The bacterium Porphyromonas gingivalis is a major etiologic agent in the pathogenesis of adult periodontitis in humans. Cysteine proteinases produced by this pathogen, termed gingipains, are considered to be important virulence factors. Among many other potentially deleterious activities, arginine-specific gingipains-R (RgpB and HRgpA) efficiently activate coagulation factors. To further expand knowledge of the interaction between gingipains and the clotting cascade, this study examined their effects on cellular components of the coagulation system. The enzymes induced an increase in intracellular calcium in human platelets at nanomolar concentrations and caused platelet aggregation with efficiency comparable to thrombin. Both effects were dependent on the proteolytic activity of the enzymes. Based on desensitization studies carried out with thrombin and peptide receptor agonists, and immunoinhibition experiments, gingipains-R appeared to be activating the protease-activated receptors, (PAR)-1 and -4, expressed on the surface of platelets. This was confirmed by the finding that HRgpA and RgpB potently activated PAR-1 and PAR-4 in transfected cells stably expressing these receptors. Cumulatively, the results indicate the existence of a novel pathway of host cell activation by bacterial proteinases through PAR cleavage. This mechanism not only represents a new trait in bacterial pathogenicity, but may also explain an emerging link between periodontitis and cardiovascular disease.
Introduction
Proteolytic modification of intracellular and extracellular proteins has recently been recognized as an important and common mechanism of regulating cell function.1,2Extracellularly, transformation of the cell surface through proteolysis plays a role in cell migration, wound healing, and tissue remodeling.3,4 In addition, a cohort of biologically active compounds, including cytokines and cytokine receptors, growth factors and growth factor receptors, cell surface adhesion molecules, Fc receptors, and G protein–coupled receptors, are known to be released from cell surfaces through proteolytic cleavage.5,6 Under physiologic conditions, it has been well established that enzymes belonging to a family of tightly regulated metalloproteases, which contain a disintegrin domain (ADAM family), mediate this ectodomain shedding.7 An alternative pathway that activates many cells operates through limited proteolysis of cell surface receptors referred to as protease-activated receptors (PARs).8 Although both pathways are tightly controlled, during bacterial infections the surface proteins and receptors may become a target for nonhost proteases, which generally escape any control by host inhibitors. Indeed, growing evidence indicates that bacterial proteases can modify the host cell surface, and in this manner, contribute significantly to microbial pathogenicity.9-17
Periodontitis, the chronic infectious disease in which uncontrolled proteolytic activity derived from both host and bacteria plays a significant role in the destruction of tooth-supporting tissues, including the alveolar bone, is the most prevalent inflammatory disease in the world. Approximately 15% of the population suffers from its most severe form and, if untreated, it may result not only in tooth loss, but also in systemic complications.18-21 The major pathogens associated with periodontitis are Porphyromonas gingivalis, Bacteroides forsythus, and Actinobacillus actinomycetemcomitans.22 Among these, P gingivalis, a gram-negative anaerobic rod, has been identified as a major etiologic agent in the pathogenesis of adult periodontitis in humans.22,23 It has also been recognized as a virulence agent that initiates the progression of periodontitis in primate and rodent models of periodontal destruction.24
Recently, a number of epidemiologic studies have linked periodontal disease to heart disease.25-29 For this reason, we have investigated molecular mechanisms that may connect the 2 pathologies, particularly with regard to the effects of proteases from P gingivalis on cellular components of the coagulation system. Gram-negative periodontal pathogens often find their way into the bloodstream (bacteremia) in patients with periodontal disease as a result of oral hygiene procedures or even chewing.30-33Furthermore, P gingivalis has not only been found in the circulation, but has been found to infect atherosclerotic plaques also.34 35
Proteases produced by P gingivalis have been shown to act as important pathogenic agents.36 Two types of cysteine proteases, responsible for the so-called trypsin-like activity of the bacterium, have been purified37,38: a lysine-specific protease, 105-kd, termed lys-gingipain (Kgp) and an arginine-specific protease referred to as arg-gingipain. The latter is present as 3 variants: 50-kd RgpAcat, 50-kd RgpB, and 95-kd HRgpA. HRgpA is the high molecular mass form of RgpAcat, formed by RgpAcat noncovalently complexed with 44-kd binding proteins that have been identified as hemagglutinins or adhesins.37,39 Curtis and colleagues40 showed that a protease preparation, termed protease I, from P gingivalis strain W83 was able to induce platelet activation, but the biochemical nature of the enzyme and the mechanism of its action were not elucidated. The questions addressed in this study are whether the highly characterized proteases RgpB and HRgpA, which are solely responsible for the hydrolysis of peptide bonds after arginine residues in P gingivalis, can elicit a cellular response in platelets that is mediated by interaction of the enzymes with platelet surface molecules.
Platelets express certain members of the PAR family on their surface that are activated by cleavage of their N-terminus by a protease, then couple to G proteins and induce cellular signals. To date, 4 PARs have been identified: PARs-1, -2, -3, and -4. The new N-terminus that is formed after receptor cleavage acts as a tethered ligand that binds to the receptor and leads to its activation. Synthetic peptide agonists corresponding to the tethered ligand of each receptor are able to activate the receptor, with the exception of PAR-3, allowing the cellular responses mediated by the different receptors to be distinguished and studied by treatment of cells with their respective agonist peptides. PAR-1 was the first receptor found and is cleaved and activated by thrombin,41 as are PAR-3 and PAR-4, the 2 thrombin receptors discovered more recently.42-44 Human platelets express PAR-1,41,45 and its agonist peptide causes activation and aggregation.41,46,47 PAR-4 is also expressed in human platelets,43,44,48 although at a lower level than that of PAR-1,48 and a cognate agonist peptide was found to induce aggregation of human platelets.48 The possible interactions of RgpB and HRgpA with the PARs expressed on the surface of platelets were investigated in this study and it was conclusively demonstrated that the enzymes are potent agonists for both PAR-1 and PAR-4, mediating platelet activation and aggregation via these receptors. This constitutes the first report of bacterial proteases demonstrably acting through these receptors to cause cellular processes that may underlie reported associations between periodontitis and cardiovascular disease.
Materials and methods
Materials
Fura-2 am was obtained from Molecular Probes (Eugene, OR). All tissue culture reagents were purchased from Gibco Life Technologies (Melbourne, Australia). TRAP (SFFLRN) and TRAP-4 (GYPGQV) (single-letter amino acid codes) peptides were synthesized by Auspep (Parkville, Australia). The rabbit anti–PAR-1 antibody was produced as previously described.49 All other materials were purchased from Sigma (Sydney, Australia).
Tissue culture
N1LF cells are immortalized murine lung myofibroblasts derived from PAR-1–deficient mice that lack functional PAR-1, PAR-2, and PAR-4.50 These cells were grown in Dulbecco modified Eagle medium (DMEM) containing glucose, 4 mM l-glutamine and 10% (vol/vol) heat-inactivated fetal calf serum (FCS), supplemented with penicillin (100 U/mL) and streptomycin sulfate (100 μg/mL). N1LF PAR-1 and N1LF PAR-4 cells stably express human PAR-1 and PAR-4, respectively,51 and were grown in the same medium as the N1LF cells with the addition of 200 μg/mL hygromycin B.
Purification of platelets from human blood
Platelets were isolated from freshly drawn human blood. Venous blood was anticoagulated by adding 6 volumes of blood to 1 volume of acid-citrate-dextrose (85 mM sodium citrate, 111 mM dextrose, and 71 mM citric acid supplemented with 50 ng/mL prostaglandin I2[PGI2] and apyrase at 0.67 U/mL). Whole blood was centrifuged at 200g for 20 minutes at room temperature to obtain the supernatant platelet-rich plasma, which was then centrifuged at 730g for 10 minutes at room temperature to sediment the platelets. The platelet pellets were resuspended in 13 mM trisodium citrate, 120 mM NaCl, and 30 mM dextrose, pH 7.0, containing 50 ng/mL PGI2, and washed twice. Platelets were resuspended in extracellular medium [EM] (12 mM NaHCO3, 138 mM NaCl, 5.5 mM glucose, 2.9 mM KCl, 10 mM Hepes, 1 mM MgCl2, pH 7.4) or Tyrode buffer (140 mM NaCl, 2.7 mM KCl, 12 mM NaHCO3, 0.76 mM Na2HPO4, 5.5 mM dextrose, 5 mM Hepes, 2 mg/mL bovine serum albumin [BSA], pH 7.4), depending on the assay performed.
Intracellular calcium measurement
Intracellular calcium levels were measured in N1LF, N1LF PAR-1, N1LF PAR-4, and platelets. N1LF, N1LF PAR-1, and N1LF PAR-4 cells were grown to 80% confluence and detached from the culture dishes by treatment with nonenzymatic dissociation solution (Sigma). All cell types were prepared for [Ca++]i measurements as described previously.52 Cells were washed and resuspended at 6 × 106 cells/mL in an EM depending on the cell type. The EM for N1LF, N1LF PAR-1, and N1LF PAR-4 consisted of 121 mM NaCl, 5.4 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 6 mM NaHCO3, 5.5 mM glucose, 25 mM Hepes, 0.1% (wt/vol) BSA, pH 7.3; the EM for platelets was as described above. In all subsequent steps the cells were protected from light.
Cells were loaded with 1 μM Fura-2 am by occasional shaking for 30 minutes at room temperature. After centrifugation at 200g for 5 minutes, they were resuspended in EM and occasionally shaken for 30 minutes at room temperature to allow hydrolysis of the intracellular Fura-2 am and then centrifuged (200g for 5 minutes). N1LF, N1LF PAR-1, and N1LF PAR-4 cells were resuspended in EM without BSA at 2 × 106 cells/mL for fluorescence measurements and platelets were resuspended in EM containing 1 mM CaCl2. [Ca2+]i was determined using a PerkinElmer LS-50 fluorometer by measuring Fura-2 fluorescence at excitation and emission wavelengths of 340:380 and 510 nm, respectively. Loaded cells were maintained at 37°C in stirred plastic cuvettes throughout the experiment. After a stable baseline was established, the agonist was added to cells and the ratio of fluorescence at the 2 excitation wavelengths was measured, which is proportional to [Ca2+]i.
Platelet aggregation studies
Platelets were collected from healthy volunteers who had not taken antiplatelet medication for 2 weeks in acid-citrate-dextrose, 6:1 vol/vol, containing 90 mM sodium citrate, 7 mM citric acid, 140 mM dextrose, pH 4.6, supplemented with 70 mM theophylline. Platelet-rich plasma (PRP) was obtained by centrifugation of whole blood at 180g for 15 minutes. The PRP was centrifuged at 2000g for 15 minutes and the platelets washed twice with buffer containing 4.3 mM Na2HPO4, 4.3 mM K2HPO4, 24.3 mM NaH2PO4, 113 mM NaCl, 5.5 mM glucose, 5 mg/mL BSA, 10 mM theophylline, pH 6.5. The final platelet preparation was resuspended in a modified Tyrode buffer, 12 mM NaHCO3, 0.32 mM NaH2PO4, 10 mM Hepes, 137 mM NaCl, 2.7 mM KCl, 0.5 mM MgCl2, and 5.5 mM glucose, pH 7.5. Platelet aggregation was measured using a 4-channel automated platelet analyzer set to 950 rpm at 37°C. Each reaction mixture (400 μL) contained washed platelets (3 × 108/mL), the indicated concentrations of HRgpA, RgpB, or thrombin (Figure 5), no exogenous fibrinogen, and 1 mM CaCl2. The rate and extent of platelet aggregation were monitored by the percentage of light transmission and presented as aggregation tracings.
Purification and assay of the bacterial proteases
HRgpA and RgpB were purified to homogeneity37,38and the amount of active enzyme determined by active site titration with F-F-R-chloromethylketone,53 all as described previously. The active site cysteine residue of the gingipains was reduced (activated) for cellular studies with 10 mM cysteine at 37°C for 10 minutes in 0.1 M Tris-HCl, 5 mM CaCl2, pH 7.4. Polymyxin B-S04 (100 μg/mL) was routinely added to inhibit any cellular stimulation by bacterial lipopolysaccharides. Gingipains were inactivated by treatment with 100 μM leupeptin or 2 μM antipain for 10 minutes.
Results
HRgpA and RgpB increase intracellular calcium levels [Ca++]i in platelets
To determine whether the gingipains were able to interact with platelet receptors and induce intracellular signals, platelets from 9 donors were isolated and individually tested for a [Ca++]i response to HRgpA, RgpB, and thrombin. As may be seen in Table 1, most donors responded similarly to HRgpA and RgpB in terms of the magnitude of the response to a defined concentration of the bacterial enzymes. HRgpA consistently induced a higher level of [Ca++]i response than the same concentration of thrombin, whereas 10-fold more RgpB consistently gave responses that were considerably lower than thrombin. HRgpA and RgpB induced a dose-dependent increase in [Ca++]i, indicated by the dose responses shown for 2 donors in comparison to thrombin (Figure 1). These data yield an enzyme concentration inducing the half maximal response (EC50) of 2.4 nM and 0.18 nM for thrombin and HRgpA, respectively, in one donor and 0.6 nM and 63 nM for HRgpA and RgpB, respectively, in a second donor. This further indicates that HRgpA is consistently a more potent platelet agonist than thrombin, whereas RgpB is much less potent than the other 2 enzymes. As indicated by the data in Table 1, the magnitude of the [Ca++]i response for thrombin, HRgpA, and RgpB varied to only a moderate extent between donors. HRgpA and RgpB, which had been inactivated by leupeptin, did not induce calcium responses in the platelets (Figure 2A,C), indicating that the [Ca++]i increase induced is due to the proteolytic activity of the enzymes.
The [Ca++]i response of platelets to different concentrations of HRgpA, RgpB, and thrombin.
Platelets from 2 donors were loaded with Fura-2 and stimulated with HRgpA, RgpB, and thrombin. The responses in one donor to HRgpA (A) and thrombin (B) and in another donor to RgpB (C) and HRgpA (D) are shown. The [Ca++]i increases at different concentrations represent the mean from 3 traces similar to those shown in Figure 2, yielding a curve that could be fitted by an equation for single site binding and generate an EC50 value with associated estimate of the SE.
The [Ca++]i response of platelets to different concentrations of HRgpA, RgpB, and thrombin.
Platelets from 2 donors were loaded with Fura-2 and stimulated with HRgpA, RgpB, and thrombin. The responses in one donor to HRgpA (A) and thrombin (B) and in another donor to RgpB (C) and HRgpA (D) are shown. The [Ca++]i increases at different concentrations represent the mean from 3 traces similar to those shown in Figure 2, yielding a curve that could be fitted by an equation for single site binding and generate an EC50 value with associated estimate of the SE.
Calcium responses in platelets are due to the proteolytic activity of the enzymes and can be desensitized with thrombin.
The [Ca++]i responses in platelets to: (A) 0.5 nM HRgpA inactivated with leupeptin; (B) 0.5 nM HRgpA, both followed by the addition of 2 μM antipain, 50 nM thrombin, and treatment with 60 μM ADP; (C) 25 nM RgpB inactivated with leupeptin, followed by 10 nM thrombin; (D) 25 nM RgpB, followed by 10 nM thrombin. The [Ca++]i responses shown are those for 1 of the 4 donors in which these effects were observed. Antipain (2 μM) was added in between the HRgpA and thrombin to ensure that the cysteine protease was not desensitizing responses by cleaving thrombin.
Calcium responses in platelets are due to the proteolytic activity of the enzymes and can be desensitized with thrombin.
The [Ca++]i responses in platelets to: (A) 0.5 nM HRgpA inactivated with leupeptin; (B) 0.5 nM HRgpA, both followed by the addition of 2 μM antipain, 50 nM thrombin, and treatment with 60 μM ADP; (C) 25 nM RgpB inactivated with leupeptin, followed by 10 nM thrombin; (D) 25 nM RgpB, followed by 10 nM thrombin. The [Ca++]i responses shown are those for 1 of the 4 donors in which these effects were observed. Antipain (2 μM) was added in between the HRgpA and thrombin to ensure that the cysteine protease was not desensitizing responses by cleaving thrombin.
The question that was then addressed was whether the increase in [Ca++]i induced by HRgpA and RgpB activity is due to cleavage of a PAR on the surface of platelets. Once a PAR is cleaved by a protease it is unable to be activated a second time by the same or another protease in a short period of time. Because cleavage of a PAR desensitizes [Ca++]i responses to subsequent protease challenges, desensitization of [Ca++]i responses by HRgpA, RgpB, and thrombin were examined. When platelets were initially activated with HRgpA, a secondary response to this enzyme was virtually eliminated (data not shown). Exposure of platelets to thrombin diminished the [Ca++]i response to a second challenge with HRgpA (Figure 3A). Platelets treated with thrombin or HRgpA still responded to a subsequent challenge with adenosine diphosphate (ADP) (Figure 2A,B), indicating that the cells could still respond to an agonist via calcium signaling pathways. The higher the thrombin concentration added to platelets, the smaller was the subsequent HRgpA response (Figure 3A). Similarly, treatment of platelets with HRgpA desensitized the response to a second challenge by thrombin (Figure 3B) and increasing concentrations of HRgpA proportionately decreased subsequent thrombin responses (Figure 3B). Similarly, when platelets were initially exposed to RgpB, a secondary response to this protease or thrombin was abolished (Figure 2D) and pretreatment with thrombin desensitized the cells to a subsequent challenge with RgpB (data not shown). It was shown that the desensitization of the responses was not due to the enzymes inactivating each other. This is illustrated, for instance, by the finding that adding 2 μM antipain (a concentration of inhibitor known to inhibit HRgpA effectively without affecting thrombin) to the cell suspension after the initial activation with HRgpA did not affect the desensitization obtained with thrombin (Figure 2A,B). The addition of the inhibitor after initially adding inactivated HRgpA also did not affect subsequent thrombin responses. These findings could be extended to all systems tested, strongly indicating that the desensitization of the platelets obtained was purely reflective of prior receptor cleavage.
Effects of desensitization of [Ca++]i responses in platelets.
Cells were exposed to: (A) 1 nM, 25 nM, or 100 nM thrombin followed by 0.5 nM HRgpA; (B) 0.05 nM, 0.1 nM, or 1 nM HRgpA followed by 42 nM thrombin. The results shown represent the desensitization effects for 1 of the 4 donors in which these effects were observed.
Effects of desensitization of [Ca++]i responses in platelets.
Cells were exposed to: (A) 1 nM, 25 nM, or 100 nM thrombin followed by 0.5 nM HRgpA; (B) 0.05 nM, 0.1 nM, or 1 nM HRgpA followed by 42 nM thrombin. The results shown represent the desensitization effects for 1 of the 4 donors in which these effects were observed.
These results suggest that HRgpA, RgpB, and thrombin activate common receptor(s) on platelets. Thrombin activates PAR-1 and PAR-4 on the surface of human platelets by cleaving after an arginine residue in their extracellular domain. Because HRgpA and RgpB have high specificity of cleavage, limited to peptide bonds after arginine residues, the calcium response observed is probably due to activation of PAR-1 and PAR-4 by these bacterial enzymes. To investigate whether the calcium increase elicited by RgpB and HRgpA is mediated by cleavage of PAR-1, platelets were incubated with rabbit anti–PAR-1 antibody49 or a control rabbit antibody (the latter serving as a control) for 15 minutes, prior to treatment with RgpB or HRgpA. Preincubation of platelets with anti–PAR-1 antibody significantly inhibited thrombin and gingipain-induced calcium increases, in comparison to pretreatment with a control antibody (Figure 4A-D).
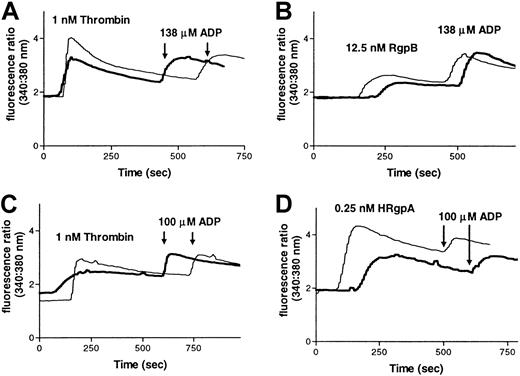
Partial inhibition of gingipain-induced calcium changes in platelets by an anti–PAR-1 antibody.
Calcium mobilization in platelets induced by RgpB or HRgpA in the presence of rabbit anti–PAR-1 antibody (bold line) or a control rabbit antibody (thin line). Platelets from one donor were incubated with the antibodies (30 μg/mL) for 15 minutes, before treatment with (A) 1 nM thrombin or (B) 12.5 nM RgpB, both followed by treatment with 138 μM ADP. Platelets from another donor were incubated with the antibodies (60 μg/mL) for 15 minutes, and then treated with (C) 1 nM thrombin or (D) 0.25 nM HRgpA, both followed by treatment with 100 μM ADP. These data are representative of 3 experiments.
Partial inhibition of gingipain-induced calcium changes in platelets by an anti–PAR-1 antibody.
Calcium mobilization in platelets induced by RgpB or HRgpA in the presence of rabbit anti–PAR-1 antibody (bold line) or a control rabbit antibody (thin line). Platelets from one donor were incubated with the antibodies (30 μg/mL) for 15 minutes, before treatment with (A) 1 nM thrombin or (B) 12.5 nM RgpB, both followed by treatment with 138 μM ADP. Platelets from another donor were incubated with the antibodies (60 μg/mL) for 15 minutes, and then treated with (C) 1 nM thrombin or (D) 0.25 nM HRgpA, both followed by treatment with 100 μM ADP. These data are representative of 3 experiments.
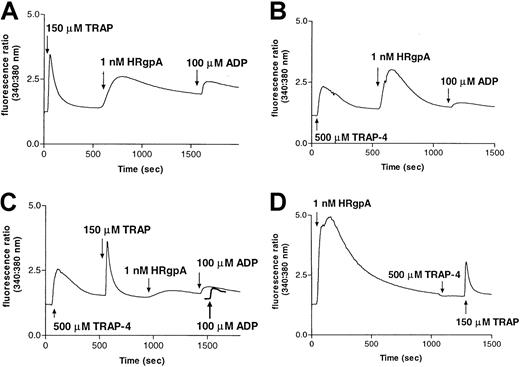
Once PARs have been activated by their cognate peptide agonist peptides, they may be rendered desensitized to subsequent exposure to protease agonists, in much the same way as the desensitization described above for treatment with sequential doses of different proteases.54 Because the agonist peptides for PAR-1 (TRAP-SFRLLN) and PAR-4 (TRAP-4–GYPGQV) are specific for their cognate receptors in the platelet context, pretreatment with these peptides may desensitize the cells to subsequent treatment with protease agonists targeting the receptors. As may be seen visually in Figure5, panels A-C, and quantitatively in Table 2, pretreatment of platelets from 3 donors with either TRAP or TRAP-4, individually, markedly reduced the [Ca++]i response to 1 nM HRgpA, whereas treatment with a combination of the peptides all but abolished the response to HRgpA. Pretreatment of the platelets with HRgpA (Figure 5D) abolished subsequent response to PAR-4–activating peptide and markedly reduced the response to TRAP. Essentially similar results were found for thrombin and RgpB (data not shown), demonstrating that the bacterial proteases most likely induce [Ca++]i responses in platelets by activating both PAR-1 and PAR-4 receptors.
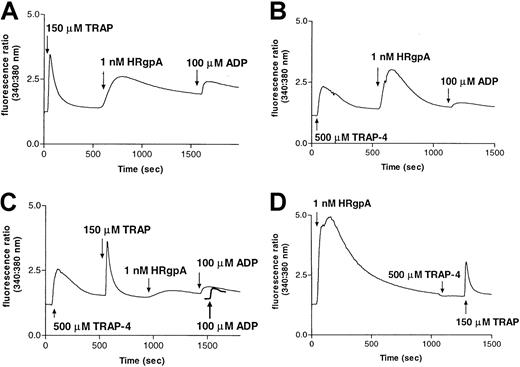
Desensitization of responses to HRgpA by prior treatment of platelets with PAR-1 and PAR-4 agonist peptides.
Calcium responses to 1 nM HRgpA and 100 μM ADP following treatment with: (A) 150 μM TRAP; (B) 500 μM TRAP-4; or (C) 500 μM TRAP-4 and 150 μM TRAP. The ADP treatment was used in each case to demonstrate that prior treatments had not significantly depleted calcium stores, so that essentially normal responses could be obtained with a noninteracting agonist. In panel C, the response to ADP prior to any additions to platelets is shown as a bold line for comparison. Pretreatment with HRgpA also desensitized responses to TRAP-4 and TRAP (D).
Desensitization of responses to HRgpA by prior treatment of platelets with PAR-1 and PAR-4 agonist peptides.
Calcium responses to 1 nM HRgpA and 100 μM ADP following treatment with: (A) 150 μM TRAP; (B) 500 μM TRAP-4; or (C) 500 μM TRAP-4 and 150 μM TRAP. The ADP treatment was used in each case to demonstrate that prior treatments had not significantly depleted calcium stores, so that essentially normal responses could be obtained with a noninteracting agonist. In panel C, the response to ADP prior to any additions to platelets is shown as a bold line for comparison. Pretreatment with HRgpA also desensitized responses to TRAP-4 and TRAP (D).
HRgpA and RgpB induce platelet aggregation
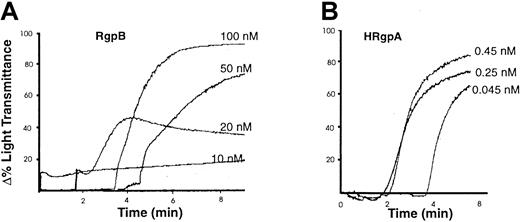
Calcium plays a key role in triggering platelet activation and is the single most important intracellular mediator of cell function. To determine whether the [Ca++]i elevation induced by the gingipains causes platelet activation, aggregation responses of these cells were investigated. Both HRgpA and RgpB induced aggregation of platelets from 6 different donors. The aggregation caused by different concentrations of HRgpA and RgpB in one preparation is shown in Figure 6, panels A and B. A consistent finding was that much higher concentrations of RgpB were required to cause the same degree of aggregation as that induced by HRgpA. To determine whether the platelet aggregation caused by the bacterial proteases was due to their proteolytic activity, the enzymes were inactivated with 100 μM leupeptin. This resulted in total inhibition of the aggregation induced by both gingipains (data not shown), indicating that the cellular effect was due to proteolysis.
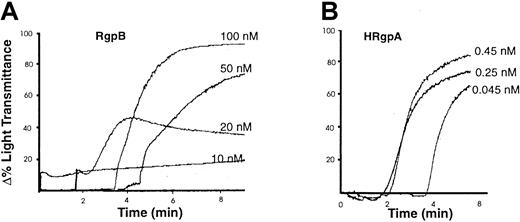
Platelet aggregation by the gingipains.
Human platelets were isolated and aggregation was induced by the indicated concentrations of (A) RgpB and (B) HRgpA (as described in “Materials and methods”). The aggregation traces presented are from one experiment, representative of 3 total.
Platelet aggregation by the gingipains.
Human platelets were isolated and aggregation was induced by the indicated concentrations of (A) RgpB and (B) HRgpA (as described in “Materials and methods”). The aggregation traces presented are from one experiment, representative of 3 total.
Agglutination of red blood cells by P gingivalis and adherence to other bacteria are thought to be mediated at least in part via the proteases of this organism.55 56 To show that the measure of aggregation observed (increase in the light transmittance level) was reflective of true platelet activation and is thus genuine aggregation, rather than an agglutination phenomenon, an inhibitor of platelet activation was used. Preincubation of platelets with 100 ng/mL PGI2 or 10 μM forskolin (inhibitors of platelet activation) at 37°C for 15 minutes completely inhibited the aggregation induced by 0.25 nM HRgpA and 20 nM RgpB (data not shown), verifying that HRgpA and RgpB indeed cause true platelet aggregation.
HRgpA and RgpB activate cells stably expressing the PAR-4 receptor
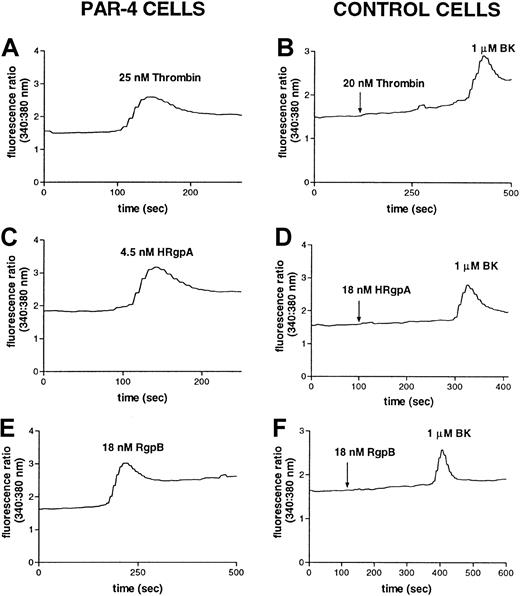
To investigate whether HRgpA and RgpB can cleave and activate PAR-4, the calcium response elicited by these 2 enzymes was studied in transfected N1LF cells stably expressing human PAR-4 (N1LF PAR-4) in comparison to nontransfected N1LF cells. Treatment of N1LF PAR-4 cells with 25 nM thrombin (Figure 7A) or 500 μM PAR-4 activating peptide (TRAP-4) (data not shown) elicited an intracellular Ca++ increase, whereas nontransfected N1LF cells did not respond to 500 μM TRAP-4 (data not shown) or 20 nM thrombin (Figure 7B). The calcium responses induced in N1LF PAR-4 cells by 4.5 nM HRgpA and 18 nM RgpB are shown in Figure 7, panels C and E, compared to the lack of response in nontransfected N1LF cells (Figure7D,F). Concentrations of HRgpA as low as 0.45 nM elicited a Ca++ increase in N1LF PAR-4 cells, whereas concentrations as high as 100 nM of HRgpA did not cause calcium mobilization in nontransfected N1LF cells (data not shown). Thus both HRgpA and RgpB specifically induced an increase in [Ca++]iin N1LF PAR-4 cells, verifying that these enzymes can activate PAR-4. The concentration dependence of such an increase is shown in Figure8, yielding an enzyme concentration inducing half the maximal response (EC50) of 1.7 nM, 4.6 nM, and 10 nM for receptor activation by HRgpA, RgpB, and thrombin, respectively.
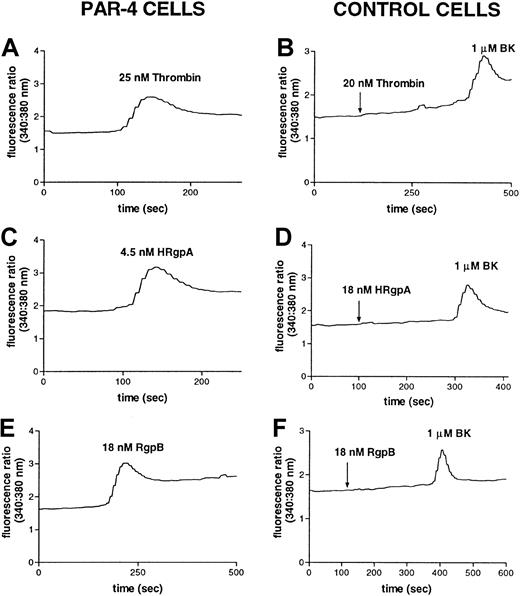
The gingipains induce responses in PAR-4–transfected cells.
Increase in intracellular calcium levels in N1LF PAR-4 cells following treatment with (A) 25 nM thrombin, (C) 4.5 nM HRgpA, or (E) 18 nM RgpB. N1LF cells were treated with (B) 25 nM thrombin, (D) 18 nM HRgpA, or (F) 18 nM RgpB, followed by treatment with 1 μM bradykinin. The traces are representative of 3 observed.
The gingipains induce responses in PAR-4–transfected cells.
Increase in intracellular calcium levels in N1LF PAR-4 cells following treatment with (A) 25 nM thrombin, (C) 4.5 nM HRgpA, or (E) 18 nM RgpB. N1LF cells were treated with (B) 25 nM thrombin, (D) 18 nM HRgpA, or (F) 18 nM RgpB, followed by treatment with 1 μM bradykinin. The traces are representative of 3 observed.
The concentration dependence of gingipain-induced changes in PAR-4–transfected cells.
The [Ca++]i response of N1LF PAR-4 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.
The concentration dependence of gingipain-induced changes in PAR-4–transfected cells.
The [Ca++]i response of N1LF PAR-4 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.
HRgpA and RgpB activate cells stably expressing the human PAR-1 receptor
To more definitively determine whether the gingipains can cleave and activate human PAR-1, the calcium response elicited by the 2 enzymes was investigated in transfected N1LF cells stably expressing human PAR-1 (N1LF PAR-1) in comparison to nontransfected N1LF cells. Treatment of N1LF PAR-1 cells with 150 μM TRAP or 20 nM thrombin elicited an intracellular Ca++ increase, whereas nontransfected N1LF cells did not respond to TRAP or to 20 nM thrombin (data not shown). Concentrations of HRgpA or RgpB as low as 2 nM elicited a Ca++ increase in N1LF PAR-1 cells, whereas concentrations of the enzymes as high as 100 nM did not cause calcium mobilization in nontransfected N1LF cells (data not shown). The nontransfected N1LF cells did respond to bradykinin, showing that G-protein–coupled receptor signaling pathways were intact. Thus, HRgpA and RgpB activate the PAR-1 receptor. The concentration dependence of the increase in [Ca++]i in N1LF PAR-1 cells induced by the different enzymes is shown in Figure9, yielding an enzyme concentration inducing half the maximal response (EC50) of 17 nM, 48 nM, and 0.26 nM for receptor activation by HRgpA, RgpB, and thrombin, respectively.
The concentration dependence of gingipain-induced changes in PAR-1 transfected cells.
The [Ca++]i response of N1LF PAR-1 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.
The concentration dependence of gingipain-induced changes in PAR-1 transfected cells.
The [Ca++]i response of N1LF PAR-1 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.
Discussion
An emerging theme in the interaction between pathogenic bacteria and the host is the ability of the microbial invader to proteolytically modify the host cell surface proteins, including various receptors. In most cases, bacterial proteases degrade receptors,9,12,13,17 or release soluble ectodomains10 11 leading to desensitization of cellular responses to their physiologic agonist. Such treatment of cells of the immune system may potentially affect antimicrobial defense mechanisms or interfere with the regulation of the inflammatory reaction. These effects can be even more deleterious if receptors such as the PARs are the target of bacterial proteases, because cleavage of these receptors will lead to uncontrolled activation of host cells, which, in the case of platelets, is normally mediated by tightly regulated thrombin cleavage of PAR-1 and PAR-4. However, despite the fact that unchecked platelet activation may have serious pathologic consequences if bacteria possessing such ability find their way into the bloodstream, the interaction between platelets and proteases from pathogenic bacteria has never been systematically investigated.
An example of bacteria that are known to escape into the bloodstream on a regular basis are the members of microbial flora forming dental plaque. This ability correlates with the severity of periodontitis when even such simple activities as chewing, flossing, and brushing cause a transient bacteremia.31-33 Fortunately, most of the bacteria are benign oral streptococci, which can cause a serious medical problem (endocarditis), only in subjects with damaged or implanted heart valves.57 In the case of severe periodontitis, however, P gingivalis can also find its way into the circulation and even infect atherosclerotic plaques.34 35 This prompted us to closely investigate the effect of arginine-specific gingipains on platelets.
HRgpA and RgpB induce an increase in [Ca++]iin human platelets, which is dependent on their proteolytic activity. Several lines of evidence strongly support the hypothesis that this increase is mediated by activation of PAR-1 and PAR-4. Exposure of human platelets to either of the gingipains desensitized the [Ca++]i response to a second challenge with the same enzyme, a phenomenon that is in agreement with the rapid desensitization that protease-activated receptors undergo after a short period of activation with a protease. Desensitization studies carried out with thrombin showed that when human platelets were activated with this enzyme, a secondary challenge with HRgpA or RgpB was virtually eliminated. Similarly, pretreatment of platelets with the bacterial enzymes desensitized the response to a second challenge with thrombin, suggesting that both enzymes activate the platelet thrombin receptors. The desensitization of the platelets to a subsequent challenge with another protease was not due to inactivation of the calcium signaling pathways, as demonstrated by the ability of the platelets to still respond to a maximal concentration of ADP.58 Antibodies specific for the sequence that occurs immediately amino-terminal to the cleavage site of PAR-1 were able to decrease the [Ca++]i increase caused by HRgpA or RgpB by approximately 50%. This finding supports the proposed mechanism that [Ca++]i mobilization by these proteases is in part mediated by cleavage of PAR-1. Further investigation of the receptors activated by the bacterial proteases used the individual agonist peptides for PAR-1 and PAR-4 to desensitize a subsequent response to the gingipains. For each of the enzymes, it could be shown that prior exposure of the platelets to a combination of the PAR-1 and PAR-4 agonist peptides almost completely abolished the response to the protease. This provides strong evidence that the bacterial proteases induce responses in the platelet by cleaving and activatingboth PAR-1 and PAR-4.
Thrombin activates PAR-1 and PAR-4 expressed on the surface of platelets by cleaving after a specific arginine residue at their respective activation sites. HRgpA and RgpB, which are absolutely specific for hydrolysis at R-X sites, were found to activate both PAR-1 and PAR-4 in transfected cells stably expressing these receptors. HRgpA cleaved the PAR-4 receptor approximately 6 times more efficiently than thrombin, based on evaluation of the enzyme concentration inducing half the maximal response (EC50) in PAR-4–transfected cells, whereas RgpB was approximately 2-fold more efficient. Thrombin was a much more efficient activator of PAR-1, however, activating the receptor 60 times more efficiently than HRgpA and 160 times more efficiently than RgpB. The greater efficiency of thrombin for cleavage of PAR-1 is expected in light of the specific association of the protease with a hirudin-like sequence that binds its fibrinogen-binding exosite.59,60 PAR-4 does not have a hirudin-like domain and this accounts for its less effective cleavage by thrombin compared to PAR-1, as seen in the present study and described elsewhere.43 44 The gingipains would also not be expected to cleave PAR-1 very efficiently in comparison to thrombin because they presumably have no mechanism for additional interactions with the receptor that are analogous to thrombin.
HRgpA exhibited higher efficiency in activating both PAR-1 and PAR-4 compared to RgpB. It was previously found that the proteolytic activity of HRgpA was 5 times higher than that of RgpB, despite the fact that both enzymes are equally active on synthetic substrates.38 Similarly, HRgpA has been found to have higher activity than RgpB in activating factor X.61 Both HRgpA and RgpB were more efficient at cleaving PAR-4 compared to PAR-1. Although these enzymes are highly specific for cleavage after arginine residues, less is known about the effect of amino acid residues around the cleavage site on the catalytic potency of HRgpA and RgpB. Investigations using synthetic substrates found HRgpA and RgpB to have similar preferences and no clear preference was observed for particular amino acid residues at the P2 or P3 position (P3-P2-R).38Elucidation of the crystal structure of RgpB revealed that, with the exception of the entrance hole to the S1 pocket, which is optimized to accommodate arginine side chains, the molecular surface around the active site of the enzyme is relatively flat, with a negative electrostatic potential.62 It is thought that this open binding site and strong binding of the arginine residue enables RgpB to cleave a multitude of R-X bonds in proteins and peptides. HRgpA is comprised of RgpA noncovalently complexed with adhesins. Because the structure of HRgpA has not been elucidated, the role of adhesins in the structure and activity of the enzyme is not known. It may be postulated that the adhesin subunits could affect the interaction of the protease with the surface of the cell by serving as an anchor and stabilizing this interaction. More stable interaction of the enzyme with the cell surface may contribute to more efficient cleavage of protease-activated receptors.
Based on the desensitization studies and the ability of HRgpA and RgpB to activate PAR-1 and PAR-4, it can be concluded that the bacterial proteases activate these receptors on the surface of platelets, resulting in calcium mobilization. HRgpA caused a [Ca++]i increase in platelets and aggregation at much lower concentrations than RgpB. This would be expected in view of the fact that HRgpA not only showed much higher efficacy than RgpB in activating PAR-1 and PAR-4 in cells stably expressing these receptors, but was also a more potent activator of PAR-4 than thrombin. In human platelets, PAR-4 messenger RNA (mRNA) has been detected at about 30% of PAR-1 mRNA levels.48 Based on studies using blocking antibodies and peptides that inhibit activation of PAR-1 and PAR-4, it appears that PAR-1 on human platelets responds to low concentrations of thrombin and PAR-4 mediates responses to high concentrations of thrombin.48 HRgpA activates platelets with similar efficiency to thrombin, and this may arise from its more potent cleavage of PAR-4 as seen in the transfected cells. PAR-4 has recently been shown to induce the majority of the calcium signal in platelets,63 thus the more potent activation of PAR-4 by HRgpA could possibly account for the similar level of platelet activation by this enzyme compared to thrombin. HRgpA, by potently activating PAR-4 and at the same time activating the more abundant PAR-1 receptor, may be causing the same overall effect induced by thrombin.
Treatment of human platelets with HRgpA or RgpB also resulted in platelet aggregation, which was dependent on the proteolytic activity of the enzymes. In platelets treated with either gingipain, the increase in light transmittance level observed was a true aggregation phenomenon because pretreatment of platelets with inhibitors of platelet activation prevented the phenomenon. The aggregation assay was carried out in the absence of exogenously added fibrinogen. In thrombin-induced platelet aggregation, fibrinogen is released from the α granules during platelet activation, which then binds to the glycoprotein IIb-IIIa complex, resulting in aggregation.64HRgpA and RgpB cause aggregation without the requirement of exogenous fibrinogen, indicating that fibrinogen is released as part of platelet activation by these bacterial enzymes. This suggests that these proteases, like thrombin, are strong agonists for platelets.
The data discussed above present compelling evidence that PAR-1 and PAR-4 on the platelet surface can be activated by R-X–specific bacterial proteases. Taken together with the activation of PAR-2 on neutrophils14 by these proteases, these data establish a new paradigm in microbial pathogenicity, specifically, that some host cell functions may be manipulated by bacterial proteases cleaving the PARs. In the case of P gingivalis, the immediate advantage of this new pathway for the pathogen may not be clearly apparent. However, uncontrolled PAR activation will certainly contribute to the deregulation of the local inflammatory reaction, which can be beneficial for the microbial community in the pathologic periodontal pocket. In addition to the PARs, several other cell surface receptors, including C5a (CD88),13 fMLP-R,12 LPS receptor (CD14),17α5β1-integrin,15 occludin, and E-cadherin16 are substrates for arginine- (Rgps) and/or lysine- (Kgp) specific gingipains. Therefore, it is conceivable that modification of molecules on the host cell surface by bacterial proteases may play an important role in the maintenance of the chronic inflammatory condition associated with periodontitis. A potentially deleterious effect of cell surface receptor cleavage was also shown for metalloproteases from Serratia marcescens, Staphylococcus aureus, Pseudomonas aeruginosa, and Listeria monocytogenes, as well as a cysteine protease from Streptococcus pyogenes.10 11 Taken together, these data firmly establish that proteolytic modification of the host cell surface by bacterial proteases represents a newly discovered virulence pathway explored by some pathogens.
Recently, periodontal diseases have been linked to cardiovascular illnesses, including heart attack, in a number of epidemiologic studies.20 The consensus viewpoint is that this correlation is an effect of sustained chronic inflammation19,65-68 of the periodontium triggered by continuous release of lipopolysaccharide from gram-negative bacteria in the subgingival dental plaque. Indeed, significantly increased levels of proinflammatory cytokines such as interleukin 1β, tumor necrosis factor-α, interleukin 6, as well as other proinflammatory active compounds including prostaglandin E2 and thromboxane B2 have been found in gingival crevicular fluid.69 Also, as in the case of many other infectious diseases, plasma fibrinogen levels and leukocyte counts are increased in individuals with periodontal disease.70,71 In addition, it was shown that high levels of factor VIII activity and its cofactors in the coagulation pathway were associated with poor dental health, linking chronic dental infections with increased thrombogenicity.72
The studies reviewed above provide a conceivable explanation for the emerging correlation between periodontitis and cardiovascular diseases. In this scheme, P gingivalis proteases would have an indirect role as factors aggravating or sustaining chronic inflammation. In light of the accumulated data, however, it is tempting to speculate that gingipains may have a more direct role in cardiovascular complications. Recently, P gingivalis has been immunolocalized in the shoulders of atherosclerotic plaque.35 If these bacterial cells still express gingipains, it is very likely that such proteases will contribute to plaque ulceration and thrombus formation through effective, uncontrolled activation of both PARs and coagulation factors.
Supported by National Health and Medical Research Council (Australia) grant 990199 and Heart Foundation of Australia grant G 99M 0336 (to R.N.P.), grant 6 PO4A 04717 from Committee of Scientific Research (KBN, Poland) (to J.P.) and National Institutes of Health grant DE 09761 (to J.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Robert N. Pike, Department of Biochemistry and Molecular Biology, Monash University, PO Box 13D, Clayton, Victoria 3800, Australia; e-mail: rob.pike@med.monash.edu.au.

![Fig. 1. The [Ca++]i response of platelets to different concentrations of HRgpA, RgpB, and thrombin. / Platelets from 2 donors were loaded with Fura-2 and stimulated with HRgpA, RgpB, and thrombin. The responses in one donor to HRgpA (A) and thrombin (B) and in another donor to RgpB (C) and HRgpA (D) are shown. The [Ca++]i increases at different concentrations represent the mean from 3 traces similar to those shown in Figure 2, yielding a curve that could be fitted by an equation for single site binding and generate an EC50 value with associated estimate of the SE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171001.jpeg?Expires=1769352924&Signature=Aggzjs-ZlX5F1hDJrxrs0FUiivaEH-bOHHkZpycU3xB5dfk2WWNvG~SraYTf5L7c~mtSI3SiBfU1vrTOOXX7i4zLQf1nlh-fBBhZ2H6utahvKuznqKQkuv8CeMFn06tUAW7vHvIGXIPblAOwfxBiv-fNah3Jo6dwCfwK~pW2hyh0mOvELYHLIUxEvFxCdDGqqj03ICM3mOLBaQ8WUA3CUGfjHm9smWnWMU8bAzcsJJVjxe8RTULANnyHdc4IpRyg4w86-WHCTNuarOg~DXhtVcoP8CXWUaE4ow2HxlCjhwhjPLK1H2rVhO2RBEE1us~HgY2IRkIrEg~~nHbbG25itg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Calcium responses in platelets are due to the proteolytic activity of the enzymes and can be desensitized with thrombin. / The [Ca++]i responses in platelets to: (A) 0.5 nM HRgpA inactivated with leupeptin; (B) 0.5 nM HRgpA, both followed by the addition of 2 μM antipain, 50 nM thrombin, and treatment with 60 μM ADP; (C) 25 nM RgpB inactivated with leupeptin, followed by 10 nM thrombin; (D) 25 nM RgpB, followed by 10 nM thrombin. The [Ca++]i responses shown are those for 1 of the 4 donors in which these effects were observed. Antipain (2 μM) was added in between the HRgpA and thrombin to ensure that the cysteine protease was not desensitizing responses by cleaving thrombin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171002.jpeg?Expires=1769352924&Signature=IDgxckZVRwgUwKb7Dz0RyzMKTyD-N9rnbIlDIaJsi-Ay68EAjbVUunFfVUdIcTsw9N7sNKK5vBklwofkXm~8Fozoxr2KlhnTLmmbhTzjz9SnGKBSlCMg3bmmJoQiWdJ5uPH67a0~EvBhrlboS-p9I9KLWZ7yg~ZW3UnB36EXgvKEFHhBD-ypKEOgx9rSFyks5a9aOo~~ZvPv--ZLRU6wKgUpiXhkRhh7D9w3O7nmmFtFyCksg-jcowYtq3HwCqATj6gpH-D-R7gYpcvQuqJ1igfgnG~rOz3SBS~Lw72TEP~wQ2dHNdCIgkg4-HaGj4~zMKndyIUt2Xld73ZYmtjFPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effects of desensitization of [Ca++]i responses in platelets. / Cells were exposed to: (A) 1 nM, 25 nM, or 100 nM thrombin followed by 0.5 nM HRgpA; (B) 0.05 nM, 0.1 nM, or 1 nM HRgpA followed by 42 nM thrombin. The results shown represent the desensitization effects for 1 of the 4 donors in which these effects were observed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171003.jpeg?Expires=1769352924&Signature=P2ofezEUuHKEirfZAyRzJB6G88-YgRjwovtxkPQLvB6nbBHTkPSXVk5MQ~9aAav1PZWbMAg-upsf7LUQWgLMZO06VjUrxh~kQ174hP70kXGNkjWImgi5rFM~SK3rs5HxpZAVOi91oEetahouqBC5ItOMrDNSIX0f9~SgpdjlwXavOLtns1oEfL3RegAR6tYy4lk-o9vP0YGNIBLINSJ8R9zqufrY97T2p887r3xVajgEAcKQpjxildzvokCqg09UkiX-V5iJ38L3cKKDJlV6E~xU112t98pWtuYvPuDSFR45BJWT-r61SrwR~buX4qpzh-80OAxzz4HoE6daPTFeHw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. The concentration dependence of gingipain-induced changes in PAR-4–transfected cells. / The [Ca++]i response of N1LF PAR-4 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171008.jpeg?Expires=1769352924&Signature=HEiJOnFSBHeEWwoGZ1PNxEAJHavSWeIjGqi89Oaf9vsLBh1Dwskx3px-kPSRxRjTy4171gnUjwpVvTYDXnLxq1XHHP8DtTkFadX8zAQrrawMWKIup4Ga2bbd8kvoPWBmGNDV7Uq1Y3dD~mc57qVyThXpTWrbgaKLKAyyZLzlP0MpXoRqxrFGJ2ZDT-aak2srlA~HJiDFyk-prWpTQFYwX01vOu6S7lOxvrW1KP5vtVqZfOf7gW63srvh0x6HibEtkVXWE~kaGY~PTdJ6rxxpOchx9TSDFAbSBw6mVqT3aL3SAsoHWOM5IQ661-kjvRJJ-0JRsKhBbwV478zjgCkq7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. The concentration dependence of gingipain-induced changes in PAR-1 transfected cells. / The [Ca++]i response of N1LF PAR-1 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171009.jpeg?Expires=1769352924&Signature=1xlhHChFWgzbpE--N6Nt9L4ZwAGkVfArLsaYw~4oMAkuZ4-YZla2x~3QbmkbP-yZ0QFYL1t3ThorMAV8if~oaABAF1nggTw1v2C38-7pCh9N2xeusL6l0hFBTDf4KuNtA6uy-QW~dVT8pkSPokCI1rTWrl40zoSZchj7JwNjMpqsFiVEUAIC5kHdKaM2fZzVV7nQOlxbdnypojN9Da84IlbT-NSxV-TtkpkihsLBplzNUli67etePZ8V22VpC3jlQSkpxgn9z8RpHw39pdnzsZvGYQ1ymXzio0wD6LHTcmfo0ncNXpPAxPB0lUun7JhbuHWv8hy2XQnRbOTWX-n8Cg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 1. The [Ca++]i response of platelets to different concentrations of HRgpA, RgpB, and thrombin. / Platelets from 2 donors were loaded with Fura-2 and stimulated with HRgpA, RgpB, and thrombin. The responses in one donor to HRgpA (A) and thrombin (B) and in another donor to RgpB (C) and HRgpA (D) are shown. The [Ca++]i increases at different concentrations represent the mean from 3 traces similar to those shown in Figure 2, yielding a curve that could be fitted by an equation for single site binding and generate an EC50 value with associated estimate of the SE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171001.jpeg?Expires=1769588804&Signature=1juKaQ4SKXA~wnLa~mz~kC2Q8Hx8rU0oYB2rnjWy6ImHcvEG7-wa8BTyZ28gGRkI-0mlvVa2fCmt2iDqrQYzhI5Yoh9kbbt5qK7~Y~Jv~Fw~P8uLoj5WkfA016u~xeNlLQXIupU84lbWAUuXrRmDn2zGkRN9oNe5hBP3qKHIOFh8kVCurHvCzS90RkFAntPOPcOX60Fvo~oI6t9oD13biQ2Od4c2lCV43PLUcIINQN~n1g-lyW3-MRjFRxnUfz0kOxe38nPeSdkxqSjC1wtNNyi~Z6wtDBz5wU-HDpj3cbYtzbaztcvCrflSBLw191SfYQ0uvOwhs-vUjZ5rD6phEg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Calcium responses in platelets are due to the proteolytic activity of the enzymes and can be desensitized with thrombin. / The [Ca++]i responses in platelets to: (A) 0.5 nM HRgpA inactivated with leupeptin; (B) 0.5 nM HRgpA, both followed by the addition of 2 μM antipain, 50 nM thrombin, and treatment with 60 μM ADP; (C) 25 nM RgpB inactivated with leupeptin, followed by 10 nM thrombin; (D) 25 nM RgpB, followed by 10 nM thrombin. The [Ca++]i responses shown are those for 1 of the 4 donors in which these effects were observed. Antipain (2 μM) was added in between the HRgpA and thrombin to ensure that the cysteine protease was not desensitizing responses by cleaving thrombin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171002.jpeg?Expires=1769588804&Signature=StpCptIBceFPpCimEnbS3UjOKOSbtD24Tjvz9ukWyvLjI9opuOBKn3xgcNS5mkYlGOFi2wrYaiqyM8qYEdOIdKsU3YscPVGx6rteHXaRn2mlEFvU0MC4rm67yjcNYwPhMGmVy2OSWQdksZ~w-IFU1iCC-5gIYeqq2h1DH-7CjRWb86uGYVn3S2plS1fE5mghsYixJnUJkv6059DwQcrt7GLxgf2JIUQKdJgowKtvg2bzgLC2CQVo-inLi1Zu5MJ3wJQmRnYKpqrlFs-kHn0XGjYTG66TzNbccZZn6mnWmm0GCCaFm2oKls604fce5FZX5oOUsfN2VcbfHztpVvZnig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Effects of desensitization of [Ca++]i responses in platelets. / Cells were exposed to: (A) 1 nM, 25 nM, or 100 nM thrombin followed by 0.5 nM HRgpA; (B) 0.05 nM, 0.1 nM, or 1 nM HRgpA followed by 42 nM thrombin. The results shown represent the desensitization effects for 1 of the 4 donors in which these effects were observed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171003.jpeg?Expires=1769588804&Signature=T9eEQuSDh73f8CxuAizOurWoj6IEQMwvdkspkdjD~Ht5OZcRpPOGNtFuGhyRHTdVz70JrTf3V7uIaFyp3GsMuEIul3pXUGE1wAbABjQ82ydgr~l0JWLw2AefBa9QyAR6tjP5ZwPR6MNvQTjmT-KUe0OnMWQTiYhfMjO~TObw3ciYaOhy8Kr1kldIh69S2wC4-vyTad-vMO1i9G7egPUrSv84YtXBdQp7Gw8Wti~mbLLBPef0NW34HHdgpNcK8bUNAp3OCH9kruhZUH6-ldClqlMAs44kx5YJBoyn74-jYg-0dXbrLTLPjnR-DuUe9NY0~JtUzXYPszRE9sWu6xh75w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 8. The concentration dependence of gingipain-induced changes in PAR-4–transfected cells. / The [Ca++]i response of N1LF PAR-4 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171008.jpeg?Expires=1769588804&Signature=VZidpRdoHlxcIEZcX3JhY4YVc39F3FwvpWg9Cbpv9qngu7L0AxMNjY6EGBYREkG8Wo~VX2Bt248G2oIoXFlJPEwyMXLLln0WnZBoIRs2x7IR1jgod~~5UcPORrX1E-PWJdh~tLVOw5M3ATcfJI0MzFiGGBWP7gLh3z4VneFYxeQeUJjr-tVvC1-CRRuUGPqsF6sMpaO7Ar-qOEaXhSR8-QSq9nYkr2vgNR8~d7pdP-BWfGLUyZvZaAFOMBnOiixGSB1O8WLYt8grpTH6Svx5SratJRJ9gmVS2CtWGjykWKWlg2-I1eF5nOxytDm87f0RKg8JIpajATW-VbYHOafMUA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. The concentration dependence of gingipain-induced changes in PAR-1 transfected cells. / The [Ca++]i response of N1LF PAR-1 cells to increasing concentrations of (A) thrombin, (B) HRgpA, and (C) RgpB.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3790/6/m_h81211171009.jpeg?Expires=1769588804&Signature=ngZ9bsdY3DYrqRoq2OXuai4RWvzGJrss4~6wjpZq1HlRIxyBFvhMkKrzjofhpo58pEyvJApf6jkw0uvw3j6PHNsfA51U46vVv3kaaRRjfFeAG4gwSnceV6NMBXRUQgr8Q7Aqw~zJOtp8sgPQQnV8DZzBil0KysUGSWWxcUFT8gn4jPG5O5vHkwqpO-DQ2N-DOQ27PwpKO0pSkvQ8hDNzTES7OSFE54K6Nr96K9EgpaqCKPwL~hnW~qMaFIcNdbvlNLFt7VGb2~22Ba~NqwbraOaw9S16HFF36PBOqeovRMCPQwZiOyqO9PpDL0DLPyMvMICBrk9Gps~R4yiSpqBbIQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)