA and ATGCC) or deletion (○, ▵). (B) Schematic representation of the reporter gene constructs used. The upper construct shows the Luc reporter plasmids consisting of the mouse Epo promoter (mP) and 3′ enhancer (mE), and the lower construct shows the β-galactosidase reporter plasmid consisting of the human Epo promoter (hP) and 3′ enhancer (hE).
The DR-2 element in the 3′ enhancer of the Epo gene is RARE in P19 cells but not in Hep3B cells.
P19 cells (A) and Hep3B cells (B) show transient expression of Luc reporter constructs. Characteristics of the plasmids are shown in Figure 3 and Table 1. After transfection, the cells were cultured in 21% oxygen for 20 hours in the presence and absence of 5 × 10−7 mol/L all-trans RA. The inducibility by RA (the ratio of Luc activity in the presence versus absence of RA) is shown. The inducibility of pWT construct was defined as 1. Each value is the mean ± SD of triplicate experiments. (C) The 3′ enhancer from the human Epo gene acts as RARE in P19 aggregates. (D) The 3′ enhancer from the human Epo gene responds to hypoxia in Hep3B cells. Permanent P19 and Hep3B cell lines harboring the β-galactosidase reporter construct were established (P19/Z and Hep3B/Z). The reporter gene is under the control of the human Epo promoter (hP) and the 3′ enhancer (hE) (Figure 3B, bottom). (C) Parental P19 cells and P19/Z cells were cultured in aggregates with or without 5 × 10−7 mol/L all-trans RA for 48 hours in 21% oxygen or were cultured for 28 hours in 21% oxygen and then for 20 hours in 2% oxygen. After 48 hours, the cells were stained with X-gal solution. (D) Parental Hep3B cells and Hep3B/Z cells were cultured with or without 5 × 10−7 mol/L all-trans RA for 24 hours in 21% or 2% oxygen. Magnifications are 25-fold.
The DR-2 element in the 3′ enhancer of the Epo gene is RARE in P19 cells but not in Hep3B cells.
P19 cells (A) and Hep3B cells (B) show transient expression of Luc reporter constructs. Characteristics of the plasmids are shown in Figure 3 and Table 1. After transfection, the cells were cultured in 21% oxygen for 20 hours in the presence and absence of 5 × 10−7 mol/L all-trans RA. The inducibility by RA (the ratio of Luc activity in the presence versus absence of RA) is shown. The inducibility of pWT construct was defined as 1. Each value is the mean ± SD of triplicate experiments. (C) The 3′ enhancer from the human Epo gene acts as RARE in P19 aggregates. (D) The 3′ enhancer from the human Epo gene responds to hypoxia in Hep3B cells. Permanent P19 and Hep3B cell lines harboring the β-galactosidase reporter construct were established (P19/Z and Hep3B/Z). The reporter gene is under the control of the human Epo promoter (hP) and the 3′ enhancer (hE) (Figure 3B, bottom). (C) Parental P19 cells and P19/Z cells were cultured in aggregates with or without 5 × 10−7 mol/L all-trans RA for 48 hours in 21% oxygen or were cultured for 28 hours in 21% oxygen and then for 20 hours in 2% oxygen. After 48 hours, the cells were stained with X-gal solution. (D) Parental Hep3B cells and Hep3B/Z cells were cultured with or without 5 × 10−7 mol/L all-trans RA for 24 hours in 21% or 2% oxygen. Magnifications are 25-fold.
We also examined RA-induced stimulation of Epo in aggregate cultures. We used human 0.2-kb promoter (hP) and human Epo 3′ enhancer (hE) in this setting and established permanent P19 cell lines (P19/Z) with the β-galactosidase reporter construct (Figure 3B, bottom). To amplify the activity of the 3′ enhancer, we ligated tandemly 4 3′ enhancers in the reporter construct. For comparison, Hep3B cell lines (Hep3B/Z) harboring the same reporter construct were made. Expression of β-galactosidase was visualized by staining the cells with X-gal. P19/Z cells were cultured in aggregates with or without all-trans RA under normoxia or hypoxia. P19/Z cells were positive for β-galactosidase only when cultured with RA (Figure 4C), and no positive cells were detected when the culture was done under hypoxia. In contrast, Hep3B/Z cells were positive only when cultured under hypoxia, whereas the RA treatment yielded no positive cells (Figure 4D). Taken together, these results clearly show that the Epo 3′ enhancer acts as a RARE in both monolayer and aggregate cultures of P19 cells and that mouse- and human-derived elements in promoter and 3′ enhancer regions are equivalent with respect to RA-induced as well as hypoxia-induced activation of Epo gene expression. Unresponsiveness of the reporter gene in P19 aggregates to hypoxia makes it very unlikely that higher stimulation of Epo production in aggregates than in monolayer cultures could be derived from the hypoxic induction of Epo production by aggregated cells.
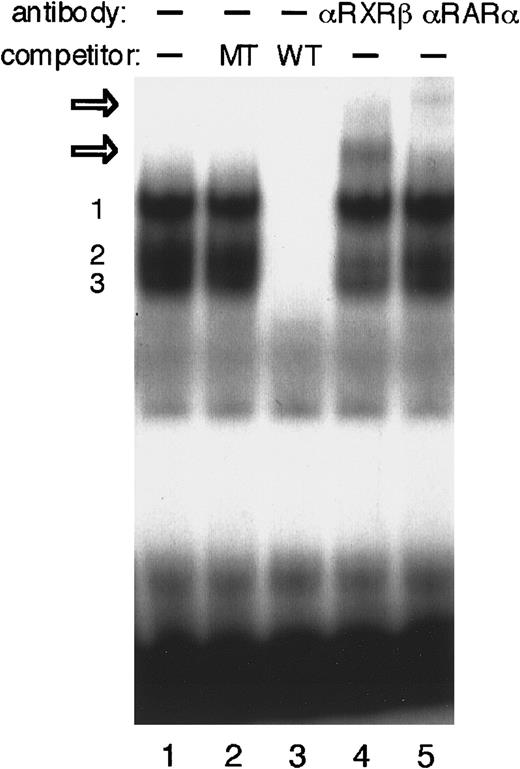
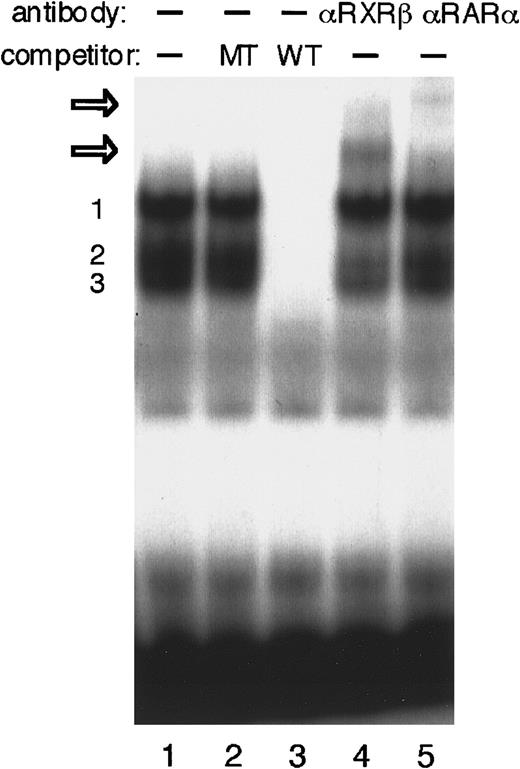
We next performed EMSAs to investigate whether the DR-2 element interacts with RA receptors in P19 cells. The DR-2 oligonucleotide probe bound to several proteins from the P19 nuclear extract (Figure5). Competition experiments with a pair of nonradiolabeled wild-type or mutant oligonucleotides showed that at least 3 specific protein–DNA complexes were formed. Both RA receptors, RAR and RXR, have 3 subtypes (α, β, and γ).46,47RARα and RXRβ have been shown to be the major subtypes in P19 cells.52 53 The addition of either anti-RARα or RXRβ antibody to the binding mixture resulted in the appearance of each further retarded complex, demonstrating that at least the heterodimer of RARα and RXRβ binds to the DR-2 element in P19 cells. Concomitantly, the second complex of 3 specific protein–probe complexes appears to be reduced.
EMSA to identify P19 nuclear proteins that interact with DR-2 in the Epo gene.
EMSA was performed using the DR-2 oligonucleotide probe. Nuclear extracts from P19 cells cultured with 5 × 10−7 mol/L all-trans RA for 20 hours were incubated with the radiolabeled wild-type DR-2 (DR-2 WT) probe in the absence of a competitor (lane 1) and in the presence of 100-fold molar excess of the unlabeled mutant DR-2 (DR-2 MT) probe (lane 2) or the unlabeled wild-type DR-2 (DR-2 WT) probe (lane 3). Three specific products (numbered 1, 2, and 3 on the left side) were found. Antibodies to RXRβ (lane 4) and RARα (lane 5) were incubated with nuclear extracts before the binding reactions were done. Two supershifted complexes appeared upon incubation with both antibodies (open arrows). Nuclear extracts from P19 cells cultured without RA produced the same results (not shown).
EMSA to identify P19 nuclear proteins that interact with DR-2 in the Epo gene.
EMSA was performed using the DR-2 oligonucleotide probe. Nuclear extracts from P19 cells cultured with 5 × 10−7 mol/L all-trans RA for 20 hours were incubated with the radiolabeled wild-type DR-2 (DR-2 WT) probe in the absence of a competitor (lane 1) and in the presence of 100-fold molar excess of the unlabeled mutant DR-2 (DR-2 MT) probe (lane 2) or the unlabeled wild-type DR-2 (DR-2 WT) probe (lane 3). Three specific products (numbered 1, 2, and 3 on the left side) were found. Antibodies to RXRβ (lane 4) and RARα (lane 5) were incubated with nuclear extracts before the binding reactions were done. Two supershifted complexes appeared upon incubation with both antibodies (open arrows). Nuclear extracts from P19 cells cultured without RA produced the same results (not shown).
Two segments, HIF-1 site and CATGG segment, in the 3′ enhancer are not required for the Epo gene to respond to RA
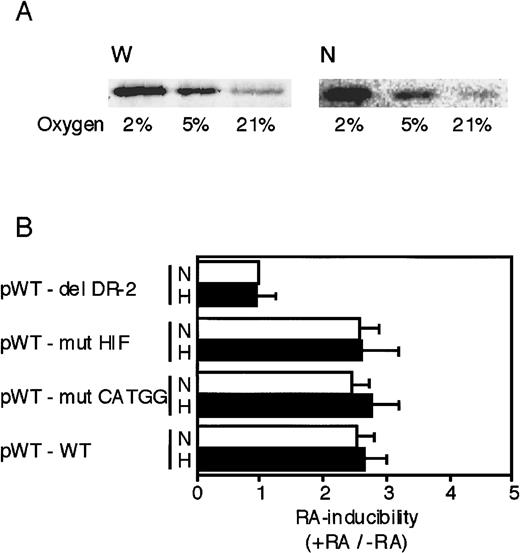
As described earlier, 3 segments—the HIF-1 site, the CATGG segment, and the DR-2 element—in the 3′ enhancer are required for the full hypoxic induction of Epo gene transcription.34-36This raises the possibility that in addition to the DR-2 element, the former 2 segments might also be important for the Epo gene to respond to RA. First, we examined whether P19 cells possess intact HIF-1α under hypoxic conditions by Western blot analysis. Whole-cell and nuclear extracts were prepared from P19 cells cultured in different oxygen concentrations. Hypoxia (2% and 5%) significantly increased HIF-1α protein in both the whole cell and the nucleus (Figure6A). Moreover, we confirmed that nuclear components derived from P19 cells cultured under hypoxia bound to the HIF-1 oligonucleotide probe but not to the mutant probe (data not shown). Second, we investigated the effects of mutations in the HIF-1 and CATGG segments on RA inducibility of the reporter gene. Each reporter plasmid containing the mutation in each site was constructed (Figure 3A, bottom) and transfected into P19 cells. Transfected cells were incubated with or without all-trans RA and also under normoxia or hypoxia to determine the effect of oxygen on RA inducibility. As shown in Figure 6B, neither the mutation in the HIF-1 binding site (pWT-mut HIF) nor the mutation in the CATGG segment (pWT-mut CATGG) affected the RA inducibility (3-fold), which was similar to that with the wild-type 3′ enhancer. The RA inducibility of the reporter gene expression was not influenced by oxygen concentration, which is consistent with the result that RA-induced accumulation of Epo mRNA is oxygen independent (Figure 2B). Thus, the 2 segments, HIF-1 and CATGG segment, in the 3′ enhancer do not work cooperatively with the DR-2 element for the RA induction of Epo gene expression in P19 cells.
The HIF-1 binding site and the middle segment (CATGG) in the 3′ enhancer are not involved in the activation by RA.
(A) Whole-cell extracts (W) and nuclear extracts (N) were prepared from P19 cells cultured in 21%, 5%, and 2% oxygen for 8 hours. The extracts were subjected to Western blot analysis using an anti–HIF-1 monoclonal antibody. (B) Transient expression of Luc reporter constructs in P19 cells was assayed. After transfection, the cells were cultured for 20 hours in 21% oxygen (N) or 2% oxygen (H) in the presence and absence of 5 × 10−7 mol/L all-trans RA. Characteristics of the plasmids are shown in Figure 3 and Table 1. The inducibility by RA (the ratio of Luc activity in the presence versus absence of RA) is indicated. The inducibility of the pWT–del DR-2 construct in 21% oxygen was defined as 1. Each value is the mean ± SD of triplicate experiments.
The HIF-1 binding site and the middle segment (CATGG) in the 3′ enhancer are not involved in the activation by RA.
(A) Whole-cell extracts (W) and nuclear extracts (N) were prepared from P19 cells cultured in 21%, 5%, and 2% oxygen for 8 hours. The extracts were subjected to Western blot analysis using an anti–HIF-1 monoclonal antibody. (B) Transient expression of Luc reporter constructs in P19 cells was assayed. After transfection, the cells were cultured for 20 hours in 21% oxygen (N) or 2% oxygen (H) in the presence and absence of 5 × 10−7 mol/L all-trans RA. Characteristics of the plasmids are shown in Figure 3 and Table 1. The inducibility by RA (the ratio of Luc activity in the presence versus absence of RA) is indicated. The inducibility of the pWT–del DR-2 construct in 21% oxygen was defined as 1. Each value is the mean ± SD of triplicate experiments.
Discussion
The 3′ enhancer responsible for the hypoxia-induced transcriptional activation of the Epo gene consists of 3 critical segments: the HIF-1 binding site, a middle segment with a function yet to be identified, and DR-2.34-36 Interaction of DR-2 with HNF-4 provides a marked hypoxia inducibility of Epo gene expression.41 One of the coactivators of gene transcription, p300/CBP,54-56 has been shown to interact with HIF-1 and HNF-4.57-59 Direct interaction between HIF-1 and HNF-4 may also be important for the hypoxic stimulation.60 These interactions are thought to form a complex on the 3′ enhancer for manifesting the full hypoxic inducibility, using p300/CBP as a scaffold.57-61
We previously showed that mouse ES and EC cells produced Epo, but their production did not respond to oxygen concentrations in cultures.45 The current study has extended this finding to understand the regulation of Epo production in embryonic cells. RA stimulated Epo production by P19 and F9 cells. The stimulation of Epo production correlated well with the accumulation of Epo mRNA, and actinomycin D inhibited the stimulatory effect of RA. EMSA demonstrated that nuclear extracts from P19 cells contained RAR and RXR that bound to DR-2. Thus, the RA-induced stimulation of Epo production in P19 cells is probably caused by transcriptional activation through binding of RA receptors to the DR-2 element in the 3′ enhancer. The requirement of p300/CBP for the transcriptional activation of target genes by RA has been amply documented.54 Reporter assays in P19 cells suggest that the HIF-1 binding site and CATGG segment in the 3′ enhancer are not required for RA inducibility. Formation of a complex between ligand-activated RA receptors and p300 on DR-2 may be sufficient for the Epo gene to respond to RA.
Although transcription of the Epo gene in P19 cells is not inducible by hypoxia, the LDHA gene, the hypoxic response of which requires HIF-1,22,23 is inducible in P19 cells,45 excluding the possibility that P19 cells lack HIF-1. This was confirmed by the current Western blot analysis (Figure6A). RA receptors expressed abundantly in P19 cells may compete with HNF-4 in the interaction with DR-2, abolishing the hypoxia inducibility of the Epo gene. This is unlikely, however, because the forced expression of functional HNF-4 conferred little hypoxic response on the Epo gene in P19 cells.45 Although the possibility that the HIF-1 region in the Epo gene is not open for HIF-1 to be accessible in P19 cells has not been completely excluded, it is noted that the HIF-1 site included in the reporter plasmids (pWT-WT in Figure 6B and P19/Z in Figure 4C) cannot exert its function in the hypoxic P19 cells. We speculate that the HIF-1/HNF-4/p300 complex on the 3′ enhancer and the RA receptors/p300 complex require different factors that link these complexes with the basal transcriptional machinery. P19 cells may be defective in a factor for the former complex.
Although further studies are clearly needed to understand the physiologic significance of RA-induced transcriptional activation of the Epo gene, RA may play a role in maintenance of the Epo level appropriate to the day-to-day production of red blood cells under normal, steady-state conditions, because RA increased the serum Epo level in vitamin A–depleted rats.44 Epo production in mouse yolk sacs is stimulated by RA,62 which may indicate that RA is also involved in the regulation of Epo production in an early stage of animal development. It thus appears that DR-2 is critical for the Epo gene to select the signals to respond in a tissue-specific or developmental stage–specific manner.
Supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan; and from Hokuto Foundation for Bioscience to T.K.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ryuzo Sasaki, Division of Integrated Life Science, Graduate School of Biostudies, Kyoto University, Kyoto 606-8502, Japan; e-mail: rsasaki@kais.kyoto-u.ac.jp.