Chronic lymphocytic leukemia (CLL) has traditionally been viewed as a neoplastic proliferation of naive B cells of the mantle zone, but this view is now changing, with reports of somatically mutated immunoglobulin (Ig) variable heavy chain (VH) genes in CLL.1,2 Recently, Hamblin et al1 and Damle et al2 have independently demonstrated that CLL comprises 2 subsets with either mutated or unmutated VH genes. The mutated CLL cases had a more favorable prognosis and required less treatment than the unmutated cases. Both studies postulated that CLL cells might originate from 2 different stages of B-cell development, ie, pre- or post-GC (germinal center) B cells. Moreover, Damle et al found a strong correlation between VH gene mutational status and CD38 expression in CLL, where unmutated cases displayed a higher percentage of CD38+ cells (> 30%) than mutated cases (< 30%).1 Damle et al proposed that both VH gene mutational status and CD38 expression could be used as novel prognostic indicators of clinical outcome in CLL. But in a follow-up study by Hamblin et al,3 they could not confirm any association between Ig mutational status and CD38 expression, although a significantly poorer prognosis was observed among the cases with more than 30% CD38+.3 We have analyzed the VH gene mutational status in 107 B-cell CLL (B-CLL) cases, and moreover, we have evaluated the CD38 expression in 48 of these cases to further clarify the correlation between VHgene mutations and CD38 expression.
DNA was extracted mainly from lymph nodes and peripheral blood lymphocytes, and VH gene family–specific polymerase chain reaction (PCR) amplification was performed as previously described.4 The majority of samples were sequenced directly using an automated DNA sequencer (ABI 377, Applied Biosystems, Foster City, CA), and the nucleotide sequences were compared to the BLASTN and V-BASE databases. Less than 98% homology to the corresponding germline gene was defined as a mutated VHgene. In a subset of cases, the surface expression of CD38 was evaluated using 2-color flow cytometry and direct conjugate antibodies (anti-CD19–FITC/anti-CD38–PE).
We amplified and sequenced 115 clonal Ig rearrangements in 107 B-CLL cases, where 8 cases displayed 2 different Ig rearrangements within the same tumor sample. In accordance with previous studies, overexpression of the VH1 family (31.3%) and, particularly, the VH1-69 gene (16.5% of all rearrangements detected) was found.1,2,4 Forty-eight cases (44.9%) showed mutated VH genes, whereas 59 cases (55.1%) were unmutated. The VH1-69 gene was exclusively expressed in unmutated cases with a high frequency (28% of unmutated VH genes), and the VH3-21 gene was mainly found expressed in the mutated cases (19% of mutated, as opposed to 3% of unmutated, VH genes). Interestingly, 1 of 8 cases displaying 2 VH rearrangements showed discrepancy in the mutation pattern between the 2 VH genes (94.4% versus 100% homology). Analysis of distribution of replacement (R) and silent mutations within the complementarity determining regions (CDRs) and framework regions (FRs) using the algorithm of Chang and Casali5 showed that 4 cases had significant clustering of R mutations in the CDRs and scarcity of R mutations in the FRs. In addition, 3 cases displayed clustering of R mutations within the CDRs, and in 6 cases evidence for preservation of the FRs was found. Thus in contrast to previous reports, only a minority of our CLL cases showed evidence of antigen-driven selection.
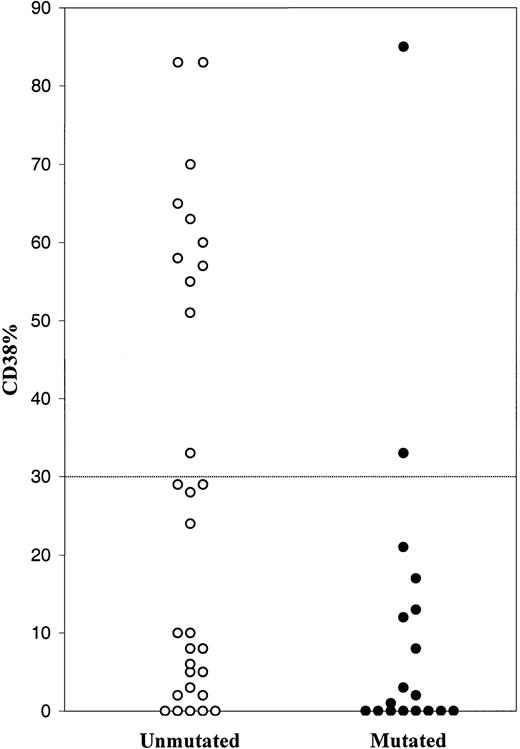
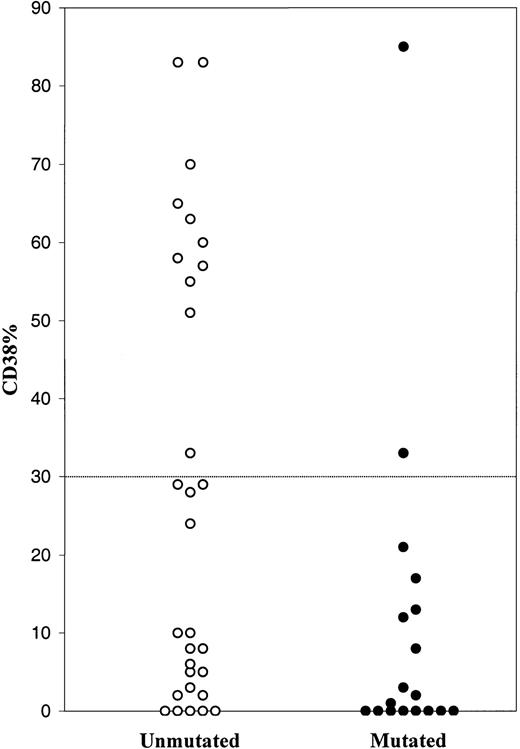
The CD38 expression was analyzed in a subset of 48 CLL patients, where 13 cases displayed more than 30% CD38+cells (27.1%) and 35 cases, less than 30% CD38+ cells (72.9%). Thirty cases had unmutated VH genes, and 18 were mutated. The unmutated cases demonstrated a variety of CD38 values (0-83%), 11 of which had more than 30% CD38+ cells (Figure 1). The majority of mutated cases showed a low level of CD38 expression, but 2 cases displayed more than 30% of CD38+ cells. Thus 11 of 13 cases with more than 30% CD38+ cells had unmutated VH genes.
Percentage of CD38+ cells among B-CLL cases with unmutated and mutated VH genes.
Preliminary survival analysis was performed in 44 of these cases with a median follow-up of 88 months (range, 24-468). All patients but one were dead at the time of analysis. Median age was 61 years at diagnosis, and there was no significant age difference concerning either CD38 status or VH gene mutational status. Kaplan-Meyer survival analysis with log-rank test was performed, and no significant difference in survival regarding CD38 status was observed in 34 CD38−and 10 CD38+ cases (P = .29). But in 26 cases with unmutated VHgenes, survival was significantly inferior compared to the 18 mutated cases (P = .0023).
Hence, the prognostic impact of VH gene mutations in CLL was confirmed by our data. In agreement with Damle et al,1 a correlation was found between unmutated VH genes and high level of CD38 expression. But a low level of CD38 expression could not predict whether the VH genes were unmutated or mutated. Moreover, we could not find any prognostic significance analyzing CD38 expression in our CLL cases. We therefore conclude that CD38 expression cannot be used as a surrogate marker for VH gene mutational status in CLL.
Immunoglobulin V genes and CD38 expression analysis in B-cell chronic lymphocytic leukemia
B-cell chronic lymphocytic leukemia (B-CLL) cases can be divided into 2 subgroups based on the presence or absence of significant numbers of mutations in the variable (V) region of the immunoglobulin (Ig) genes utilized by the malignant cells. This idea was first suggested by a retrospective analysis of the available Ig V gene sequences1-1 and was definitively demonstrated by a study of a large cohort of randomly chosen B-CLL patients by Fais et al.1-2 Since then, other groups1-3 have confirmed this observation, and it is now accepted that at least 50% of B-CLL cases express mutated Ig V genes.
This finding became especially important when it was recognized that a highly significant correlation existed between the clinical course of B-CLL patients and the presence or absence of Ig V somatic mutations.1-3,1-4 Specifically, cases expressing unmutated V genes have a worse clinical course in terms of overall survival, requirement for treatment, and progression to more advanced stages than those cases expressing mutated V genes.1-3-1-5
Moreover, Damle et al1-4 showed that the expression of surface membrane CD38 by more than 30% of B-CLL cells was another independent indicator of poorer prognosis as measured by overall survival and requirement for treatment. Another component of this study was the observation that, in most of the cases analyzed, an inverse correlation existed between CD38 expression and presence of Ig V mutations. This latter finding suggested that these 2 markers usually identified the same cases (accuracy 92% in Damle et al1-4and 90% in a later communication1-6).
The notion that CD38 expression may be used as a prognostic marker has been reinforced in Hamblin et al,1-7 showing that CD38 surface expression was even a better marker of poor prognosis than unmutated Ig V genes. But these authors did not find significant association between CD38 expression and Ig V gene status, although their technical approach to the issue differed considerably from that of Damle et al.1-6 1-7
Thunberg et al show that, although the Ig V region sequence represents a reliable prognostic marker, this may not be the case for CD38 expression as assessed in a subset (n = 44) of the cohort of patients analyzed for V gene mutations (n = 107). Moreover, in contrast with Damle et al's data,1-4 the expression of low levels of CD38 (< 30%) seems to distribute independently of Ig V gene status. Most striking are the data that CD38 expression and poor outcome do not correlate,1-4 since this observation has been corroborated in an independent study of another well-characterized B-CLL cohort.1-7
Unfortunately, some technical details are not reported in Thunberg et al's letter, and therefore the causes for the discrepancies are matters of speculation. One finding, however, is immediately apparent and clearly contrasts with the observations of other groups: the percentage of cases falling within the CD38+group is below the expected value. Thus, 13 of 48 patients were classified as CD38+ (27%), whereas the percentage was 47% in Damle et al1-4 and 44% in Hamblin et al.1-7
In addition, in many of the cases investigated by Thunberg et al, lymph nodes, rather peripheral blood cells, were studied. So far, to our knowledge no detailed analysis on potential differences of surface phenotypes has been carried out between cells derived from different sources. Furthermore, the immunofluorescent methodology used by Thunberg et al may be less precise since a double-staining approach was employed (compared to the triple staining procedure reported previously by our groups1-4 1-6). In addition, no information is provided on the CD38 monoclonal antibody employed.
As a final note, we would like to add 2 considerations about Ig V gene analyses. In Damle et al,1-4 not only the VH but also VL genes were analyzed in each case. Because about 5% of B-CLL cells may have mutations restricted to the VLgenes (Damle et al1-4 and our unpublished observations, 2000), this may be a reason for the discrepancy between our and their results. In addition, in our study that first identified the 2 B-CLL subgroups based on V gene mutation differences,1-2,1-4 care was taken to exclude cases that express more than one VH or VL gene (ie, cases that lack allelic exclusion1-8). This was done because such double expressers can differ in the mutation status of the 2 alleles, thereby making it difficult to decide into which subgroup to assign these cases. This approach has not been followed by Thunberg et al.
In our hands1-6 CD38 expression still remains a reliable prognostic marker that more often than not correlates with Ig V gene mutation status. We hope that future comparisons of this issue will employ similar technological approaches (3-color immunofluorescence using comparable monoclonal antibodies) in B-CLL cases that have been studied at similar molecular levels (VH and VLgene sequence analyses in cases that maintain allelic exclusion). This will permit the investigative community to understand more precisely the degree to which CD38 expression and Ig V gene mutation status independently predict clinical course and the frequency at which they identify the same B-CLL cases.