Abstract
Following stress signals, the p53 tumor suppressor protein plays a critical role in regulation of cell proliferation, mainly through induction of growth arrest or apoptosis. Therefore, this protein needs to be strictly regulated and numerous studies have shown that the MDM2 protein is an essential element for p53 regulation in normal cells and, most importantly, that overexpression of MDM2 is responsible for p53 inactivation in various types of tumors. A previous study showed that this is the case in some Burkitt lymphoma (BL) cell lines, where enhanced translation of mdm2 messenger RNA results in overexpression of the protein that complexes and inactivates wild-type p53. To further investigate the role of the p53/MDM2 complex in these BL cells, as well as in other lymphoid cells that do not overexpress MDM2, this study used antisense oligodeoxynucleotides directed either against mdm2 or against p53. Results show that the mdm2 antisense oligodeoxynucleotide induces apoptosis of cells that express a high or low level of MDM2 protein, only if they contain wild-type p53. Moreover, apoptosis is independent of the accumulation of p53 following mdm2 antisense treatment. Finally, the p53 antisense oligodeoxynucleotide, which inhibits the expression of wild-type p53, also induces a decrease of the MDM2 level in cells, whether or not they overexpress this protein, and causes apoptosis of these cells. These results indicate that decreasing the MDM2 protein level by directly or indirectly targeting its biosynthesis is a potent tool for the induction of apoptosis.
Introduction
The tumor suppressor p53 is a multifunctional protein that has a critical role in various pathways controlling cellular responses to stress signals.1,2 The main mechanism whereby this molecule controls cell growth is through transactivation of genes involved either in cell cycle arrest (p21WAF1, 14-3-3 ς, orGADD45) or in induction of apoptosis (Bax or thePIGs genes) (reviewed in reference 3). It has been amply demonstrated that the loss of p53 function is a key event in tumorigenesis, and abnormalities of the p53 gene, which generally result in inactivation of the protein, are among the most common molecular events in human malignancies (reviewed in references 4 and 5). Moreover, in several tumor types, p53 inactivation can also occur through complex formation with either virally encoded proteins (SV40 T, adenovirus E1A, HPV E6) or the cellular protein MDM2.1
MDM2 was originally identified as one of the genes amplified on double minute particles in a spontaneously transformed mouse 3T3 cell line.6 The oncogenic potential of this molecule was then confirmed, when it was shown that specific overexpression of themdm2 gene increases the tumorigenic properties of mouse or rat tumor cell lines7 8 and when it was observed that the mdm2 gene is amplified in a variety of human tumors, the highest frequency being in soft-tissue tumors and osteosarcomas (reviewed in reference 9).
Although recent data obtained with mdm2 transgenic, p53-null mice indicate that MDM2 could be oncogenic by itself,10,11 it is generally assumed that the transforming properties of this molecule are due to its ability to inhibit the tumor suppressor function of p53 (reviewed in references 12-14). MDM2 negatively regulates p53 function through its ability to bind to the N-terminal transactivation domain of p53,15,16 but recent reports have also shown that it is involved in the degradation of p53. Indeed, MDM2 is apparently able to transport p53 from the nucleus to the cytoplasm17,18 where its ubiquitin-ligase activity19 contributes to the proteasome-mediated degradation of p53. Because themdm2 gene itself is inducible by p53,20 a functional feedback loop exists between these 2 molecules, with p53 inducing expression of MDM2 and MDM2 inhibiting p53 function.21 In addition to being oncogenic when overexpressed, the ability of MDM2 to strictly regulate p53 makes it a critical element for the fate of normal cells, as demonstrated by studies showing that mdm2-deficient mice can only develop in a p53-null background.22 23
Burkitt lymphoma (BL) is a B-cell neoplasm characterized by chromosomal translocations affecting the c-myc locus and leading to the constitutive activation of this oncogene (reviewed in references 24 and 25). In addition to the activation of c-myc, the inactivation of p53 seems to be an important step in the development of this tumor. Indeed, studies have shown that, among hematologic malignancies, one of the highest incidences of p53 mutations is observed in BL (33%-40% of biopsies26,27 and 60%-83% of BL-derived cell lines28,29), and that these mutations result in the inactivation of the protein.30 It was, therefore, proposed that p53 inactivation contributes to the transformation of BL cells, most probably at a stage subsequent to c-myc activation.31 This hypothesis is supported by our recent finding that, in some BL cell lines, inactivation of p53 can also result from the overexpression of MDM2. Of 20 BL cell lines studied, 4 exhibited a wild-type p53 protein associated with high levels of MDM2 and these cells, similarly to p53 mutated BL cell lines, neither showed a G1 arrest nor underwent apoptosis following γ-radiation, but rather showed a strong G2/M arrest.32
To further investigate the role of the p53/MDM2 complex we decided to use antisense oligodeoxynucleotides directed against the corresponding transcripts. These experiments were performed in cells presenting different p53 and MDM2 status: BL cells containing wild-type p53 and overexpressing MDM2; cells with inactivated p53 (p53 mutated BL cells) or without p53 expression (osteosarcoma cells) and no overexpression of MDM2; and lymphoblastoid cells with wild-type p53 and no overexpression of MDM2. Here, we report that the mdm2 antisense (mdm2 AS) induces apoptosis only in cells carrying a wild-type p53 and that the cell death does not necessarily correlate with an accumulation of p53. Moreover, we also demonstrate that treatment of cells containing wild-type p53 with the p53 antisense (p53AS) induces a decrease in both p53 expression and MDM2 level (independent of the basal level of expression of this protein) and causes apoptosis of these cells.
Materials and methods
Cell culture and transfections
The Ramos BL cell line and the Priess lymphoblastoid cell line were kindly given by Pr G. Klein (Stockholm, Sweden); the BL2 cell line was a kind gift of Dr G. Lenoir (Lyon, France). These cell lines were cultured in RPMI 1640 medium (GIBCO-BRL, Paisley, Scotland). The Saos-2 osteosarcoma cell line was obtained from Dr J. Feunteun (Villejuif, France) and was maintained in Dulbecco modified Eagle medium (DMEM, GIBCO-BRL). Two mM l-glutamine, 1 mM pyruvate, 20 mM glucose, 20μg/mL gentamicine, and 5% heat-inactivated fetal calf serum (FCS) were added to these culture media.
For transfection experiments, Saos-2 cells were seeded at 1 × 106 cells per 10-cm dish and after 18 to 24 hours they were transfected by the calcium phosphate method33 with the pCMV-mdm2 expression plasmid34 or the pCMV empty vector (both kindly supplied by Dr D. Trouche (Toulouse, France), which harbor the G418resistance gene. The cells were collected 48 hours after transfection and seeded in 96-well plates (at 1000 and 10 000 cells/well) in DMEM containing G418 (600 μg/mL). Expression of MDM2 was tested by Western blot analysis of cells obtained from wells where only one colony was growing.
mdm2 and p53 oligodeoxynucleotides
All the oligodeoxynucleotides used were phosphorothioate modified to protect them from the cell nucleases. They were obtained from Eurogentec (Seraing, Belgium) or Pharmacia Biotech (Orsay, France). The mdm2 AS is a 15-mer (5′-GTTGGTATTGCACAT-3′) complementary to a sequence beginning at the ATG initiation codon of mdm2 complementary DNA (cDNA).35Its control oligodeoxynucleotide (5′-TCGATGTACGTGATT-3′) was chosen to be a scrambled sequence of the AS to ensure identical nucleotide content and minimize differences potentially attributable to nucleic acid content. The p53 AS is an 18-mer targeting the region of the initiation codon (6 base pairs immediately before the first codon and the first 4 coding codons): 5′-CGGCTCCTCCATGGCAGT-3′. This AS was previously used by Bi and coworkers.36 The p53 control oligodeoxynucleotide (5′-CTGAGGCTCTACCGCTGC-3′) was also a scrambled sequence of the p53 AS.
Electropermeabilization
Electropermeabilization of the cells was performed with a PS-15 electropulsator (Jouan, Saint-Herblain, France). Electrodes were 2 stainless steel plates that were 2 mm apart. Cells were electroporated in 50μL RPMI 1640 medium containing 1% FCS, at a concentration of 2 × 106 cells/50μL, with 80μM oligodeoxynucleotides. Cells were deposited between the electrodes and immediately exposed to 8 square-wave electric pulses of 100 μs and 280 V delivered at a frequency of 1 Hz. Immediately after electroporation, cells were resuspended in 4 mL RPMI 1640 medium containing 5% FCS and cultured at 37°C.
Western blot analysis
Cell pellets, resuspended in lysis buffer (50 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 100 mM DTT, 1 mM aprotinin), were sonicated and then incubated for 10 minutes at 4°C. Lysates were then diluted in loading buffer (50 mM Tris-HCl, pH 7.4, 2% SDS, 9% glycerol, 0.7 M β-mercaptoethanol, 0.005% bromophenol blue) and proteins were separated on a SDS-7.5% polyacrylamide gel electrophoresis [PAGE]. After electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA) by a semidry electroblotting system (Millipore). After blocking with phosphate-buffered saline (PBS) containing 3% nonfat dry milk and 2% glycine, blots were probed with either an anti-MDM2 monoclonal antibody (mAb) (IF2, Oncogene Research, Boston, MA), or an anti-p53 mAb (DO1, Santa Cruz Biotechnology, Santa Cruz, CA) or an anti-PARP mAb (C2-10, Oncogene Research) or an anti-β actin mAb (AC-74, Sigma, L'isle d'Abeau Chesnes, France). All these antibodies were used at a concentration of 2 μg/mL. After incubation with a rabbit antimouse IgG (ICN, Costa Mesa, CA), the blots were revealed with [125I]-protein A (Amersham, Orsay, France) and submitted to autoradiography.
Induction of apoptosis
Apoptosis was either induced by cytotoxic agents or by γ-irradiation. Saos-2 parental cells (5 × 105) or the various stable mdm-2 transfectants were taken in 4 mL DMEM containing okadaı̈c acid at 20 nM or doxorubicin at 100 ng/mL and incubated for 48 hours at 37°C ; 5 × 105cells were irradiated at room temperature (8 Gy) using a137Cs source delivering 1.57 Gy/min and then incubated for 72 hours at 37°C. All cells (adherent and detached) were then collected and cell viability was measured.
Cell viability
After treatment, cells were centrifuged, resuspended in HEPES buffer (10 mM HEPES/NaOH, pH7.4, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2) containing 2.5 μg/mL fluorescein isothiocyanate (FITC)-labeled annexin V (Euromedex, Mundolsheim, France) and incubated at 4°C for 5 minutes. They were then washed twice in HEPES buffer and propidium iodide (10 μg/mL) was added immediately before analyzing the cells with a FACSCalibur (Becton Dickinson, San Jose, CA) cytofluorometer. The percentage of apoptotic cells is: % annexin V (+) PI(−) + % annexin V(+) PI(+).
Results
Effects of mdm2 AS on MDM2 and p53 protein levels
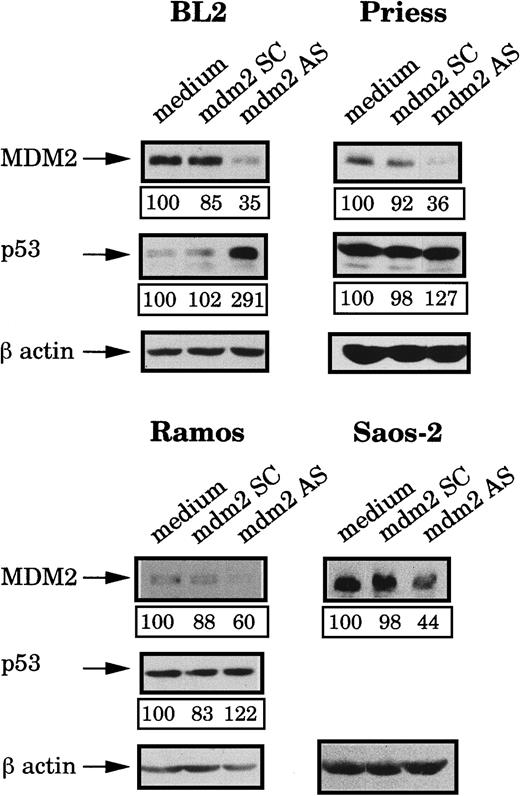
We have previously reported that p53 is functionally inactivated in 4 BL cell lines (BL2, Ly47, Ly91, and Seraphina), which overexpress MDM2 protein.32 To further study the function of MDM2, we then decided to treat, with an mdm2 AS oligodeoxynucleotide, various cell lines having different MDM2 and p53 status: BL2 (BL cells, MDM2 high, wild-type p53), Ramos (BL cells, MDM2 low, mutant p5329), Priess (lymphoblastoid cells, MDM2 low, wild-type p53), and Saos-2 (osteosarcoma cells, MDM2 low, p53-null37). The levels of expression of MDM2 and p53 were tested by Western blot analysis and are shown in Figure1. As expected, the mutant p53 is greatly accumulated in Ramos cells.
Expression of MDM2 and p53 protein in BL2, Saos-2, Priess, and Ramos cell lines.
Equal quantities of protein extracts obtained from the indicated cell lines were separated on 7.5% SDS-PAGE, transferred to PVDF membranes, and probed with either an anti-MDM2 mAb (IF2) or anti-p53 mAb (DO1). After incubation with a rabbit antimouse IgG, the blots were revealed with [125I]-protein A and submitted to autoradiography.
Expression of MDM2 and p53 protein in BL2, Saos-2, Priess, and Ramos cell lines.
Equal quantities of protein extracts obtained from the indicated cell lines were separated on 7.5% SDS-PAGE, transferred to PVDF membranes, and probed with either an anti-MDM2 mAb (IF2) or anti-p53 mAb (DO1). After incubation with a rabbit antimouse IgG, the blots were revealed with [125I]-protein A and submitted to autoradiography.
An AS phosphorothioate oligodeoxynucleotide targeting the AUG initiation codon of mdm2 messenger RNA (mRNA) (mdm2 AS) and the corresponding scrambled control oligodeoxynucleotide (mdm2 SC), were introduced into cells by electropermeabilization. Our protocol using square-wave electric pulses, which is a modification of the classical electroporation technique, was chosen because it allows efficient delivery of various types of molecules, including oligonucleotides,38 into different types of cells with minimal damages.39-41 Before electropermeabilization, cells were taken in 50μL RPMI 1640 containing 1% FCS, mixed with mdm2 AS or SC (at a concentration of 80 μM) and immediately submitted to the electric pulses (8 pulses of 100μs at 280 V). They were then resuspended in 4 mL RPMI 1640 containing 5% FCS (being therefore in contact with the oligodeoxynucleotides at a concentration of 1 μM).
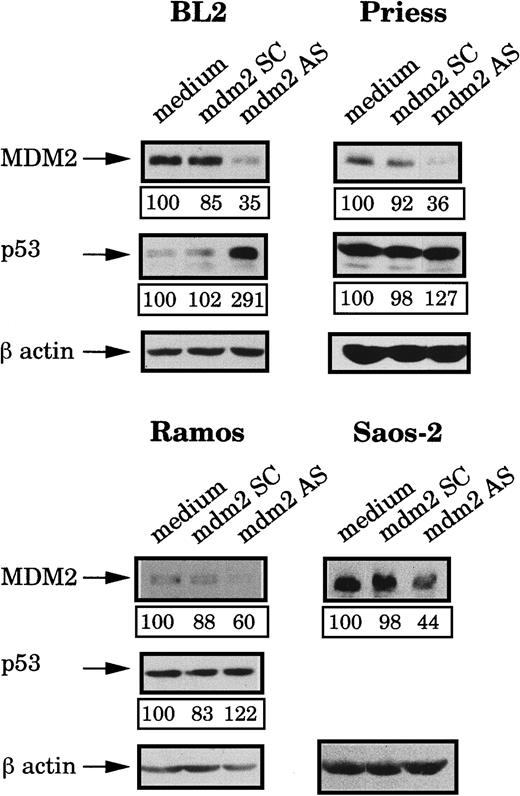
After 4 hours of incubation at 37°C, the MDM2 protein expression was analyzed by Western blot using IF2 mAb and bands were quantified by densitometry (following normalization for β-actin expression). Figure2 shows that, in comparison to cells electroporated with medium alone or with the mdm2 SC, MDM2 expression was inhibited in the 4 cell lines treated with the mdm2 AS. The level of MDM2 protein then gradually increased until 10 hours after electropermeabilization where initial levels were restored (data not shown). Expression of p53 in the treated cells was also tested by Western blot analysis using DO1 mAb. As shown in Figure 2, treatment of the cells with the mdm2 AS for 4 hours, induced various modifications of the p53 level, as compared to the cells electroporated with medium or with the mdm2 SC. A strong increase of the p53 level was observed in BL2 cells, whereas no real accumulation could be seen in Priess or Ramos cells.
Analysis of MDM2 and p53 protein levels after mdm2 AS electropermeabilization.
BL2, Priess, Ramos, and Saos-2 cells were electroporated with either mdm2 AS, a control scrambled (SC) oligonudeoxycleotide, or medium and cultured for 4 hours before being analyzed by Western blot using IF2 anti-MDM2 mAb or DO1 anti-p53 mAb. The blots were subsequently hybridized with AC-74 anti-β-actin mAb to verify loading. The level of protein expression indicated below each band was obtained by densitometry following normalization for β-actin expression.
Analysis of MDM2 and p53 protein levels after mdm2 AS electropermeabilization.
BL2, Priess, Ramos, and Saos-2 cells were electroporated with either mdm2 AS, a control scrambled (SC) oligonudeoxycleotide, or medium and cultured for 4 hours before being analyzed by Western blot using IF2 anti-MDM2 mAb or DO1 anti-p53 mAb. The blots were subsequently hybridized with AC-74 anti-β-actin mAb to verify loading. The level of protein expression indicated below each band was obtained by densitometry following normalization for β-actin expression.
Effects of mdm2 AS on cell viability
We then examined the effects of the mdm2 AS on cell viability. At various times after electropermeabilization (8, 12, 18, and 24 hours), cells were labeled with annexin V-FITC and propidium iodide (PI) and analyzed with a FACSCalibur flow cytometer. No effects on cell viability were observed after 8, 12, and 18 hours (data not shown), whereas apoptotic cells could be seen in some samples 24 hours after treatment. For BL2 cells, it can be seen in Table1 that treatment with the mdm2 AS strongly increased the number of apoptotic cells, which increased from 20% ± 3% (for the cells electroporated with medium) and 25% ± 8% (for the mdm2 SC-treated cells) to 54% ± 9%. Because the Priess cells do not overexpress MDM2 and do not accumulate p53 after AS treatment, we were surprised to see that the mdm2 AS also increased the mortality of the Priess cells (from 28% ± 1% and 33% ± 3% in the control cells to 44% ± 2%). In contrast, no effect of the mdm2 AS could be observed on the viability of Ramos or Saos-2 cells.
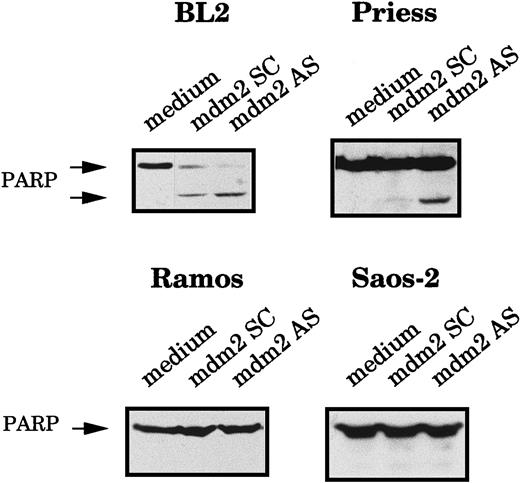
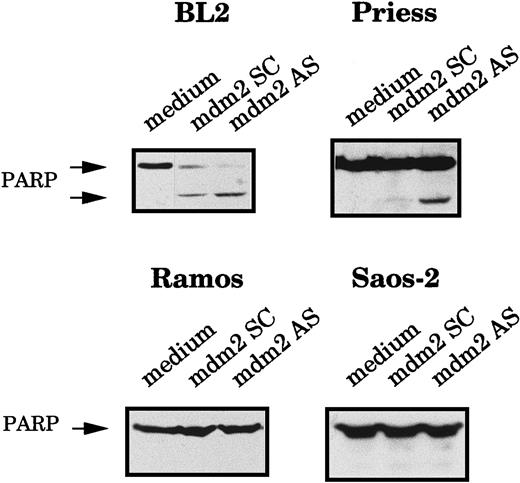
To confirm that the AS-treated cells were truly dying by apoptosis, we also measured the cleavage of the poly-ADP ribose polymerase (PARP), an enzyme involved in DNA repair, which is generally cleaved during apoptosis.42 The expression of the 116- kd native PARP protein and its 85-kd cleavage product were analyzed by Western blot using C2-10 mAb. The PARP cleavage product was clearly visible in BL2 and Priess cells electroporated with mdm2 AS, whereas it was not detected in Ramos and Saos-2 cells (Figure3).
Analysis of the PARP cleavage after mdm2 AS electropermeabilization.
BL2, Priess, Ramos, and Saos-2 cells were electroporated with either mdm2 AS, a control scrambled (SC) oligodeoxynucleotide, or medium and cultured for 20 hours before being analyzed by Western blot using C2-10 anti-PARP mAb.
Analysis of the PARP cleavage after mdm2 AS electropermeabilization.
BL2, Priess, Ramos, and Saos-2 cells were electroporated with either mdm2 AS, a control scrambled (SC) oligodeoxynucleotide, or medium and cultured for 20 hours before being analyzed by Western blot using C2-10 anti-PARP mAb.
These results indicate that mdm2 AS induces apoptosis of the cells that contain MDM2 protein (whether this protein is overexpressed or not) and wild-type p53 (whether this protein accumulates after AS treatment or not) but have no effects on the viability of the cells that contain MDM2 and mutated p53 or MDM2 and no p53.
Effects of p53 AS on p53 and MDM2 protein levels
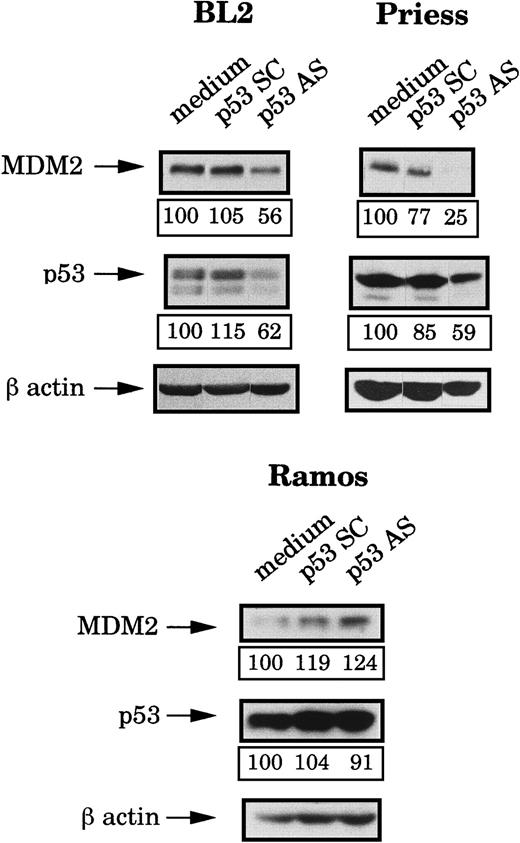
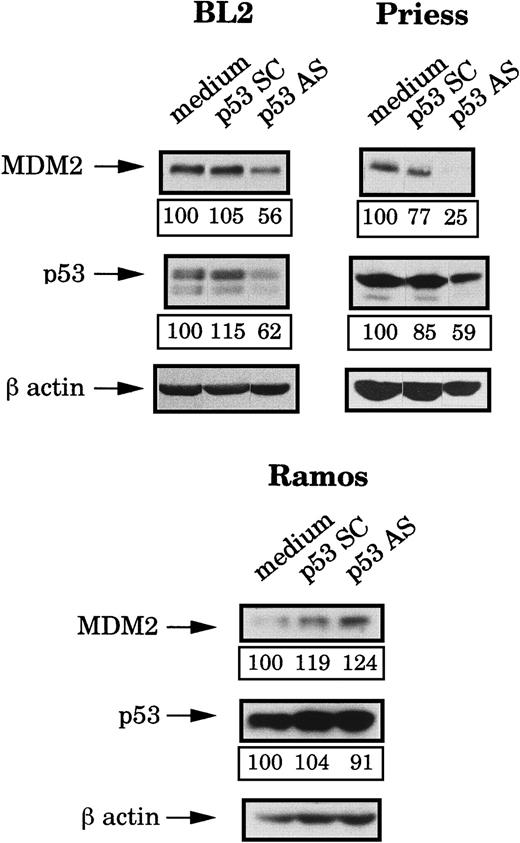
Because the mdm2 AS-induced apoptosis occurred only in wild-type p53 lymphoid cell lines but seemed to be independent of the level of this protein, we decided to use p53 AS to further investigate the role of this protein. First, we tested the effects of a previously described p53 AS36 on the p53 and the MDM2 protein levels. Four hours after electropermeabilization of p53 AS, p53 SC (each at 80 μM during the electroporation and then at 1 μM) or medium, p53 and MDM2 expression were tested by Western blot analysis and the bands were quantified by densitometry (following normalization for β-actin expression). It can be seen in Figure 4that p53 AS substantially reduced (as compared to cells electroporated with medium or p53 SC) the expression of p53 in BL2 and Priess cells but had no effect in Ramos cells. It is possible, however, that this apparent lack of inhibition was only due to the strong accumulation of p53 in these cells, which could mask an effect of the p53 AS on the newly synthesized protein. In Figure 4, it can also be seen that the p53 AS induced a clear decrease of the MDM2 protein level in BL2 and Priess cells. In contrast, no specific change in MDM2 level could be observed after p53 AS in Ramos cells.
Analysis of the p53 and MDM2 protein levels after p53 AS electropermeabilization.
BL2, Priess, and Ramos cells were electroporated with either p53 AS, a control scrambled (SC) oligodeoxynucleotide, or medium and cultured for 4 hours before being analyzed by Western blot using IF2 anti-MDM2 mAb and DO1 anti-p53 mAb. The blots were subsequently hybridized with AC-74 anti-β-actin mAb to verify loading. The level of protein expression indicated below each band was obtained by densitometry following normalization for β-actin expression.
Analysis of the p53 and MDM2 protein levels after p53 AS electropermeabilization.
BL2, Priess, and Ramos cells were electroporated with either p53 AS, a control scrambled (SC) oligodeoxynucleotide, or medium and cultured for 4 hours before being analyzed by Western blot using IF2 anti-MDM2 mAb and DO1 anti-p53 mAb. The blots were subsequently hybridized with AC-74 anti-β-actin mAb to verify loading. The level of protein expression indicated below each band was obtained by densitometry following normalization for β-actin expression.
Effects of p53 AS on cell viability
The viability of the cells treated for 24 hours with p53 AS and p53 SC was then examined by labeling the cells with annexin V-FITC and PI. It can be seen in Table 2 that the p53 AS increased the mortality of BL2 and Priess cells (43% ± 2% and 61% ± 1%, respectively) as compared to cells treated with p53 SC (28% ± 3% and 41% ± 4%) or medium (14% ± 1% and 27% ± 2%). In contrast, no changes were observed for Ramos cells. From these results, it can be concluded that p53 AS induces apoptosis of the cells when a concomitant decrease of wild-type p53 and MDM2 protein levels are observed.
Susceptibility to apoptosis of MDM2-overexpressing Saos-2 cells
Our results show that a decrease of the MDM2 protein level triggers an apoptotic process that is not correlated to the level of the p53 protein but is apparently dependent of the p53 activity. To confirm these data, we decided to check whether the level of MDM2 by itself could modulate the susceptibility of the cells to apoptosis by submitting a series of mdm2 transfectants made in the p53-null Saos-2 cells to various apoptotic stimuli. Saos-2 cells were transfected with an expression plasmid containing the human mdm2 cDNA (pCMV-neo-mdm2) or with the empty vector and after 5 weeks of culture in the selection media (containing G418) expression of MDM2 was tested by Western blot analysis. Among the 20 MDM2-transfectants tested, 5 were chosen because they greatly overexpressed MDM2. None of the control transfectants overexpressed MDM2 (data not shown). Stability of the MDM2 expression was then checked by Western blot every 2 weeks. Cells were treated for 48 hours with okadaic acid (20 nM) or doxorubicin (100 ng/mL) or they were exposed to 8 Gy of γ-radiation and then cultured for 72 hours. As shown in Table 3, okadaic acid, doxorubicin, and γ-radiation induced similar rates of apoptosis in the MDM2-overexpressing transfectants and in the control transfectants. The PARP cleavage was tested for most of the transfectants treated by okadaic acid, doxorubicin, or γ-radiation. Similar data were obtained, whether or not the cells overexpressed MDM2 (data not shown). These results indicate that overexpression of MDM2 in the absence of p53 does not confer to cells an increased resistance to various types of apoptotic stimuli.
Discussion
In this study, we have shown that an antisense oligodeoxynucleotide, which targets the mdm2 gene and is able to specifically reduce the level of the protein, induces apoptosis of cells only when they contain wild-type p53. We have also shown that the inhibition of MDM2 may or may not correlate with an accumulation of p53. We observed that treatment with the mdm2 AS induces accumulation of p53 in BL2 cells, which overexpress MDM2, but not in Priess lymphoblastoid cells, which express a low level of MDM2. It is therefore likely that these results are due to the ability of MDM2 to target p53 degradation. Indeed, it has been shown that, when the 2 proteins are complexed, MDM2 promotes the rapid degradation of p53 via the proteasome pathway.43,44 The exact mechanism is not known, but recent reports indicate that MDM2 is apparently able to fulfill 2 different functions: it shuttles p53 from the nucleus to the cytoplasm17,18 where it participates in the ubiquitination of p53 via its ubiquitin-ligase activity.19 It can be assumed that in BL2 cells that contain a large amount of MDM2, p53 is actively degraded and that a decrease in the level of MDM2 will slow down this degradation process and result in the accumulation of p53. This hypothesis is supported by our observation that, in BL2 cells, p53 accumulation is not due to a transcriptional mechanism because p53 mRNA level was not increased after mdm2 AS treatment (data not shown). This hypothesis also implies that in Priess cells where the basal level of MDM2 is low, its decrease does not modulate the degradation of p53.
Previous reports have also examined the level and the functionality of p53 after AS inhibition of MDM2 expression. Chen and coworkers reported the accumulation/activation of p53 in various tumor cell lines, whether or not these cells overexpressed MDM2,45 and Teoh and associates46 observed an activation of p53 without concomitant stabilization in cells overexpressing MDM2. Together with ours, these results suggest that the regulation of p53 level after AS inhibition of MDM2 expression depends on the cell type and that this regulation probably involves other proteins, such as, for example, p19ARF, Rb, or p300, which are known to form complexes with p53 and MDM2 and to modulate their stability and function.47-49
Our data showing that a decrease in the MDM2 level only results in apoptosis of cells containing wild-type p53, as well as other studies demonstrating that disruption of the p53/MDM2 complex (triggered by peptide homologues of p53 that bind to MDM250,51) also induces growth arrest or apoptosis, certainly reinforce the hypothesis that MDM2 is an important modulator of the p53-mediated apoptosis. Furthermore, our results demonstrate that in absence of p53 the MDM2 level cannot influence the apoptotic process, because we showed that various transfectants of a p53-null cell line, whether or not they overexpress MDM2, are similarly sensitive to various apoptotic stimuli. Because it has been shown recently that MDM2 can interact with p73α (which is expressed in Saos-2 cells) and impede its translational activity,52 our results also indicate that p73α is probably not involved in apoptosis induced by the stimuli that we used.
Finally and most interestingly, we have also demonstrated that, if apoptosis triggered by inhibition of MDM2 certainly depends on p53 activation, this activation is not always correlated with an accumulation of the protein and can even be observed after a decreased expression of p53. Indeed, we have shown that treatment of cells containing wild-type p53 with a p53 AS results in a specific inhibition of p53 expression but also in a decreased level of MDM2 and, subsequently, in the apoptosis of the cells.
In normal cells, p53 is a transcription factor that has an important role in response to stress signals. After DNA damage or hypoxia, for example, p53 induces cell cycle arrest or apoptotic cell death and thus contributes to the elimination of cells containing damaged DNA (reviewed in references 3 and 53). Oncogenic transformation, therefore, often involves the inactivation of this molecule, either by mutation or by formation of complexes with other proteins such as MDM2.1,4,5 Among the multitude of studies carried out to better understand the role of p53 protein in transformation, several have been performed with antisense oligodeoxynucleotides in cells containing either a mutated or a wild-type p53. Although some authors showed that inhibition of p53 expression results in enhanced proliferation of certain tumor cell lines,36,54 several others reported that treatment with p53 AS induces growth inhibition or cytotoxicity.55,56 Very interestingly and unexpectedly, most of these reports underlined that these effects were observed in both mutant and wild-type p53-containing cells.57-59Unfortunately, none of these studies analyzed the effects on the p53 AS on the level of MDM2, but our data suggest that, in cells expressing wild-type p53, it is the decreased level of MDM2 induced by the inhibition of p53 that is responsible for the growth inhibition or the cell death.
The observation that inhibition of p53 expression is accompanied, in BL2 and Priess cells, by a decreased level of MDM2, suggests that in these cells the level of MDM2 is, at least partly, regulated by p53. For BL2 cells, where we have shown previously that p53 is inactivated (following γ-radiation cells neither arrest in G1 nor enter into apoptosis), most probably because it is in complex with the overexpressed MDM2 protein,32 this result was unexpected. However, these apparently conflicting results can be reconciled by keeping in mind that DNA damage leads to activation of p53 by inducing modifications (ie, phosphorylation, acetylation, or redox modulation) of the protein,1,2 whereas p53 AS acts at the mRNA level to prevent the biosynthesis of the protein. Furthermore, in these cells the overexpression of MDM2 is due to enhanced translation.32 It can, therefore, be assumed that, in BL2 cells, p53 normally regulates mdm2 at the transcriptional level but that the p53 protein is unable to respond to a DNA-damaging agent due to the high amount of MDM2.
Taken together our results obtained after exposure to γ-radiation and treatment with antisense oligodeoxynucleotides in BL2 cells indicate that a decrease of MDM2 level induces activation of p53 and apoptosis of the cells, whereas DNA damage does not. These results certainly strengthen the notion that the balance between MDM2 and p53 is of great physiologic importance, and suggest that disrupting this balance in tumor cells overexpressing MDM2 could be more efficient, for eliminating these cells, than treatment with a DNA-damaging agent. However, it must also be considered that inhibition of MDM2 expression in Priess lymphoblastoid cells, which contain a level of MDM2 similar to that in normal lymphoid cells, also results in p53-dependent apoptosis.
Acknowledgments
We are grateful to Annick Harel-Bellan, Christian Jaulin, and Marc Lipinski for helpful discussions. We also thank Colin Robert Gardner for his careful editing of the manuscript.
Supported in part by grants from the Ligue Nationale Française contre le Cancer, the Association pour la Recherche sur le Cancer (ARC 6915), the Fondation pour la Recherche Medicale, and the Fondation de France (comité leucémie).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Joëlle Wiels, Interactions moléculaires et cancer CNRS UMR 1598, Institut Gustave Roussy, 39 rue Camille Desmoulins, 94805 Villejuif Cedex, France; e-mail:wiels@igr.fr.

![Fig. 1. Expression of MDM2 and p53 protein in BL2, Saos-2, Priess, and Ramos cell lines. / Equal quantities of protein extracts obtained from the indicated cell lines were separated on 7.5% SDS-PAGE, transferred to PVDF membranes, and probed with either an anti-MDM2 mAb (IF2) or anti-p53 mAb (DO1). After incubation with a rabbit antimouse IgG, the blots were revealed with [125I]-protein A and submitted to autoradiography.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/4/10.1182_blood.v97.4.1043/5/m_h80410708001.jpeg?Expires=1768185964&Signature=paErh3Ep7SpAhzDUveFGfGKgTHQKwnyFVXkC4mMPaPJWUjYLskxw9iW25Aji-M-7qwO6L9VO7mCmBFEeLcCS~pfFc6pJGMaBR4aT9Yp~zhttVtezgs~9FHOMBNIfK~nKAgDA2V7VE3i3q~RLb1jPF0ze1NwGtXf8zyef55jGG8HOZjx9~p~AGl2WKQ~MDm0DzJAgPEQjOHtyjPfhD3HmsLTQJdkL9nbs0xgkXN6~f5abgZ-RPxIhMbp24H2rsPRTmM1nEolCabzUNgCYI4rKfrwD-ULdHGf4BkuE1X5bfmH54eXQJ3d2p~b55hQ3rOMdSmuI-94OQY1Gumnq3N3rxg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 1. Expression of MDM2 and p53 protein in BL2, Saos-2, Priess, and Ramos cell lines. / Equal quantities of protein extracts obtained from the indicated cell lines were separated on 7.5% SDS-PAGE, transferred to PVDF membranes, and probed with either an anti-MDM2 mAb (IF2) or anti-p53 mAb (DO1). After incubation with a rabbit antimouse IgG, the blots were revealed with [125I]-protein A and submitted to autoradiography.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/4/10.1182_blood.v97.4.1043/5/m_h80410708001.jpeg?Expires=1768502260&Signature=sdVguzNxPxZrT~8wRKj7-fEzpANWiV238IpIJOHs89kOk6t4w-aixfMb0j~WRSkyZWZDM3eNubQbgMUZzfqVqD9~C8iYYtwMxZI5~ke1BXxe1LPpOolnXKe8WVDDLm32WrgpLXEzdsJf7wy53Xcd8cb10m6aiAGjwvKtTBFTKrTJUYbB6TtgcBj2ntvSxXLU~lRfwfYT9PMwKpv~oNzmYUgZ0rcYW9n8Ak8E2M42iZnR6CRL5zgD6oObGEyLxMOziaGE28tUjCua9~99hNAKuwLRssI5-Q8gJVMsRw1DcLvMBS0hUPDVi2Aqd5liqYfMapXzBAPQHzt6RaklcjvRGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)