Exposure of U937 human leukemic cells to the phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) induces their differentiation into monocyte/macrophage-like cells. This terminal differentiation is associated with a resistant phenotype to apoptosis induced by the topoisomerase II inhibitor etoposide. The inhibition occurs upstream of the mitochondrial release of cytochrome c and the activation of procaspase-2, -3, -6, -7, -8, and -9. By using cell-free systems, it was demonstrated that the mitochondrial pathway to cell death that involves mitochondrial membrane depolarization, cytochrome c release and cytosolic activation of procaspases by cytochrome c/dATP remains functional in TPA-differentiated U937 cells. Accordingly, 2 drugs recently shown to target the mitochondria, namely lonidamine and arsenic trioxide, bypass the resistance of TPA-differentiated U937 cells to classical anticancer drugs. Cell death induced by the 2 compounds is associated with mitochondrial membrane depolarization, release of cytochrome c and Smac/Diablo from the mitochondria, activation of caspases, poly(ADP-ribose) polymerase cleavage and internucleosomal DNA fragmentation. Moreover, the decreased glutathione content associated with the differentiation process amplifies the ability of arsenic trioxide to activate the mitochondrial pathway to cell death. Similar results were obtained by comparing undifferentiated and TPA-differentiated human HL60 leukemic cells. These data demonstrate that mitochondria-targeting agents bypass the resistance to classical anticancer drugs induced by TPA-mediated leukemic cell differentiation.
Introduction
Chemotherapeutic drugs can kill cultured leukemic cell lines by inducing apoptosis.1 This mode of cell death may be clinically relevant because apoptotic blast cells have been detected in the peripheral blood of patients with acute leukemia who received chemotherapeutic drugs.2 Hallmarks of apoptosis in leukemic cells include characteristic morphologic changes such as cell shrinkage, membrane blebbing, and cell fragmentation into apoptotic bodies, internucleosomal DNA fragmentation, and limited proteolytic cleavage of selective intracellular proteins such as the nuclear enzyme poly(ADP-ribose)polymerase (PARP). This cell death phenotype is induced by activation of a constitutively expressed apoptotic machinery that involves a family of cysteine proteases known as caspases, organized in a branched proteolytic cascade. These enzymes are synthesized as inactive proenzymes (procaspases) whose proteolysis at internal aspartate residues generates large and small subunits.3 Heterodimerization of these subunits is required to form the active enzyme (caspase) that cleaves intracellular substrates, either a downstream procaspase or other cellular proteins.4
Activation of the caspase cascade in response to specific damage induced by chemotherapeutic drugs is the consequence of a disruption of the mitochondrial membrane barrier function.5 Although the exact mechanism remains controversial, mitochondrial changes lead to the release of soluble apoptogenic proteins such as cytochrome c,6 apoptosis-inducing factor (AIF),7 and Smac/Diablo8,9 from the mitochondrial intermembrane space to the cytosol. Once in the cytosol, cytochrome c, in the presence of adenosine triphosphate (ATP), induces conformational changes of an adaptor molecule designated Apaf-1 that recruits and activates procaspase-9 molecules.10,11 In turn, caspase-9 activates the downstream caspase cascade that involves caspase-3 and other effector enzymes.12 Smac/Diablo was recently shown to suppress the inhibition of caspases by proteins known as inhibitor of apoptosis proteins (IAPs), thereby increasing their sensitivity to the cytochrome c/ATP pathway of activation.8,9 Once activated, effector caspases cleave a limited set of essential cellular proteins, leading to cell dismantling.13-15 Mitochondrial function during apoptosis is controlled by the Bcl-2 family of proteins that could prevent the opening of the permeability transition pore or stabilize the barrier function of the outer mitochondrial membrane and prevent the release of apoptogenic molecules.5 16
How the specific damage induced by chemotherapeutic drugs leads to mitochondrial changes and activation of the caspase cascade under the control of Bcl-2–related proteins remains poorly understood. We have shown previously that differentiation of human myeloid leukemia cell lines along a macrophagic pathway induced by exposure to the phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) correlated with a resistance phenotype to apoptosis induced by a series of anticancer drugs with distinct primary intracellular targets.17,18 This inhibition occurs downstream of specific damage, measured as cleavage complexes, induced by topoisomerase poisons.17 In addition, TPA-differentiated cells are resistant to apoptosis induced by exposure to Fas-ligand, a specific pathway that was proposed as an amplifier system in drug-induced cell death.19 A resistance to drug-induced and TRAIL-mediated apoptosis was also reported in vitamin D3- and dimethyl sulfoxide (DMSO)-differentiated HL60 cells, respectively.20 21
Identification of the molecular mechanisms that modulate cell sensitivity to chemotherapeutic drugs in differentiated cells could help to define strategies combining differentiating and cytotoxic agents for treating acute leukemias. The present study further explores the level of inhibition of drug-induced apoptosis in TPA-differentiated cells. By using cell-free systems, we demonstrate that the pathway involving mitochondria and the downstream caspase cascade remains functional in terminally differentiated cells. Based on this observation, we tested the ability of 2 chemotherapeutic drugs recently shown to target the mitochondria, namely the indazole-3-carboxylic acid derivative lonidamine and the metalloid arsenic trioxide, to induce apoptosis in these cells.
Materials and methods
Drugs and chemical reagents
Etoposide (VP-16) and TPA were obtained from Sigma-Aldrich Laboratories (St Quentin Fallavier, France). Lonidamine was kindly provided by Dr M. F. Poupon (Institut Curie, Paris, France). Stock solutions were prepared by diluting these reagents in DMSO and storing them at −20°C. Further dilutions were made in culture medium just before use. The final concentration of DMSO in culture medium never exceed 0.1% (vol/vol), which was nontoxic to the cells. Arsenic trioxide was kindly provided by Dr G. Q. Chen (Shangai Institute of Hematology, China). [2-14C]-Thymidine (56 mCi/mM) was obtained from Amersham (Les Ulis, France). All other chemicals were of reagent grade and were purchased from local sources.
Antibodies
The tested mouse monoclonal antibodies (mAbs) included antihuman HSC70 from Santa Cruz Biotechnology (Santa Cruz, CA), cytochrome c, caspase-2, caspase-6, and caspase-8 from Pharmingen (San Diego, CA), caspase-3 and caspase-7 from Transduction Laboratories (Lexington, KY), and cytochrome oxidase subunit IV (COX IV) from Molecular Probes (Eugene, OR). We also used the polyclonal rabbit antibodies raised against human caspase-3-p17 (kindly provided by Donald W. Nicholson, Merk-Frost, Toronto, ON, Canada), caspase-9 (Pharmingen), caspase-7 (StressGen, Victoria, BC, Canada), PARP (Boehringer-Mannheim, Germany), and Smac/Diablo (kindly provided by Xiadong Wang, Howard Hughes Medical Institute, Dallas, TX).
Cell culture and differentiation
The human leukemic cell lines U937 and HL60 (American Type Culture Collection, Rockville, MD) were grown in suspension in RPMI 1640 medium with glutamax-I (Gibco BRL, Life Technologies, Cergy Pontoise, France) supplemented with 10% (vol/vol) fetal bovine serum (BioWhittaker, Fontenay-sous-bois, France) in an atmosphere of 95% air and 5% CO2 at 37°C. Cell viability was determined by using a trypan blue dye exclusion assay. To ensure exponential growth, cells were resuspended at a density of 0.5 × 106cells/mL in fresh medium 24 hours before each treatment. To induce differentiation, cells were culture in the presence of 20 nM TPA for 24 hours (HL60) or 72 hours (U937) as previously described.17 18 After treatment, adherent cells were harvested using a cell scraper. U937 and HL60 cell clones containing the full-length human bcl-2 cDNA were kindly provided by Jacqueline Bréard (INSERM 461, Chatenay-Malabry, France) and Michele Allouche (CNRS-UPCM, CHU Purpan, Toulouse, France), respectively.
DNA fragmentation
DNA fragmentation was quantified by using a previously reported filter elution assay.22 Briefly, exponentially growing cells were incubated with 0.02 μCi/mL [2-14C]-thymidine and cultured at 37°C for 2 days. Then, cells were chased in isotope-free medium overnight, resuspended in fresh medium, and treated with different agents before or after TPA treatment. Approximately 1.0 × 106 treated or untreated labeled cells were loaded onto a protein-absorbing filter (polyvinylidene difluoride filters, 0.65 μm pore size, 25 mm diameter; Durapore membrane, Millipore, Saint-Quentin en Yvelines, France). Cells were then washed once with ice-cold phosphate-buffered saline (PBS) and subsequently lysed in 0.2% sodium sarkosyl, 2 M NaCl, 0.04 M EDTA, pH 10.0. Filters were washed with 0.02 M EDTA, pH 10.0. DNA was depurinated by incubation of filters in 1 M HCl at 65°C for 45 minutes, then released from the filters with 0.4 M NaOH for 45 minutes at room temperature. Radioactivity was counted by liquid scintillation spectrometry in each fraction (wash, lysis, EDTA wash, and filter). DNA fragmentation was measured as the fraction of disintegrations per minute (dpm) in the lysis fraction plus EDTA wash relative to the total intracellular dpm. For analyzing DNA fragmentation by agarose gel electrophoresis, cellular DNA was extracted by a previously described salting-out procedure,23 and electrophoresis was performed in 1.8% agarose gel in Tris-borate–EDTA buffer (pH 8.0) at 20 V for 14 hours. After electrophoresis, DNA was visualized by ethidium bromide staining.
Western blot analysis
After treatment, whole cell lysates were prepared by lysing the cells in boiling buffer (1% SDS, 1 mM Na-vanadate, 10 mM Tris pH 7.4) in the presence of protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride [PMSF, 2.5 μg/mL pepstatin, 10 μg/mL aprotinin, 5 μg/mL leupeptin). Viscosity of the samples was reduced by several passages through a 26-gauge needle. Protein concentration was measured by the Bio-Rad DC protein assay kit (Ivry sur Seine, France). Thirty micrograms protein was incubated in loading buffer (125 mM Tris-HCl, pH 6.8, 10% β-mercaptoethanol, 4.6% SDS, 20% glycerol, and 0.003% bromophenol blue), separated by sodium dodecyl sulfate–polyacrylamide gel (SDS-PAGE), and electroblotted to polyvinylidene difluoride membrane (Bio-Rad). After blocking nonspecific binding sites overnight by 5% nonfat milk in TPBS (0.1% PBS-Tween 20), the membrane was incubated for 2 hours at room temperature with primary antibody. After 2 washes in TPBS, the membrane was incubated with horseradish peroxidase–conjugated goat antimouse or antirabbit antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 minutes at room temperature, then washed twice in TPBS. Immunoblot was revealed using enhanced chemiluminescence detection kit (Amersham) by autoradiography.
Cell fractionation
Cytosolic fractions were prepared by resuspending the cells in ice-cold buffer A (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 17 μg/mL PMSF, 8 μg/mL aprotinin, 2 μg/mL leupeptin [pH 7.4]) before passing them through an ice-cold cell homogenizer. Unlysed cells and nuclei were pelleted by a 10-minute, 750g spin. This step was repeated twice. The supernatant was centrifuged at 10 000g for 25 minutes, and the resultant supernatant was further centrifuged at 100 000g for 1 hour. The supernatant (cytosolic fraction) was frozen at −80°C. Nuclei-free, mitochondria-free cytosolic extracts (cell-free extracts) were generated as previously described.24 Briefly, cells (1-2 × 108) were pelleted and washed twice with PBS, pH 7.2, followed by a single wash with 4 mL ice-cold cell extract buffer (20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 100 μM PMSF, 2 μg/mL aprotinin, 10 μg/mL leupeptin [pH 7.4]). Two volumes ice-cold extract buffer were added to 1 vol packed cell pellet before the cellular suspension was transferred to a 2-mL Dounce homogenizer. Cells were allowed to swell under the hypotonic condition for 20 minutes, then were disrupted with 30 strokes of a B-type. Lysis was confirmed with the trypan blue dye exclusion test before centrifugation of lysates at 16 000g for 15 minutes at 4°C. Caspase activation by the cytochrome c/dATP combination was tested in these clarified supernatants.
Purification of mitochondria
Purified mitochondria were isolated as previously described.24 Briefly, cells (1 × 108) were resuspended in buffer H (300 mM saccharose, 5 mM N-tris(hydroxymethyl)methyl-2-amino-ethanesulfonic acid, 200 μM EGTA, pH 6.9), homogenized in a Potter-Thomas homogenizer, then centrifuged for 10 minutes at 760g. The supernatant was recovered while the pellet was resuspended in H buffer and centrifuged to recover the supernatant as above. Both supernatants were mixed and centrifuged for 10 minutes at 8740g before the mitochondria pellet was resuspended in buffer H.
Activation of cell-free apoptosis
Cell-free extracts (10 μL, 5-10 mg/mL protein) were incubated at 37°C with 5 μM horse heart cytochrome c (Sigma) and 1 mM dATP (Pharmacia). Then caspase activation was determined by Western blot analysis. For cell-free reaction activated by atractyloside, purified mitochondria were exposed to 5 mM atractyloside for 30 minutes and pelleted by a 10 000g spin for 10 minutes before cytochrome c release was determined by Western blot in both the supernatant and the mitochondria pellet.
Analysis of mitochondrial membrane potential
Mitochondrial membrane depolarization was analyzed using the DePsipher kit (R&D Systems, Abington, United Kingdom) according to the manufacturer's instructions. Briefly, 106 cells were incubated for 20 minutes at 37°C in the presence of 5 μg/mL DepSipher (R&D Systems) solution and then washed 2 times in PBS. In some cases, cells were treated for 30 minutes with the uncoupling agent carbonyl cyanide m-chlorophenylhydrazone (100 μM; Sigma-Aldrich). Analysis was performed by the use of a FACscan cytometer (Becton Dickinson, Le Pont de Claix, France).
Quantification of intracellular glutathione and reactive oxygen species content by flow cytometry
The level of cellular glutathione (GSH) was determined by flow cytometry after staining with monochlorobimane (Molecular Probes) as previously described.25 Briefly, 200 μM monochlorobimane [syn-(ClCH2, CH3)-1,5-diazabicyclo-[3.3.0]-octa-3,6-dione-2,8-dione)] was added to cell suspensions for 30 minutes at 37°C. After 2 washes in PBS, cells were resuspended in PBS and analyzed on a Bio-Rad flow cytometer (Hercules, CA). For reactive oxygen species (ROS) measurements, cells were incubated for 15 minutes at 37°C in the presence of 6.6 μM dihydro-ethidium (Sigma-Aldrich) and analyzed by flow cytometry.
Results
TPA-induced differentiation of U937 human leukemic cells prevents etoposide-induced apoptosis
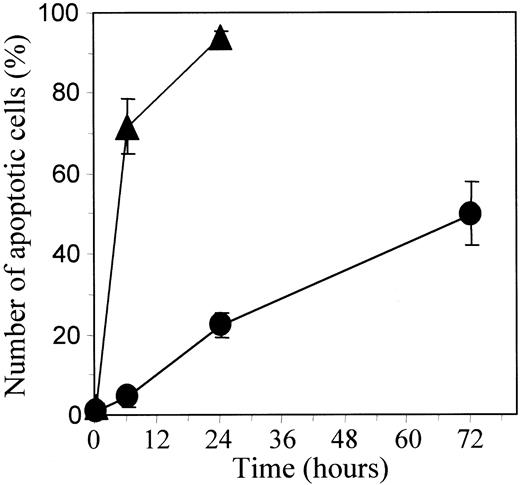
U937 leukemic cells were induced to differentiate along a macrophagic pathway on exposure to 20 nM TPA. After a 48- to 72-hour exposure to TPA, more than 90% cells demonstrate a differentiated phenotype including adhesion to the culture flask and increased expression of the glycoprotein CD11b at the cell surface. On continuous exposure to 50 μM etoposide, most of the parental U937 cells undergo apoptosis in 6 hours, as demonstrated by staining nuclear chromatin with Hoechst 33342, whereas TPA-differentiated cells strongly resist drug-induced cell death (Figure1).
TPA-mediated differentiation of U937 cells delays etoposide-induced apoptosis.
Undifferentiated (▴) and TPA-differentiated (●) U937 cells were treated with 50 μM etoposide for indicated times. Percentages of apoptotic cells were determined by fluorescent microscopy after Hoechst staining. Each value is the mean ± SD of triplicate determinations.
TPA-mediated differentiation of U937 cells delays etoposide-induced apoptosis.
Undifferentiated (▴) and TPA-differentiated (●) U937 cells were treated with 50 μM etoposide for indicated times. Percentages of apoptotic cells were determined by fluorescent microscopy after Hoechst staining. Each value is the mean ± SD of triplicate determinations.
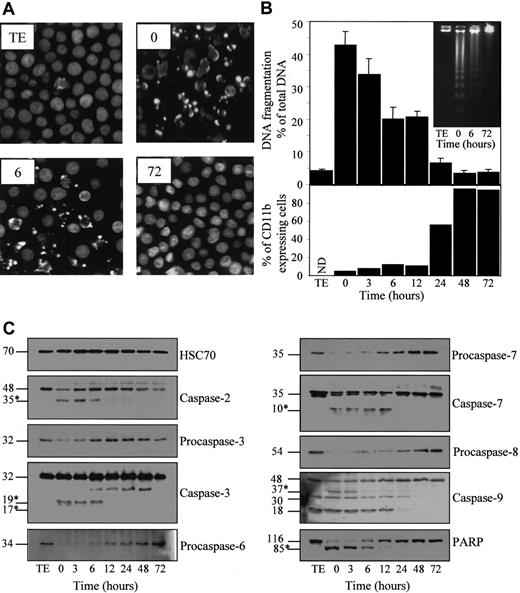
To link the resistance phenotype to the differentiation process, we exposed the cells for 4.5 hours to 50 μM etoposide at various times after the beginning of TPA treatment (20 nM) (Figure2). Apoptosis was identified by Hoechst staining of nuclear chromatin (Figure 2A) and quantified by measuring nuclear DNA fragmentation (Figure 2B). Exposure to TPA provoked a time-dependent decrease of etoposide-induced morphologic changes and DNA fragmentation that were almost completely inhibited 48 to 72 hours after the beginning of TPA exposure. Accordingly, by using agarose gel electrophoresis, the characteristic nucleosome-sized ladder fragmentation of nuclear DNA observed in parental undifferentiated cells exposed for 4.5 hours to the drug was not detected in TPA-differentiated cells treated in the same conditions (Figure 2B, insert).
Inhibition of etoposide-induced apoptosis in U937 cells induced to differentiate by TPA.
U937 cells were untreated (TE) or preincubated for indicated times with 20 nM TPA, washed, then treated for 4.5 hours with 50 μM etoposide. (A) Nuclear morphologic changes associated with apoptosis were visualized by fluorescent microscopy after Hoechst staining. (B) (upper panel) Apoptotic DNA fragmentation was quantified using a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (inset) Visualization of apoptotic DNA fragmentation by agarose gel electrophoresis. (lower panel) Percentage of differentiated cells according to CD11b expression measured before etoposide treatment. (C) Indicated caspases and PARP were analyzed in whole-cell extracts by immunoblot. Numbers are molecular weights in kilodaltons. “Procaspase” and “caspase” are used to design the studied enzymes, depending on whether the tested antibody does not or does detect the cleavage fragments (∗), respectively. A unique series of cell lysates was used for these experiments. Loading of each strip was checked by using an antihuman HSC70 mAb (one representative control is shown).
Inhibition of etoposide-induced apoptosis in U937 cells induced to differentiate by TPA.
U937 cells were untreated (TE) or preincubated for indicated times with 20 nM TPA, washed, then treated for 4.5 hours with 50 μM etoposide. (A) Nuclear morphologic changes associated with apoptosis were visualized by fluorescent microscopy after Hoechst staining. (B) (upper panel) Apoptotic DNA fragmentation was quantified using a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (inset) Visualization of apoptotic DNA fragmentation by agarose gel electrophoresis. (lower panel) Percentage of differentiated cells according to CD11b expression measured before etoposide treatment. (C) Indicated caspases and PARP were analyzed in whole-cell extracts by immunoblot. Numbers are molecular weights in kilodaltons. “Procaspase” and “caspase” are used to design the studied enzymes, depending on whether the tested antibody does not or does detect the cleavage fragments (∗), respectively. A unique series of cell lysates was used for these experiments. Loading of each strip was checked by using an antihuman HSC70 mAb (one representative control is shown).
Etoposide treatment of parental cells also induced a decrease of procaspase-2, -3, -6, -7, -8, and -9 expression, as determined by Western blot analysis (Figure 2C). By using antibodies reacting with the cleavage fragments that characterize active enzymes, decreased expression of the 48-kd procaspase-2, the 32-kd procaspase-3, the 35-kd procaspase-7, and the 48-kd procaspase-9 correlated with the appearance of a p35, a p19/p17, a p10, and a p37 active fragment, respectively. The 30-kd fragment identified with the anti–caspase-9 antibody might correspond to the previously described caspase-9b isoform.26 The significance of the 18-kd band remains unknown. All these etoposide-induced events were progressively inhibited as U937 cells progressed toward the differentiation phenotype on TPA exposure. This inhibition became identifiable between 6 and 12 hours after the beginning of TPA exposure, depending on the experiment. In addition, TPA-induced differentiation was associated with a progressive inhibition of the 116-kd DNA repair enzyme PARP cleavage into an 85-kd N-terminal fragment (Figure 2C).
Caspases remain able to be activated in cell-free extracts from TPA-induced differentiated cells
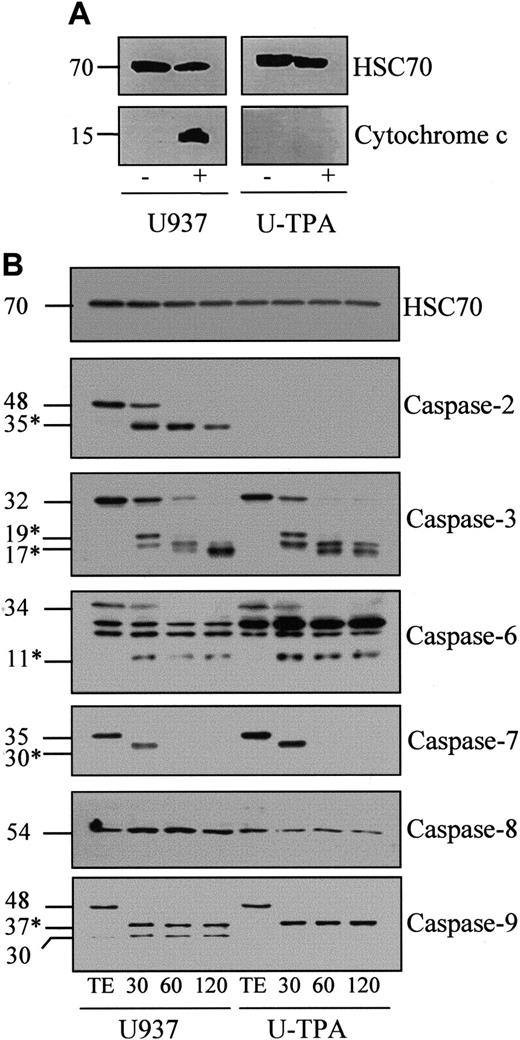
Exposure of undifferentiated U937 cells to 50 μM etoposide for 4.5 hours triggered the appearance of cytochrome c in the cytosol. This event was prevented by TPA-induced differentiation (Figure3A). To determine whether the cytochrome c–mediated activation of the caspase cascade was influenced by the differentiation process, we used a previously described cell-free system.24 Exposure of cell-free extracts from undifferentiated U937 cells to 5 μM cytochrome c and 1 mM dATP induced a time-dependent decrease of the expression of procaspase-2, -3, -6, -7, and -9, suggesting their cleavage into active fragments. Accordingly, immunoblot experiments detected the time-dependent appearance of the 35-kd active fragment of caspase-2, the 19- and 17-kd fragments of caspase-3, the 11-kd fragment of caspase-6, the 30-kd fragment of caspase-7, and the 37-kd fragment of caspase-9 (Figure 3B). In this time-course, procaspase-8 expression remained unchanged. TPA-mediated differentiation of U937 cells induced some changes in the subcellular localization of procaspases, mainly the disappearance of procaspase-2 from the cytosol (O.S. et al, manuscript in preparation). The other procaspases remained sensitive to activation by the cytochrome c–dATP combination with kinetics similar to that observed in cytosolic extracts from undifferentiated cells (Figure 3B). Thus, TPA-induced differentiation does not significantly influence procaspase activation by cytochrome c/dATP.
Cytochrome c-mediated activation of caspases in cell-free extracts.
(A) Western blot analysis of cytochrome c in cytosolic fractions from undifferentiated (U937) and TPA-differentiated (U-TPA) U937 cells treated (+) or not treated (−) with 50 μM etoposide for 4.5 hours. (B) Cell-free extracts from U937 and U-TPA cells were left untreated for 2 hours (TE) or were incubated for indicated times (minutes) in the presence of 5 μM cytochrome c and 1 mM dATP. Processing of indicated caspases was analyzed by Western blot. Numbers are molecular weights in kilodaltons. *Cleavage products. Loading was checked with the use of an antihuman HSC70 mAb.
Cytochrome c-mediated activation of caspases in cell-free extracts.
(A) Western blot analysis of cytochrome c in cytosolic fractions from undifferentiated (U937) and TPA-differentiated (U-TPA) U937 cells treated (+) or not treated (−) with 50 μM etoposide for 4.5 hours. (B) Cell-free extracts from U937 and U-TPA cells were left untreated for 2 hours (TE) or were incubated for indicated times (minutes) in the presence of 5 μM cytochrome c and 1 mM dATP. Processing of indicated caspases was analyzed by Western blot. Numbers are molecular weights in kilodaltons. *Cleavage products. Loading was checked with the use of an antihuman HSC70 mAb.
Mitochondria from TPA-induced differentiated U937 cells remain reactive
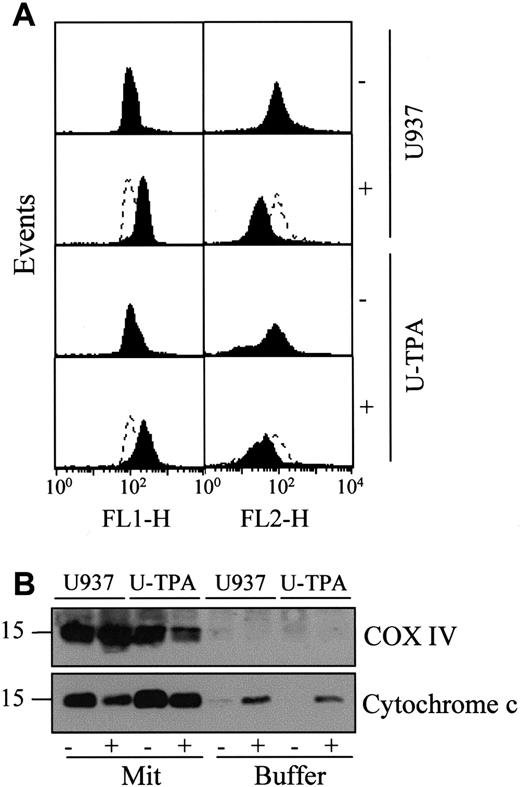
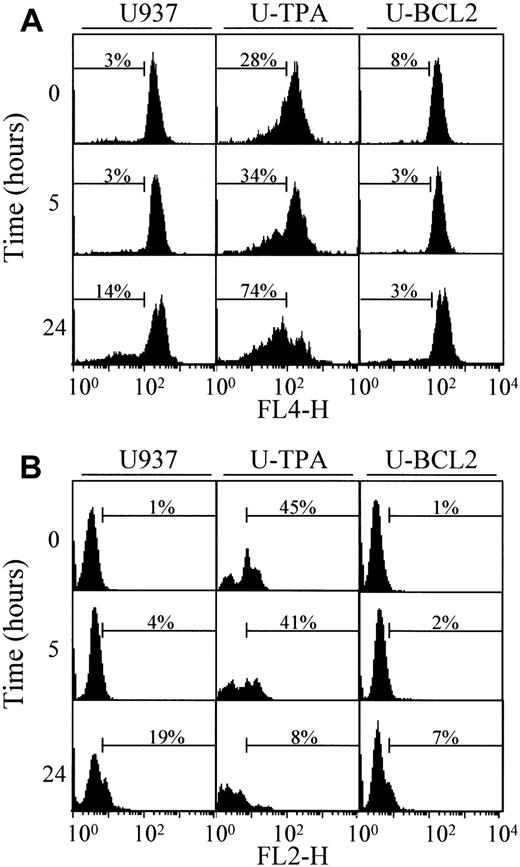
The mechanisms that account for the mitochondrial release of cytochrome c are still controversial. One of the proposed mechanisms involves a decrease of the mitochondrial membrane potential Δψm.5 To determine whether TPA-induced differentiation was associated with a decreased reactivity of mitochondria to uncoupling agents, we exposed undifferentiated and TPA-differentiated U937 cells for 30 minutes to 100 μM of the uncoupling agent carbonyl cyanide m-chlorophenylhydrazone (mCICCP). This treatment induced a switch from the mitochondrial aggregate (FL2-H) to the cytosolic monomer (FL1-H) that was similar in mitochondria from both undifferentiated and TPA-differentiated cells indicating mitochondrial depolarization (Figure 4A). A mechanism proposed to account for the mitochondrial release of cytochrome c was the opening of the megachannel that can be induced by exposing purified mitochondria to atractyloside (5 mM, 30 minutes). This compound induced the release of cytochrome c from mitochondria of undifferentiated and differentiated U937 cells with a similar efficacy (Figure 4B). In the presence of cell-free extracts from undifferentiated U937 cells, the effect of atractyloside on purified mitochondria resulted in cleavage of the DEVD-AFC substrate, suggesting caspase activation (data not shown). Together these results suggested that the mitochondrial pathway to cell death may remain functional in TPA-differentiated U937 cells.
TPA-induced differentiation of U937 cells does not alter mitochondrial reactivity.
(A) Flow cytometry analysis of mitochondrial potential in undifferentiated (U937) and TPA-differentiated (U-TPA) U937 cells, treated (+) or not treated (−) with 100 μM m-chlorophenylhydrazone for 30 minutes. Mitochondrial membrane depolarization is indicated by the decreased mitochondrial aggregate (FL2-H) and the increased cytosolic monomer (FL1-H). Dashed lines indicate the controls. (B) Purified mitochondria isolated from U937 and U-TPA cells were treated (+) or not treated (−) with 5 mM atractyloside for 30 minutes. Cytochrome c expression was analyzed by Western blot in mitochondrial (Mit) and supernatant (Buffer) fractions. Lack of supernatant contamination by mitochondria extracts was checked by using an antihuman cytochrome oxidase subunit IV (COX IV) mAb.
TPA-induced differentiation of U937 cells does not alter mitochondrial reactivity.
(A) Flow cytometry analysis of mitochondrial potential in undifferentiated (U937) and TPA-differentiated (U-TPA) U937 cells, treated (+) or not treated (−) with 100 μM m-chlorophenylhydrazone for 30 minutes. Mitochondrial membrane depolarization is indicated by the decreased mitochondrial aggregate (FL2-H) and the increased cytosolic monomer (FL1-H). Dashed lines indicate the controls. (B) Purified mitochondria isolated from U937 and U-TPA cells were treated (+) or not treated (−) with 5 mM atractyloside for 30 minutes. Cytochrome c expression was analyzed by Western blot in mitochondrial (Mit) and supernatant (Buffer) fractions. Lack of supernatant contamination by mitochondria extracts was checked by using an antihuman cytochrome oxidase subunit IV (COX IV) mAb.
Mitochondria-targeting agent lonidamine induced apoptosis of TPA-differentiated cells
The previous observations suggested that drugs shown to directly target the mitochondria might overcome the resistance of TPA-differentiated cells to etoposide and various other classical anticancer drugs.18 We first tested the mitochondria-targeting drug lonidamine.27 The ability of this compound to induce apoptosis of TPA-differentiated cells was only slightly delayed in TPA-differentiated compared to undifferentiated U937 cells, as demonstrated by quantifying DNA fragmentation (Figure5A) and by counting the percentage of cells demonstrating nuclear chromatin condensation after a 24- to 48-hour exposure to 200 μM lonidamine (Table1). Lonidamine-induced apoptotic DNA fragmentation and chromatin condensation were completely inhibited by stable overexpression of Bcl-2 in U937 cells (Figure 5A; Table 1). In undifferentiated and TPA-differentiated cells, lonidamine induced mitochondrial depolarization (Figure 5C), cytochrome c release in the cytosol (Figure 5B), procaspase-3 cleavage in its active fragments, and PARP cleavage (Figure 5D). All these events were prevented by Bcl-2 overexpression. The recently described Smac/Diablo protein8 9 was only slightly released in the cytosol on lonidamine exposure, an effect that was not prevented by Bcl-2 overexpression (Figure 5B).
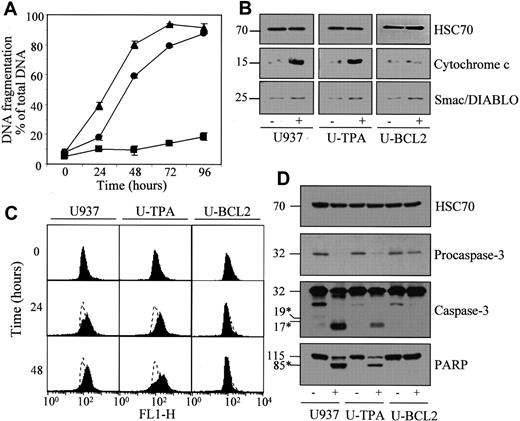
Lonidamine induces apoptosis in TPA-differentiated U937 cells.
Undifferentiated (▴, U937), TPA-differentiated (●, U-TPA), and Bcl-2–overexpressing U937 cells (▪, U-BCL2) were treated for indicated times with 200 μM lonidamine. (A) Apoptotic DNA fragmentation was quantified using a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (B) Western blot analysis of cytochrome c and Smac/Diablo expression in cytosolic fractions from U937, U-TPA, and U-BCL2 cells treated (+) or not treated (−) with 200 μM lonidamine for 48 hours. (C) Mitochondrial membrane depolarization was visualized by flow cytometry in U937, U-TPA, and U-BCL2 cells treated with 200 μM lonidamine for indicated times. Mitochondrial depolarization was identified by an increased level of cytosolic green monomer (FL1-H; dashed lines indicate the controls). (D) Western blot analysis of procaspase-3 and caspase-3 active fragments and PARP expression in whole-cell extracts from U937, U-TPA, and U-BCL2 cells treated (+) or not treated (−) with 200 μM lonidamine for 48 hours. Numbers are molecular weights in kilodaltons. *Cleavage products. Loading was checked with the use of an antihuman HSC70 mAb.
Lonidamine induces apoptosis in TPA-differentiated U937 cells.
Undifferentiated (▴, U937), TPA-differentiated (●, U-TPA), and Bcl-2–overexpressing U937 cells (▪, U-BCL2) were treated for indicated times with 200 μM lonidamine. (A) Apoptotic DNA fragmentation was quantified using a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (B) Western blot analysis of cytochrome c and Smac/Diablo expression in cytosolic fractions from U937, U-TPA, and U-BCL2 cells treated (+) or not treated (−) with 200 μM lonidamine for 48 hours. (C) Mitochondrial membrane depolarization was visualized by flow cytometry in U937, U-TPA, and U-BCL2 cells treated with 200 μM lonidamine for indicated times. Mitochondrial depolarization was identified by an increased level of cytosolic green monomer (FL1-H; dashed lines indicate the controls). (D) Western blot analysis of procaspase-3 and caspase-3 active fragments and PARP expression in whole-cell extracts from U937, U-TPA, and U-BCL2 cells treated (+) or not treated (−) with 200 μM lonidamine for 48 hours. Numbers are molecular weights in kilodaltons. *Cleavage products. Loading was checked with the use of an antihuman HSC70 mAb.
TPA-induced differentiation sensitizes U937 cells to arsenic trioxide–induced apoptosis
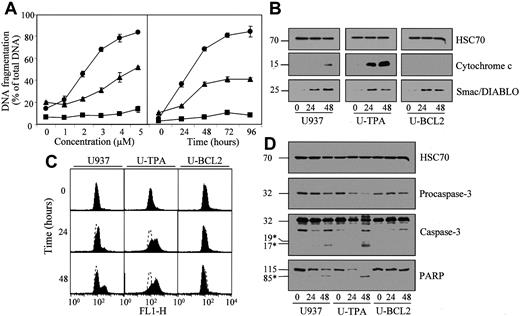
As2O3 is another member of this novel class of chemotherapeutic agents that induce apoptosis, at least in part, by acting on the mitochondria.28,29As2O3 induced apoptosis of undifferentiated U937 cells in a dose- and time-dependent manner (Figure6A; Table 1). TPA-induced differentiation did not prevent, and even sensitized, U937 cells to As2O3-induced apoptosis. The differentiation process also increased the ability of As2O3 to induce mitochondrial depolarization (Figure 6C), cytochrome c release from the mitochondria to the cytosol (Figure 6B), procaspase-3 cleavage in its active fragments and PARP cleavage (Figure 6D). All these events were prevented by Bcl-2 overexpression in U937 cells (Figure 6A-D; Table 1). Smac/Diablo protein was released in the cytosol on As2O3 treatment more efficiently than on lonidamine exposure. Again, this effect was not prevented by Bcl-2 overexpression (Figure 6B). Because As2O3-induced apoptosis has been shown to be modulated by the cellular GSH redox system, with increased intracellular levels of reduced GSH having an inhibitory effect,30 we measured the influence of TPA-induced differentiation on cellular GSH and ROS content in untreated and As2O3-treated cells. By using flow cytometry methods, we observed that the differentiation process was associated with a decrease in the basal level of GSH (Figure7A) and an increase in the basal ROS content in the cells (Figure 7B). Arsenic exposure decreased GSH content and increased ROS level in parental cells. These effects were prevented by Bcl-2 overexpression. In differentiated cells, As2O3 treatment further decreased GSH content without increasing the cellular ROS content, as measured by using dihydro-ethidium (Figure 7A-B).
TPA-induced differentiation sensitizes U937 cells to arsenic trioxide (As2O3)-induced differentiation.
(A) Undifferentiated (▴), TPA-differentiated (●), and Bcl-2 overexpressing U937 cells (▪, U-BCL2) were treated with increasing concentrations of As2O3 for 72 hours (left panel) or with 4 μM As2O3 for indicated times (right panel). Apoptotic DNA fragmentation was quantified with the use of a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (B) Western blot analysis of cytochrome c and Smac/Diablo expression in cytosolic fractions from undifferentiated (U937) and TPA-differentiated (U-TPA) U937 cells and U937 cells overexpressing Bcl-2 (U-BCL2) treated with 4 μM As2O3 for indicated times. (C) Mitochondrial membrane depolarization was visualized by flow cytometry in U937, U-TPA, and U-BCL2 cells treated with 4 μM As2O3 for indicated times. Mitochondrial depolarization was identified by an increased level of cytosolic green monomer (FL1-H; dashed lines indicate the controls). (D) Western blot analysis of procaspase-3 and caspase-3 active fragments and PARP expression in whole-cell extracts from U937, U-TPA, and U-BCL2 cells treated with 4 μM As2O3 for indicated times. Numbers are molecular weights in kilodaltons. *Cleavage products. Loading was checked with the use of an antihuman HSC70 mAb.
TPA-induced differentiation sensitizes U937 cells to arsenic trioxide (As2O3)-induced differentiation.
(A) Undifferentiated (▴), TPA-differentiated (●), and Bcl-2 overexpressing U937 cells (▪, U-BCL2) were treated with increasing concentrations of As2O3 for 72 hours (left panel) or with 4 μM As2O3 for indicated times (right panel). Apoptotic DNA fragmentation was quantified with the use of a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (B) Western blot analysis of cytochrome c and Smac/Diablo expression in cytosolic fractions from undifferentiated (U937) and TPA-differentiated (U-TPA) U937 cells and U937 cells overexpressing Bcl-2 (U-BCL2) treated with 4 μM As2O3 for indicated times. (C) Mitochondrial membrane depolarization was visualized by flow cytometry in U937, U-TPA, and U-BCL2 cells treated with 4 μM As2O3 for indicated times. Mitochondrial depolarization was identified by an increased level of cytosolic green monomer (FL1-H; dashed lines indicate the controls). (D) Western blot analysis of procaspase-3 and caspase-3 active fragments and PARP expression in whole-cell extracts from U937, U-TPA, and U-BCL2 cells treated with 4 μM As2O3 for indicated times. Numbers are molecular weights in kilodaltons. *Cleavage products. Loading was checked with the use of an antihuman HSC70 mAb.
Intracellular GST and ROS levels.
Analysis by flow cytometry of (A) intracellular GST and (B) ROS levels of undifferentiated (U937), TPA-differentiated (U-TPA), and Bcl-2 overexpressing (U-BCL2) U937 cells treated with 4 μM As2O3 for indicated times.
Intracellular GST and ROS levels.
Analysis by flow cytometry of (A) intracellular GST and (B) ROS levels of undifferentiated (U937), TPA-differentiated (U-TPA), and Bcl-2 overexpressing (U-BCL2) U937 cells treated with 4 μM As2O3 for indicated times.
Lonidamine and arsenic trioxide also bypass the resistance of TPA-differentiated HL60 cells to classical chemotherapeutic drugs
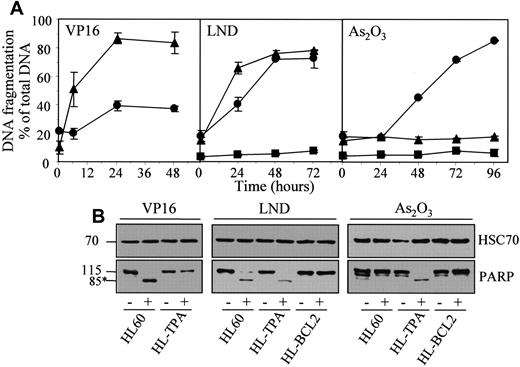
The results obtained in U937 cells were reproduced in HL60 cells whose exposure to TPA also induces a macrophagic-like differentiation process with a resistant phenotype to classical anticancer drugs.17 TPA-differentiated HL60 cells, which were resistant to etoposide-induced apoptosis, demonstrated a sensitivity to lonidamine that was only slightly delayed compared to that of parental cells, whereas their sensitivity to As2O3-induced cell death was greatly increased (Figure 8; Table 1).
Lonidamine and arsenic trioxide bypass the resistance of TPA-differentiated HL60 cells to etoposide.
Undifferentiated (▴, HL60), TPA-differentiated (●, HL-TPA), and Bcl-2 overexpressing (▪, HL-BCL2) HL60 cells were treated with 50 μM etoposide (VP16), 200 μM lonidamine (LND), or 4 μM arsenic trioxide (As2O3) for indicated times. (A) Apoptotic DNA fragmentation was quantified with the use of a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (B) Western blot analysis of PARP cleavage in whole-cell extracts from HL60, HL-TPA, and HL-BCL2 cells, either untreated (−) or treated (+) with 50 μM VP16 for 6 hours, 200 μM LND for 48 hours, or 4 μM As2O3 for 48 hours. Numbers are molecular weights in kilodaltons. *Cleavage product. Loading was checked with the use of an antihuman HSC70 mAb.
Lonidamine and arsenic trioxide bypass the resistance of TPA-differentiated HL60 cells to etoposide.
Undifferentiated (▴, HL60), TPA-differentiated (●, HL-TPA), and Bcl-2 overexpressing (▪, HL-BCL2) HL60 cells were treated with 50 μM etoposide (VP16), 200 μM lonidamine (LND), or 4 μM arsenic trioxide (As2O3) for indicated times. (A) Apoptotic DNA fragmentation was quantified with the use of a filter elution assay. Values from 1 of 2 independent experiments are shown (mean ± SD of triplicate samples). (B) Western blot analysis of PARP cleavage in whole-cell extracts from HL60, HL-TPA, and HL-BCL2 cells, either untreated (−) or treated (+) with 50 μM VP16 for 6 hours, 200 μM LND for 48 hours, or 4 μM As2O3 for 48 hours. Numbers are molecular weights in kilodaltons. *Cleavage product. Loading was checked with the use of an antihuman HSC70 mAb.
Discussion
Classical chemotherapeutic drugs induce apoptosis by compromising mitochondrial function in an indirect fashion—for example, by inducing DNA damage that triggers p53 activation and interferes with cell cycle progression31 by stimulating the Fas-dependent pathway,19,32 by triggering ceramide production,33 and by inducing shifts in redox potentials.1 A series of new chemotherapeutic drugs, many of them currently tested in clinical trials, have been shown to act, at least in part, by directly targeting the mitochondria, as demonstrated by their ability to activate the permeability transition pore complex and to trigger the release of soluble intermembrane proteins from purified mitochondria in vitro.16 This observation led to the theoretical suggestion that these compounds may bypass the resistance to anticancer drugs induced by alterations of indirect death-inducing pathways. The current study demonstrates that TPA-mediated differentiation of leukemic cells reproduces a situation in which the pathway to cell death, triggered by a classical anticancer drug such as etoposide, is inhibited upstream of the mitochondrial events. Analysis of this model system confirms the hypothesis that mitochondria-targeting drugs can overcome such a resistant phenotype.
TPA-mediated inhibition of etoposide-induced apoptosis increased as leukemic cells progressed toward the macrophagic differentiation phenotype. Short-term effects of TPA are mainly the activation of several protein kinase C (PKC) isoenzymes.34-36 The differentiation process involves several events downstream of the transient activation of PKCs.37 Thus, the differentiation-related resistance of leukemic cells to etoposide-induced cell death might be distinguished from the direct consequences of PKC activation. A differentiation-related resistance to apoptosis induced by various stimuli has been described in leukemic cells exposed to either 1,25-dihydroxyvitamin D338,39 or DMSO.21 A resistance to growth factor deprivation–induced cell death was also identified in differentiated myocytes.40 Thus, several differentiation processes in different cell types were associated with a decreased sensitivity to apoptosis induced by a series of distinct stimuli.
Most pathways to apoptosis converge on the mitochondrial release of apoptogenic molecules to the cytosol and the activation of the caspase cascade.1,4,5 The current study confirms that etoposide activates a variety of caspases while inducing apoptosis of undifferentiated U937 cells. Various mechanisms negatively interfere with this final pathway, including overexpression of antiapoptotic proteins of the Bcl-2 family that prevents the cytosolic release of mitochondrial soluble molecules7,41 and overexpression of heat-shock proteins such as HSP27 and HSP70 that prevent cytochrome c–mediated activation of the caspase cascade by interacting with cytochrome c24 and the Apaf-1 adaptor molecule,42 respectively. Despite the fact that the TPA-induced differentiation process induces some changes in the subcellular localization of procaspases (O.S. et al, manuscript in preparation) and modulates the expression of inhibitory proteins—for example, by inducing transient changes in the expression of the Bcl-2–related protein Mcl-118 43—our data, obtained by using cell-free systems, demonstrate that the differentiation process does not significantly influence the mitochondrial pathway to cell death.
Lonidamine (1-[(2,4-dichlorophenylmethyl) methyl]-1H-indazole-3-carboxylic acid]) is an antineoplastic drug derived from indazole-3-carboxylic acid. This compound, which was shown to exacerbate the response of human tumor cells to cisplatin, irradiation, doxorubicin, or cyclophosphamide, has been tested in phase II and III trials of metastatic breast cancer and ovarian cancer.44-48 Although several distinct mechanisms were proposed to account for lonidamine cytotoxic effects,49,50recent data suggest that mitochondria could be the main subcellular target of this compound. Lonidamine directly acts on the permeability transition pore complex when tested on purified organelles, an effect inhibited by Bcl-2.27 Since we have shown that the mitochondrial pathway to cell death remains unchanged in TPA-differentiated cells, lonidamine may have demonstrated a similar efficacy on undifferentiated and TPA-differentiated leukemic cells. Actually, TPA-induced differentiation slightly, but reproducibly, delays lonidamine-induced apoptosis in U937 and HL60 cells, suggesting a negative effect of the differentiation process on some other potential consequences of lonidamine treatment, such as cytosolic calcium increase and intracellular lactate accumulation. This effect of the differentiation process on lonidamine-induced cell death remains limited compared with the strong inhibition of apoptosis triggered by classical anticancer drugs.
As2O3 is another therapeutic compound that was suggested to directly target the mitochondria.28,29Although As2O3-induced apoptosis was initially described in acute promyelocytic cell lines,51,52 this compound induces the death of many other leukemia cell types.53-56 The hypothesis that As2O3 directly targets the mitochondria is based on the observation that exposure of purified organelles to arsenic triggers opening of the permeability transition pore and the release of soluble intermembrane proteins.28 In whole cells, As2O3 induces disruption of the mitochondrial transmembrane potential Δψm51 and caspase activation,56,57 2 events that are inhibited by Bcl-2 overexpression.28As2O3-induced apoptosis was also related to Bcl-2 down-regulation,52,56,58 Bax up-regulation,59 suppression of the apoptosis inhibitory Ras/MAP kinase cascade,60,61 and enhanced translocation of PML or PML/RARα chimeric proteins to nuclear bodies,62but these events depend on As2O3 dose, cell type, and cellular environment. We show here that TPA-induced differentiation of leukemic cells increases the ability of As2O3 to target the mitochondria, to induce the release of cytochrome c and Smac/Diablo, to activate downstream caspases, and to induce apoptotic cell death. A similar synergy was described between As2O3 and retinoic acid, another differentiating agent, in an arsenic-resistant NB4 subline.63,64 The differentiation-induced modulation of leukemic cell sensitivity to As2O3-induced apoptosis could account for the conflicting reports concerning the sensitivity of cell lines such as HL-60. Arsenic toxicity, including its mitochondrial effects, presumably results from forming reversible bonds with the thiol groups of proteins.65 Intracellular level of GSH, which potentially titrates intracellular arsenic by formation of transient As(GS)3 complexes, is a major determinant of As2O3-triggered cell death system, with increased intracellular levels of GSH having an inhibitory effect.30,66 The proton donor GSH prevents arsenic from attacking its targets and increases the GSH peroxidase-catalyzed breakdown of H2O2.67 Thus, the differentiation-related decrease in GSH cellular content may contribute to the sensitization of leukemic cells to As2O3-mediated cell death.
In conclusion, we have shown that the TPA-mediated differentiation of human myeloid leukemic cell lines induces a resistance phenotype to classical anticancer drugs such as etoposide without inhibiting the mitochondrial pathway to cell death. The differentiation process has a limited influence on lonidamine-induced cell death and sensitizes leukemic cells to As2O3-mediated apoptosis (Figure 9). Although the As2O3 doses required for inducing apoptosis in differentiated cells remain high compared with clinically achieved concentrations,68 these observations suggest that chemotherapeutic drugs that directly or rapidly target the mitochondria in vitro warrant further clinical investigation in the treatment of acute leukemias. These drugs may represent an effective strategy to bypass the resistance of leukemic cells to classical anticancer drugs, or they may be used in combination with differentiating agents.
Influence of TPA-induced differentiation on drug-induced cell death.
TPA-mediated differentiation of leukemic cells strongly decreases the ability of classical anticancer drugs, such as those inducing DNA damage, to activate the mitochondrial pathway to cell death. In contrast, the differentiation process has limited influence on cell death induced by the mitochondria-targeting agent lonidamine and even sensitizes the cells to arsenic-mediated activation of the mitochondrial pathway.
Influence of TPA-induced differentiation on drug-induced cell death.
TPA-mediated differentiation of leukemic cells strongly decreases the ability of classical anticancer drugs, such as those inducing DNA damage, to activate the mitochondrial pathway to cell death. In contrast, the differentiation process has limited influence on cell death induced by the mitochondria-targeting agent lonidamine and even sensitizes the cells to arsenic-mediated activation of the mitochondrial pathway.
We thank M. F. Poupon, X. Wang, and D. G. Chen for providing lonidamine, anti-Smac antibody, and As2O3, respectively.
Supported by grants from the Ligue National Contre le Cancer (Equipe labelisée “la ligue”) and the Association Régionale pour l'Enseignement et la Recherche Scientifique. O.S. is the recipient of a grant from the Fondation pour la Recherche Médicale.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Eric Solary, Faculty of Medicine and Pharmacy, INSERM U517, 7 boulevard Jeanne d'Arc, 21033 Dijon, France; e-mail: esolary@u-bourgogne.fr.