Abstract
The myeloma plasma cell is a postgerminal center, isotype-switched B cell. Chromosomal translocations into immunoglobulin heavy chain (IgH) switch regions, recombination sites in isotype switching, were initially demonstrated in myeloma cell lines but only a limited number of primary tumors. Molecular cytogenetics have since been applied to a series of primary tumors, in which IgH translocations accounted for many recurrent aberrations, among numerous nonrecurrent changes of unknown significance. This study, therefore, examined primary myeloma for IgH switch translocations using an established Southern blot assay that detected illegitimate switch recombinations. Sensitivity of the method was established by confining the analysis to 21 samples (4 stable, 17 progressive disease) with demonstrable legitimate isotype switches, of a total of 60 samples. Illegitimate recombinations were found in 12 or 57% (1 stable, 11 progressive) of 21 samples, comparable with estimates by molecular cytogenetics. The presence of switch translocations was supported by demonstrating up-regulated expression in myeloma marrow of cyclin D1 and fibroblast growth factor receptor 3 (FGFR3), candidate oncogenes on chromosomes 11q13 and 4p16, respectively. Illegitimate switches were detected most frequently in Sμ, with more than one region involved in 6 cases. Although these results confirmed the presence of switch translocations in primary myeloma, their absence in 43% of cases may imply heterogeneity of pathogenesis. In progressive disease, there was no significant difference between patients with and without illegitimate switches in survival, nor the prognostic indicators of β2microglobulin (β2m) and serum thymidine kinase (STK). Hence IgH switch translocations as a single entity are unlikely to be a feature of disease progression or have prognostic significance.
Introduction
Myeloma is a malignancy of the plasma cell, the terminally differentiated B cell, in which immunoglobulin genes have undergone variable region recombination, followed by isotype switching and then somatic hypermutation in the germinal center. As in other B-cell malignancies, chromosomal translocations into the immunoglobulin loci have been found. In myeloma they involve mainly the immunoglobulin heavy chain (IgH) genes on chromosome 14q32, at the switch regions upstream of each constant region gene.1-3 The translocations may have occurred during the recombination of switch regions in isotype switching. Candidate oncogenes dysregulated by these translocations have been localized.2-9
Conventional cytogenetics are relatively insensitive in the detection of switch translocations in myeloma, with an overall positivity rate of between 10% and 40%.10-12 Using more sensitive molecular techniques, translocation breakpoints were initially identified in myeloma cell lines and confirmed in a limited number of primary tumors.2,4-9 The most common partner chromosomes and their candidate oncogenes are 11q13 (cyclinD1/bcl-1),4 4p16 (FGFR3 andMMSET or WHSC1),5,7,9,13 6p25 (IRF4),8 and 16q23 (c-maf).6 More recently,myeov and WWOX have been proposed as possible candidate genes for dysregulation on chromosomes 11 and 16, respectively.14,15 These translocations account for approximately 70% of the cell lines analyzed, with the remaining translocations involving a large array of other partners.1,2 More recently, molecular cytogenetic techniques such as fluorescent in situ hybridization (FISH) and spectral karyotyping (SKY) have been applied to a series of primary tumor samples, in which switch translocations have been demonstrated in up to 74%.16,17 However, these represent only a small proportion of the total number of chromosomal aberrations detectable,17,18 many of which are non-recurrent and do not affect the IgH locus. Thus, despite the increased sensitivity of FISH and SKY, individual changes were often found to occur at low frequency, whereas the recurrent defects, which are more likely to be relevant to pathogenesis and clinical outcome, were predominantly those detectable by less sensitive methods such as Southern blotting. For example, t(11;14) was found in 5 (12%) of 43 primary tumor samples,16 and t(4;14) in approximately 15% of 30 patients19 by FISH, whereas t(14;16) was reported in 6 of 50 (12%) tumors by SKY.18
We have analyzed a series of primary myeloma marrow samples, using a Southern hybridization method, previously established by Bergasagel et al,2 to demonstrate “illegitimate recombinations” (see “Materials and methods”) as an indication of IgH switch translocations. The main aims of our study were (1) to demonstrate the presence of illegitimate switch recombinations in primary myeloma tumor by this relatively simple molecular method (readily accessible to a diagnostic molecular laboratory), (2) to assess their frequency of occurrence, and (3) to determine whether there is a relationship between illegitimate recombinations as a single entity and disease behavior. The sensitivity of the assay in each sample was first established by the detection of the physiologic “legitimate” isotype switch. Although illegitimate switches were present in over half of the primary tumor samples, their absence in the remaining 43% may suggest heterogeneity in the molecular pathogenesis of the disease. Analysis of the relationships between the presence of illegitimate recombinations, known prognostic indicators, and overall survival revealed that IgH translocations when analyzed as a single entity do not appear to have prognostic significance and are unlikely to be a feature of disease progression.
Materials and methods
Marrow samples
Sixty myeloma marrow samples from 59 patients were analyzed, together with samples from 2 patients with monoclonal gammopathy of uncertain significance (MGUS). Their disease status was classified as stable or progressive by clinical assessment and laboratory investigations. The following parameters were assessed: lytic lesions, full blood count, serum calcium, serum creatinine and creatinine clearance, paraprotein level, Bence Jones proteinuria, serum β2 microglobulin (β2m), and STK.20
Southern hybridization
Mononuclear cells were purified from marrow samples by Ficoll-Hypaque density centrifugation. DNA was extracted, digested byHindIII, SphI, and BglII, electrophoresed on 0.8% agarose gels, and transferred to nylon membranes by 6× standard sodium citrate (SSC). Five pairs of probes, complementary to sequences 5′ and 3′ of the switch regions and the ςμ/ςδ sequences—Sμ (5′Sμ, 3′Sμ), Sγ (5′Sγ,3′Sγ), Sα (5′Sα,3′Sα), Sε (5′Sε,3′Sε), and ςμ/ςδ (5′ςμ,3′ςδ)—were amplified by polymerase chain reaction (PCR) from normal genomic DNA and gel purified; PCR conditions and primer sequences were as previously published.2 Legitimate switches, which occur during normal isotype switching and involve the recombination of 2 switch regions, are indicated by a recombinant fragment hybridized by 2 switch probes from different regions; that is, the recombinant fragments are “matched.” In contrast, an “illegitimate switch,” defined as involving only one switch region, can result from the recombination of the IgH switch region with other nonimmunoglobulin sequences in a chromosomal translocation. This is indicated by a recombinant fragment hybridized by only one switch probe.
For each sample, the following hybridizations were first performed:HindIII— 5′Sμ, 3′Sμ, 5′Sγ, 3′Sγ; SphI— 5′Sα, 3′Sα, 5′Sε, 3′Sε; BglII— 5′ςμ, 3′ςδ. To determine whether a recombinant fragment represented a legitimate (matched) or illegitimate (unmatched) recombination, the same restriction digest was then hybridized by all the other probes that had demonstrated recombinant bands in the “screening” blots. Germline controls of genomic DNA from peripheral blood leukocytes were similarly analyzed and compared with the tumor, to determine whether extra bands were nongermline and tumor specific. Only those samples in which a legitimate switch was demonstrated were amenable to analysis. In 7 samples, HindIII/BamHI-digested DNA was hybridized with a JH region probe (5.4-kbBamHI/HindIII fragment flanking J1-J6 sequences) to detect clonal JH rearrangements. Where present they were found to correlate with a detectable legitimate isotype switch.
To confirm the illegitimate nature of a recombinant band, DNA was digested with the 2 other enzymes referred to above and hybridized by the same probe. Where the enzymes chosen were not informative (depending on the presence of the restriction site on the unknown translocated fragment), 2 steps were taken: (1) for recombinations in the Sμ region, probes within the same region—5′Sμ/5′ςμ and 3′Sμ/3′ςδ—were used to demonstrate the presence of the same band and (2) digestion by the informative enzyme was repeated at least twice to confirm veracity of the recombinant fragment, thereby excluding the possibility that any extraneous bands were the result of partial DNA digestion.
Analysis of cyclin D1 and FGFR3 expression
Cyclin D1 real-time quantitative reverse transcription-PCR.
Real-time quantitative reverse transcription (RT)-PCR was performed on the ABI Model 7700 Sequence Detector using the double-stranded DNA specific fluorophore Sybr Green I.21 Messenger RNA (mRNA) transcripts were quantitated in “real time” as the PCR products were formed, by incorporating and monitoring fluorescence at every cycle of amplification. Total RNA was extracted by standard methods.22 One microgram total RNA was reverse transcribed into complementary DNA (cDNA) using an oligo(dT)15 primer and avian myeloblastosis virus (AMV) RT in a buffer containing ribonuclease inhibitor 20 U, 1 mM each of the 4 deoxynucleotides, 5 mM MgCl2, 10 mM Tris-HCl pH 8.8, 50 mM KCl, and 0.1% Triton X-180 in a 20-μL volume, by incubation at 42°C for 15 minutes followed by inactivation of the RT at 99°C for 5 minutes. All reagents were supplied by Promega (Southampton, United Kingdom).
Cyclin D1 cDNA comprising exons 3 and 4 (196 bp) was amplified by 200 nM primers (5′-AAC AGA TCA TCC GCA AAC AC-3′ and 5′-TCA CAC TTG ATC ACT CTG GA-3′) in a PCR reaction with 200 mM dNTPs, 3 mM MgCl2, 0.5 × Sybr Green I (Perkin Elmer, Foster City, CA), Perkin Elmer buffer A (Perkin Elmer) containing the passive reference fluorochrome 6-carboxy-X-rhodamine and Amplitaq Gold 1.25 U (Perkin Elmer) in a total volume of 25 μL. Forty cycles of amplification of standard and patient samples were performed, with denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 1 minute. Using the ABI Sequence Detector, fluorescence data were acquired at 72°, 83°, 86°, and 89°C, and the temperature at which the least primer-dimer formation occurred was chosen for analysis. β-Actin cDNA was amplified in separate reactions using primers 5′-TCG ACA ACG GCT CCG GCA TGT TGC AAG-3′ and 5′-AGC CAC ACG CAG CTC ATT GTA GAA G-3′ to give a 263-bp product as an internal control, using the same conditions except that the reaction mixture contained 2 mM MgCl2 and annealing was performed at 67°C. DNA standards were prepared by amplifying and purifying cyclin D1 and β-actin cDNA from K562 human erythroleukemia cell line RNA, quantitated by spectrophotometry, from which standard preparations of 103 to 107 copies were prepared. All samples were measured in duplicate. Fluorescence was plotted against PCR cycle number, from which the threshold cycle (at which a statistically significant increase in fluorescence occurred) was derived for each sample, corresponding to an exponential increase in PCR product. From a standard curve, the initial copy number of each sample was determined from its threshold cycle, and the ratio of target to control β-actin cDNA calculated. The PCR products were gel electrophoresed to confirm specific bands and purified for sequencing.
Detection of FGFR3 cDNA by radioactive RT-PCR.
Total RNA from myeloma marrow samples and normal controls was reverse transcribed as described above. FGFR3 cDNA was identified either by a radiolabeled primer or by hybridization with an internal oligoprobe. In the former, the reverse primer (5′-GTG GTG TGT TGG AGC TCA TG-3′) was first 5′-end labeled with [γ-32P]dATP using T4 polynucleotide kinase (Amersham, Amersham, United Kingdom) in a total volume of 50 μL, containing 10 pmol oligonucleotide, 6 μL [γ-32P]dATP in a buffer of 50 mM Tris-HCl pH 7.6, 10 mM MgCl2, and 10 mM β-mercaptoethanol, at 37°C for 30 minutes. Then, 2 pmol labeled primer (10 μL of the above reaction) was combined with 18 pmol of unlabeled primer for the PCR reaction in a total volume of 100 μL reaction containing 50 mM KCl, 10 mM Tris, 2 mM MgCl2, 2 mM of each dNTP and 0.2 μM of the forward primer (5′-CGG CAG ACG TAC ACG CTG-3′) to produce a 598-bp product. Denaturation at 94°C for 1 minute, annealing at 65°C for 1 minute, and extension at 72°C for 1 minute, was carried out for 25 to 35 cycles with the initial denaturation and extension increased to 4 minutes. RNA extracted from K652 erythroleukemia cell line (from whichFGFR3 was initially cloned)23 was used as positive control. The PCR products were electrophoresed and the gel exposed to film. Second, nonradioactive PCR products were transferred to a positively charged membrane with 0.5 M NaOH and 1.5 M NaCl and hybridized by a [γ-32P]dATP -labeled oligoprobe (5′-GAG TCC AAC GCG TCC ATG AGC-3′) at the 3′ end of the cDNA product, in a buffer of 7% polyethylene glycol (PEG)/10% sodium dodecyl sulfate (SDS) at 42°C.
Results
Of the 60 myeloma marrow samples studied, 24 were from patients with stable disease, 34 from progressive phase, and 2 of unknown clinical status. Two samples from patients with MGUS were also analyzed. Legitimate recombinations were demonstrated in 21 of the 60 myeloma samples (and not in the MGUS cases), comprising 4 stable and 17 progressive disease samples. Characteristics of the patients are presented in Table 1. Marrow samples taken in progressive phase were more heavily infiltrated than those with stable disease and were associated with higher mean levels of β2m and STK. The remaining “uninformative” marrow samples had a lower myeloma cell content, with over half containing less than 25% plasma cells.
Illegitimate switch recombinations were detected in 12 of 21 (57%) informative samples, comprising one of the 4 stable disease samples, and 11 of 17 from patients in progressive phase. These positive cases included 7 of the 12 IgG myeloma marrows, all 3 cases of IgA myeloma, and one case each of κ light chain and IgM myeloma. In 6 of the 12 positive samples, illegitimate recombinations occurred in more than one switch region. The most common switch region involved was Sμ, affecting 11 of 12 cases. Sγ and Sα translocations were detected in 5 and 3 cases, respectively. The myeloma isotypes and switch regions involved in the illegitimate recombinations are shown in Table2. Of the 19 illegitimate recombinant fragments detected, their illegitimate nature was confirmed by using more than one restriction enzyme in 11 of 19 cases, or probes in proximity to each other (5′Sμ/5′ςμ or 3′Sμ/3′ςδ) in 4 of 19 cases (of which 3 were also confirmed by multiple enzymes). In the remaining 7 recombinant fragments where the use of 2 other enzymes was uninformative, the veracity of these bands was confirmed by repeating the digestion and hybridization at least twice. As noted earlier, a legitimate switch results from the recombination of 2 switch regions, such that a legitimate recombinant fragment must be hybridized by the “partner” switch region probe. Thus, the demonstration of hybridization of a recombinant fragment by only one switch probe and not by any others effectively excluded the possibility of a legitimate isotype switch. The most likely cause is an illegitimate switch; although IgH mutations, polymorphisms, deletions, or internal rearrangements are also possible, in practice they are much less likely as indicated by previous analyses of myeloma cell lines (see “Discussion”).2 4-9
Cyclin D1 and FGFR3 are 2 candidate genes previously found to be dysregulated in myeloma cell lines and several primary tumors carrying t(11q13;14q23) and t(4p16;14q23), respectively, 2 of the most frequent switch translocations in myeloma lines.4,5,9,24,25 To confirm the occurrence of switch translocations in the primary myeloma marrow samples, the expression of these 2 genes was examined. Using real-time PCR, up-regulated cyclin D1 expression was detected in 2 of 7 patients examined, with cyclin D1/β-actin cDNA ratios of 15 and 7.4, respectively, compared with the other 5 patients with a mean cyclin D1/β-actin cDNA ratio of 0.2 (range 0.002-0.3). One of these 2 patients provided informative data in the Southern blot assay and was found to have illegitimate recombinations involving Sμ and Sα (Table 2). FGFR3 expression was examined by RT-PCR. Because there is no FGFR3 expression in normal bone marrow, detection of FGFR3 mRNA is indicative of up-regulation.9 23 Of a total of 9 patients examined, 2 were positive. One of the 2 patients had 2 Sα illegitimate recombinations in the Southern blot assay. No illegitimate switch was detected in the other patient (see “Discussion”).
In patients with progressive disease, no significant difference in survival was found between patients with and without illegitimate switch recombinations (Figure 1). There were also no significant differences in β2m between patients with illegitimate recombinations (5.6 ± 7.1 mg/L, range 2-24 mg/L) and patients without illegitimate recombinations (7.5 ± 5.2 mg/L, range 3.2-16.2 mg/L), nor in STK—12.7 ± 7.2 U/L (range 6.1-25 U/L) and 7.0 U/L ± 2.7 U/L (range 2.0-10 U/L), respectively.
Patient survival.
Survival of patients in progressive phase, with (closed boxes) and without (closed triangles) illegitimate switch recombinations. No significant difference was found (P = NS).
Patient survival.
Survival of patients in progressive phase, with (closed boxes) and without (closed triangles) illegitimate switch recombinations. No significant difference was found (P = NS).
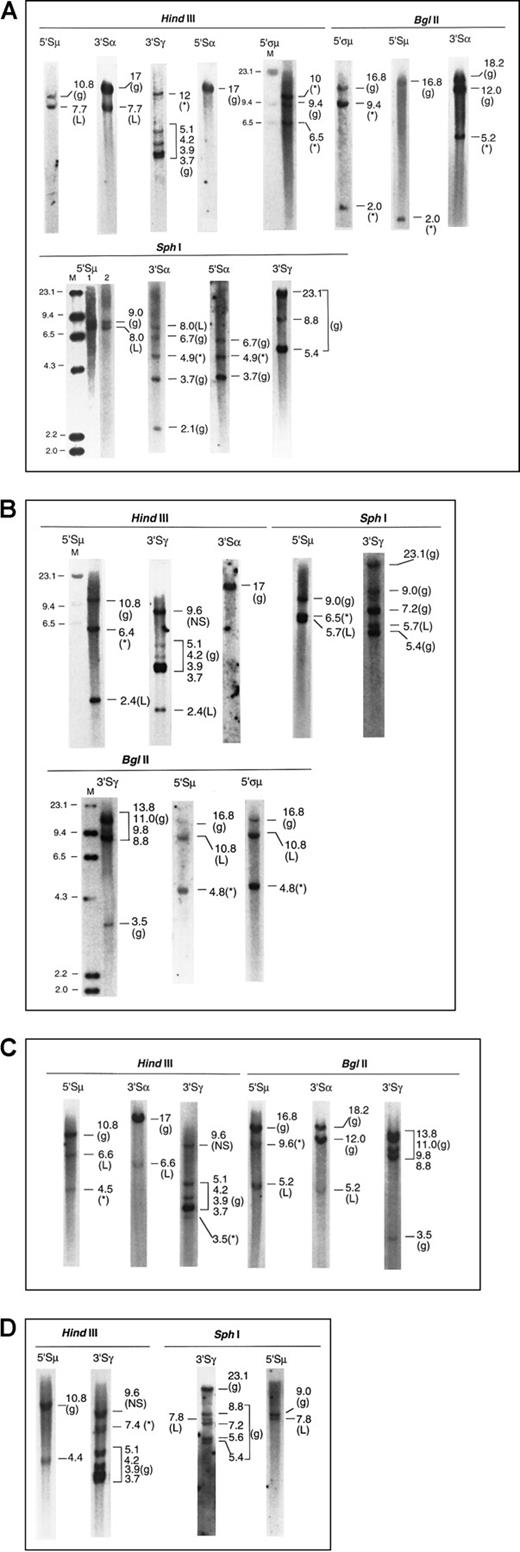
Four examples of the Southern blot analysis are presented in Figure2—2 cases of IgG and 2 of IgA myeloma. Details of the analysis are summarized in the legend.
Southern blots of 4 myeloma marrow samples with illegitimate recombinations.
The g denotes germline; L, legitimate recombination; *, illegitimate recombination; M, λHindIII molecular size marker, sizes indicated in kb. (A) Patient 1. IgAκ myeloma (1) μ to α isotype switch detected by matched 7.7-kb fragments inHindIII-5′Sμ and 3′Sα, and 8.0-kb fragments inSphI-5′Sμ and 3′Sα. Lanes 1 and 2 ofSphI-5′Sμ represent 2 hybridizations of separateSphI digests. (2) Illegitimate fragments detected in (i)HindIII-3′Sγ, 12 kb; absent in HindIII-3′Sα and 5′Sα, (ii) SphI-3′Sα and 5′Sα, 4.9 kb, suggesting a translocation upstream of 5′Sα or downstream of 3′Sα; absent inSphI-5′Sμ and 3′Sγ. The 5.2-kb band shown inBglII-3′Sα is also unmatched in BglII-5′ςμ or 5′Sμ, 5′Sα, and 3′Sγ (latter two not shown). Thus the presence of an illegitimate recombination in Sα has been confirmed by 2 restriction enzymes. (iii) BglII-5′ςα showed 9.4-kb and 2.0-kb fragments (the latter also detected by the 5′Sμ probe in close proximity to it), not present in Bgl II-3′Sα, 5′Sα, and 3′Sγ (latter two not shown). These recombinant bands are confirmed by 10 and 6.5 kb HindIII fragments detected by 5′ςμ. (NB: The 5′ςμ probe is located upstream of the HindIII site 5′ of the Sμ region, such that the 5′ςμ and 5′Sμ probes are located 5′ and 3′ of the HindIII site, respectively. Hence, the HindIII germline fragment detected by 5′ςμ does not contain the Sμ switch region.). (B) Patient 2. IgGκ myeloma (1) μ to γ isotype switch detected by 2.4-kb HindIII and 5.7-kbSphI fragments hybridized by 5′Sμ and 3′Sγ. Of the two recombinant BglII fragments (10.8 and 4.8 kb) hybridized by the 5′Sμ and 5′ςμ probes, the 10.8-kb fragment is likely to represent the legitimate switch, a matched fragment hybridized by 3′Sγ being obscured by the germline bands. (2) Illegitimate 6.4-kbHindIII fragment hybridized by 5′Sμ; absent inHindIII-3′Sγ and 3′Sα. This is confirmed by the 6.5-kbSphI fragment hybridized by 5′Sμ and the 4.8-kbBglII fragments hybridized by both 5′Sμ and 5′ςμ. Thus, the illegitimate recombination involving Sμ has been confirmed by 3 restriction enzymes. A nonspecific (NS) HindIII fragment that was hybridized by 3′Sγ was detected in both myeloma and germline control digests. (C) Patient 3. IgAκ myeloma (1) μ to α isotype switch detected by matched 6.6-kb HindIII fragments and 5.2-kb BglII fragments hybridized by 5′Sμ and 3′Sα. (2) Illegitimate 4.5-kb HindIII fragment hybridized by 5′Sμ but not by 3′Sα or 3′Sγ. This may be confirmed by the 9.6-kbBglII fragment detected by the 5′Sμ but not the 3′Sα probe, although a matching fragment hybridized by the 3′Sγ probe may be obscured by the BglII germline bands. (3) Illegitimate 3.5-kb HindIII fragment hybridized by 3′Sγ; not detected by the 5′Sμ and 3′Sα probes. (D) Patient 4. IgGκ myeloma (1) μ to γ isotype switch demonstrated by matched 7.8-kb SphI fragments hybridized by 5′Sμ and 3′Sγ. The 4.4-kbHindIII fragment hybridized by 5′Sμ probably represents the same legitimate switch, but a matching 3′Sγ fragment would be obscured by the germline fragment of similar size. (2) Illegitimate 7.4-kb HindIII fragment hybridized by 3′Sγ; absent in hybridization with 5′Sμ, 3′Sμ, 5′Sγ, 5′Sα, and 3′Sα. A nonspecific (NS) HindIII fragment that was hybridized by 3′Sγ was detected in both myeloma and germline control digests.
Southern blots of 4 myeloma marrow samples with illegitimate recombinations.
The g denotes germline; L, legitimate recombination; *, illegitimate recombination; M, λHindIII molecular size marker, sizes indicated in kb. (A) Patient 1. IgAκ myeloma (1) μ to α isotype switch detected by matched 7.7-kb fragments inHindIII-5′Sμ and 3′Sα, and 8.0-kb fragments inSphI-5′Sμ and 3′Sα. Lanes 1 and 2 ofSphI-5′Sμ represent 2 hybridizations of separateSphI digests. (2) Illegitimate fragments detected in (i)HindIII-3′Sγ, 12 kb; absent in HindIII-3′Sα and 5′Sα, (ii) SphI-3′Sα and 5′Sα, 4.9 kb, suggesting a translocation upstream of 5′Sα or downstream of 3′Sα; absent inSphI-5′Sμ and 3′Sγ. The 5.2-kb band shown inBglII-3′Sα is also unmatched in BglII-5′ςμ or 5′Sμ, 5′Sα, and 3′Sγ (latter two not shown). Thus the presence of an illegitimate recombination in Sα has been confirmed by 2 restriction enzymes. (iii) BglII-5′ςα showed 9.4-kb and 2.0-kb fragments (the latter also detected by the 5′Sμ probe in close proximity to it), not present in Bgl II-3′Sα, 5′Sα, and 3′Sγ (latter two not shown). These recombinant bands are confirmed by 10 and 6.5 kb HindIII fragments detected by 5′ςμ. (NB: The 5′ςμ probe is located upstream of the HindIII site 5′ of the Sμ region, such that the 5′ςμ and 5′Sμ probes are located 5′ and 3′ of the HindIII site, respectively. Hence, the HindIII germline fragment detected by 5′ςμ does not contain the Sμ switch region.). (B) Patient 2. IgGκ myeloma (1) μ to γ isotype switch detected by 2.4-kb HindIII and 5.7-kbSphI fragments hybridized by 5′Sμ and 3′Sγ. Of the two recombinant BglII fragments (10.8 and 4.8 kb) hybridized by the 5′Sμ and 5′ςμ probes, the 10.8-kb fragment is likely to represent the legitimate switch, a matched fragment hybridized by 3′Sγ being obscured by the germline bands. (2) Illegitimate 6.4-kbHindIII fragment hybridized by 5′Sμ; absent inHindIII-3′Sγ and 3′Sα. This is confirmed by the 6.5-kbSphI fragment hybridized by 5′Sμ and the 4.8-kbBglII fragments hybridized by both 5′Sμ and 5′ςμ. Thus, the illegitimate recombination involving Sμ has been confirmed by 3 restriction enzymes. A nonspecific (NS) HindIII fragment that was hybridized by 3′Sγ was detected in both myeloma and germline control digests. (C) Patient 3. IgAκ myeloma (1) μ to α isotype switch detected by matched 6.6-kb HindIII fragments and 5.2-kb BglII fragments hybridized by 5′Sμ and 3′Sα. (2) Illegitimate 4.5-kb HindIII fragment hybridized by 5′Sμ but not by 3′Sα or 3′Sγ. This may be confirmed by the 9.6-kbBglII fragment detected by the 5′Sμ but not the 3′Sα probe, although a matching fragment hybridized by the 3′Sγ probe may be obscured by the BglII germline bands. (3) Illegitimate 3.5-kb HindIII fragment hybridized by 3′Sγ; not detected by the 5′Sμ and 3′Sα probes. (D) Patient 4. IgGκ myeloma (1) μ to γ isotype switch demonstrated by matched 7.8-kb SphI fragments hybridized by 5′Sμ and 3′Sγ. The 4.4-kbHindIII fragment hybridized by 5′Sμ probably represents the same legitimate switch, but a matching 3′Sγ fragment would be obscured by the germline fragment of similar size. (2) Illegitimate 7.4-kb HindIII fragment hybridized by 3′Sγ; absent in hybridization with 5′Sμ, 3′Sμ, 5′Sγ, 5′Sα, and 3′Sα. A nonspecific (NS) HindIII fragment that was hybridized by 3′Sγ was detected in both myeloma and germline control digests.
Discussion
In this study, a Southern blot method was used to examine a cohort of 60 primary myeloma marrow samples. The assay was considered sufficiently sensitive to provide reliable information on 21 samples, based on the detection of one or more legitimate recombinations. Illegitimate recombinations were found in 12 of these 21 samples (57%), of which one was from a patient in stable phase and 11 from patients with progressive disease.
The sensitivity of the assay appeared to depend on the number of malignant plasma cells in the marrow sample. Thus, by confining our analysis to samples in which legitimate switches were demonstrated, it is unlikely that the absence of an illegitimate recombination was due to insensitivity of the method. Not surprisingly, most of the informative samples were derived from patients in progressive phase, with greater myeloma cell content. IgH translocations may not be detected if they occur outside the switch regions, or if the restriction sites are not appropriate for a translocation. Thus, the frequency reported here may be an underestimate. Although some illegitimate recombinations may represent mutations, polymorphisms, deletions, or internal rearrangements of IgH, these are likely to represent a minority. In our series, 12 of the 19 illegitimate recombinant fragments were confirmed to be illegitimate by multiple enzymes and/or more than one probe in the same region, whereas the remaining 7 fragments were examined by repeated DNA digestion and hybridization. Moreover, all the illegitimate recombinations detected in a series of 21 myeloma cell lines were shown previously to be switch region translocations.2 4-9
Despite the relatively small numbers of informative samples, our results confirmed the presence of illegitimate recombinations as an indication of IgH switch translocations in primary myeloma tumor. Using this basic molecular assay, the detection rate of 57% compared favorably with results from conventional cytogenetics (frequency of detection 10-40%),10-12 for which insensitivity has been attributed to the difficulty in obtaining metaphases from infrequently dividing cells, and the telomeric location of the IgH genes and breakpoints. A different Southern blot assay designed to detect differences in migration of J and C fragments yielded a slightly lower rate of 25%.9 Until the advent of molecular cytogenetics, switch translocations in myeloma cell lines had been confirmed in only a limited number of primary tumors. In contrast, larger series of primary tumors were examined by FISH and SKY.16-18,26Nishida et al found an incidence of 74% and 51% in interphase and metaphase FISH, respectively.16 More recently, Avet-Loiseau et al demonstrated IgH translocations in 47% cases by FISH.26 Our rate of detection is comparable with the latter estimates of incidence. When a screen of the entire karyotype was performed by chromosome painting in SKY,18 IgH translocations were found in 17 of 50 patients (34%) excluding the most frequent t(11;14) translocation, which is readily detectable by conventional karyotype, among a large number of nonrecurrent abnormalities. Recombinations with 4 known partners—t(11;14), t(6;14), t(8;14), and t(14;16)—all of which had been detectable by Southern blot,2,4-9 accounted for 13 of the 17 IgH translocations. It would therefore appear that most if not all the recurrent IgH rearrangements are detectable by basic molecular techniques such as the method used here, which is readily applicable to a diagnostic laboratory. In another SKY study of 8 marrow biopsy specimens, none of the changes found were recurrent.17
The finding here of up-regulated expression of cyclin D1 andFGFR3, candidate genes in the t(11;14) and t(4;14) translocations, is consistent with the presence in our primary myeloma samples of switch translocations involving the most frequent chromosomal partners detected in myeloma cell lines. Why one of the subjects had up-regulated FGFR3 expression without evidence of an illegitimate switch is not known. As noted earlier, it is possible for translocations to occur outside the switch regions, or up-regulation may have occurred by mechanisms other than a switch translocation. However, taken together, our results on cyclin D1 and FGFR3 expression lend additional support to our conclusions and the potential usefulness of this accessible molecular assay in the evaluation of patients.
Our results also show that a substantial number of primary myeloma tumors (43%) did not contain illegitimate recombinations, when the clonal isotype switch recombinations were clearly demonstrated. The absence of IgH translocations in myeloma tumors has also been noted in several molecular cytogenetics studies.16,18,27 In the multistep transformation hypothesis, IgH translocations are considered to be an early event.1 3 From these and previous results, a significant proportion of tumors would appear to have arisen in the absence of IgH rearrangement, suggesting possible heterogeneity in pathogenesis.
In our series, illegitimate recombinations involving more than one switch region occurred in 6 of 12 positive samples. This may be consistent with the finding of complex translocations in molecular cytogenetic studies. Whether these changes are cumulative or are the result of treatment or genetic instability is not known, although it is interesting to note that in one of the 6 cases, the multiple changes were detected at diagnosis (patient 1, Figure 2A).
Our results do not suggest any relationship between the frequency of illegitimate recombinations and disease status or prognosis, because there was no difference in survival or the known prognostic indicators of β2m or STK between patients with and without illegitimate recombinations in the progressive disease group. In contrast, some investigators have suggested that IgH translocations may be a feature of active disease, because most of them were initially identified in myeloma cell lines propagated from aggressive myeloma tumor. Moreover, structural chromosomal aberrations detected by conventional karyotype were also found to correlate with poor disease outcome, with abnormalities involving chromosomes 11 and 13 conferring the most unfavorable prognosis.28,29 However, in concurrence with our findings, subsequent analyses by molecular cytogenetics have failed to demonstrate any correlation between chromosomal aberrations detectable by FISH and disease stage or β2m, or a significant difference between tumors at diagnosis and relapse.27 Although we have shown that as a single entity IgH switch translocations are not prognostically significant, it is possible that different partner loci may confer different prognostic value. This will be clarified by further analysis of chromosomal partners as the basis of subgroup analysis of disease behavior.
Acknowledgments
We would like to acknowledge the assistance of Prof G. McCaughan and his laboratory for the use of the ABI Model 770 Sequence Detector.
Supported in part by grants from the Leo and Jenny Leukaemia and Cancer Foundation, and the Royal Australasian College of Physicians.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
P. Joy Ho, Institute of Haematology, Royal Prince Alfred Hospital, Missenden Road, NSW 2050, Australia; e-mail:j.ho@centenary.usyd.edu.au.