Abstract
The cellular localization of human cytokeratin 1 (CK1), urokinase plasminogen activator receptor (uPAR), and gC1qR, high-molecular-weight kininogen (HK)-binding proteins on endothelial cells, was determined. CK1 was found on the external membrane of nonpermeabilized endothelial cells by immunoperoxidase staining, immunofluorescence, and transmission electron microscopy using immunogold. Human umbilical vein endothelial cells (HUVECs) had 7.2 ± 0.2 × 104specific CK1 membrane sites/cell by125I-F(ab′)2 anti-CK1 antibody binding. Flow cytometry studies confirmed the presence of CK1, uPAR, and gC1qR on HUVECs. On laser scanning confocal microscopy and transmission electron microscopy, CK1 and uPAR, but not gC1qR, colocalized on the cell surface of HUVECs. The HUVEC surface distribution of these proteins was distinctly different from that for von Willebrand factor. In competitive inhibition experiments, anti-CK1, anti-uPAR, or anti-gC1qR blocked both biotin-HK binding and prekallikrein (PK) activation on HUVECs with an inhibitory concentration of 50% (IC50) of 300 to 350 nM, 50 to 60 nM, or 35 to 100 nM, respectively. Also, antibodies to uPAR and gC1qR each inhibited 86% of kallikrein-mediated, 2-chain urokinase plasminogen activation, whereas antibodies to CK1 only inhibited 24% of plasminogen activation. On HUVECs, CK1 and uPAR, but not gC1qR, colocalized to be a multiprotein receptor complex for HK binding, PK activation, and 2-chain urokinase plasminogen activation.
Introduction
Recent investigations indicate that the plasma proteins, high- and low-molecular-weight kininogens, have multiple protein binding sites, putative receptors, on the surface of human umbilical vein endothelial cells (HUVECs).1 These proteins include gC1qR, urokinase plasminogen activator receptor (uPAR), and cytokeratin 1 (CK1).1-5 Some controversy has developed as to whether gC1qR is, in fact, expressed on the plasma membrane of endothelial cells to contribute to kininogen binding.6-8 However, others believe that some portion of the total cellular pool of gC1qR is expressed on the external membrane of endothelial cells.9,10 High-molecular-weight kininogen (HK) binds to CK1 on endothelial cells by a portion on domain 3 on its heavy chain and domain 5 on its light chain.5,11,12 By a region on its light chain, HK also interacts with domains 2 and 3 on uPAR.3,13 Other studies also suggest that CK1 mediates kininogen heavy chain binding by domain 3 and gC1qR mediates HK's domain 5 cell binding.10 Kininogen's domain 3 does not bind to gC1qR.1
It is unknown how HK interacts with one or all of its membrane-binding proteins, putative receptors, on endothelial cells and how these interactions modulate HK-dependent activities on endothelial cells. Using commercially available antibodies, one laboratory found that 30% of HK binding was inhibited by an antibody to CK1, 72% of HK binding was inhibited by an antibody to gC1qR, and both antibodies combined inhibited binding 86%.10 Alternatively, it is possible that these membrane-binding proteins may colocalize on the endothelial cell surface to form a multiprotein receptor complex for HK. Recent information indicates that uPAR and Mac-1, αMβ2 integrin, are physically associated on the membrane of granulocytes.14,15 Also, HK has been shown to bind to Mac-1 expressed on transfected cells.16 The current investigations indicate that human CK1 expresses 2 epitopes on the external membrane of endothelial cells. Further, human CK1 and uPAR, but not gC1qR, colocalize on the membrane of these cells. Antibodies to all of these proteins modulate HK binding, prekallikrein (PK) activation, and prourokinase (ProUK)-mediated plasminogen activation. The kininogen-binding site, putative receptor, on endothelial cells appears to be a multiprotein complex consisting of at least CK1 and uPAR. gC1qR is present on endothelial cell membranes, but it does not colocalize with either CK1 or uPAR.
Materials and methods
Materials
Carrier-free Na 125I was obtained from ICN (Irvine, CA). Iodogen (chloroamide, 1,3,4,6-tetrachloro-3α-6(diphenylglycoluril), biotinylation kit, Immunopure F (ab′)2 preparation kit, and Immunopure streptavidin horseradish peroxidase dihydrochloride (turbo-TMP) were supplied by Pierce Chemical (Rockford, IL). Prestained and low-molecular-weight standards, nitrocellulose, and polyacrylamide were purchased from Bio-Rad (Richmond, CA). HUVECs, endothelial cell growth medium (EGM), trypsin-EDTA, and trypsin neutralizing buffer were purchased from Clonetics (San Diego, CA).
Proteins, peptides, and antibodies
Single-chain HK with a specific activity of 13 U/mg in 4 mM sodium acetate-HCl and 0.15 M NaCl, pH 5.3, was purchased from Enzyme Research Laboratories (South Bend, IN). HK was biotinylated according to the procedure of Pierce Chemical.12 Briefly, 5 mg HK dissolved in 1 mL 0.01 M sodium phosphate, 0.15 M NaCl, pH 7.4, was dialyzed against the same buffer. Sulfo-NHS-LC-Biotin was added to HK to give 5-fold molar excess of Sulfo-NHS-LC-Biotin to HK. After incubation for 2 hours in ice, the sample was then loaded onto 10-mL Econo-Pac 10 DG column (Pierce Chemical). Biotinylated-HK (biotin-HK) was monitored by absorbance at 280 nm using an extinction coefficient of 7.0 for HK and a protein assay (Bio-Rad). Biotin-HK had a specific activity of 11 U/mg. PK that was purchased from Enzyme Research Laboratories had a specific activity of 22 to 27 U/mg in 4 mM sodium acetate-HCl, 0.15 M NaCl, pH 5.3.
Peptide acetyl-R239RYDQLKSDQSRLDSELC256-amide (RRY18) and peptide acetyl-G143PVCSPGGIQEVTINQSLLQ162-amide (GPV20) (single-letter amino acid codes) were used to immunize goats for the production of antisera (anti-RRY18 and anti-GPV20) at Quality Controlled Biochemicals (Hopkinton, MA). Peptides GPV20 and RRY18 are both unique to CK1 and are coded by exons 1 and 2, respectively.5,17 Both regions have been shown to be exposed on the membranes of HUVECs.4,5 Antisera produced were affinity purified on a peptide column. Anti-RRY18 F(ab′)2 was prepared from anti-RRY18 antibody using an ImmunoPure F(ab′)2 preparation kit from Pierce Chemical. The anti-RRY18 F(ab′)2 was iodinated by the iodogen technique with specific activity of 20 to 45 μCi/μg, as previously described.18,19 Anti-uPAR monoclonal antibody 3B10 was previously described to block urokinase binding to cells.20 Anti-gC1qR monoclonal antibody (clone 74.5.2) was purchased from Covance Research Products (Richmond, CA).
Gelelectrophoresis and immunoblot analysis
Proteins were separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes at 100 V for 1 hour. The electroblots then were incubated in blocking buffer (5% [wt/vol] dry milk, 20 mM Tris-HCl, 0.15 M NaCl pH 7.4 containing 0.05% Tween 20) for 1 hour.21 Then the membranes were incubated with goat anti-GPV20 at 20 μg/mL overnight. After washing, the nitrocellulose membrane was incubated with an antigoat antibody conjugated with horseradish peroxidase (Sigma Chemical, St Louis, MO) at 1:2000 dilution for 1 hour. The specific reactivity of antibodies with the electroblotted proteins was detected with a chemiluminesence system from Amersham (Arlington Heights, IL). All steps were carried out at room temperature.
Endothelialcell culture
The HUVECs were obtained from Clonetics and cultured according to the recommendations of the manufacturer with their reagents. Cells between the first and fifth passage were subcultured onto fibronectin-treated 96-well plates (1 μg/well) 24 hours before the start of the experiment. Cell viability was determined using trypan blue exclusion. Cell numbers were determined by counting on a hemocytometer. The endothelial cell line EA.hy926 was kindly provided by Dr Cora-Jean S Edgell (Department of Pathology, University of North Carolina, Chapel Hill, NC).22 These cells were cultured using Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 μM hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine (Gibco BRL, Grand Island, NY) and a mixture of penicillin, streptomycin, and amphotericin (Sigma).
Binding of 125I-anti-RRY18 F(ab′)2 to endothelial cells
125I-anti-RRY18 F(ab′)2 (20 nM) in Hepes-Tyrode buffer (0.135 M NaCl, 2.7 mM KCl, 11.9 mM NaHCO3, 0.36 mM NaH2PO4, 14.7 mM Hepes [N-2-hydroxyethylpiperazine-N-2-ethanosulfonic acid]) containing 50 μM ZnCl2 was incubated with HUVECs in the absence or presence of a 50-fold molar excess unlabeled anti-RRY18 antibody for variable intervals at room temperature.23 After incubation, the cells were washed twice and dislodged from the plate with 1 N NaOH.24 The solubilized cells were then transferred into polypropylene tubes and counted in a gamma counter. Specific binding of the cell-associated radioactivity was defined as the difference between total and nonspecific binding, that is, binding of the125I-anti-RRY18 F(ab′)2 in the presence of a 50-fold molar excess of anti-RRY18 antibody. Specific binding for each experiment is expressed as nanograms 125I-anti-RRY18 F(ab′)2 bound per 106 HUVECs. The amount of125I-anti-RRY18 F(ab′)2 bound was determined from the specific radioactivity of the iodinated protein and the amount of radioactivity associated with the washed cells.23
Flowcytometry
Flow cytometry was performed with washed HUVECs (5 × 106 cells/mL). Detached HUVECs (100 μL) were incubated with either affinity purified goat anti-GPV20 (300 μg/mL), goat anti-RRY18 (62 μg/mL), mouse anti-uPAR (8 μg/mL) antibodies, mouse anti-gC1qR (8 μg/mL), goat IgG at the same concentrations as the immune antibodies, or mouse IgG (8 μg/mL) in Hepes-carbonate buffer (137 mM NaCl, 3 mM KCl, 12 mM NaHCO3, 14.7 mM Hepes, 5.5 mM dextrose and 0.1% gelatin, pH 7.4) containing 1 mg/mL human γ-globulin with occasional gentle mixing for 1 hour at 37°C.4 After washing by centrifugation at 400g 3 times, the HUVECs were resuspended in the same buffer and incubated with a 1:40 dilution of fluorescent isothiocyanate (FITC)-labeled sheep antigoat or goat antimouse secondary antibodies, respectively, for 1 hour at room temperature in the dark. The bound FITC-labeled secondary antibody to HUVECs was monitored with an Epics-C flow cytometer (Coulter Electronics, Hialeah, FL). Light scatter and fluorescence channels were set at logarithmic gain. Laser excitation was at 488 nm and the green fluorescence was observed through a 525-nm band pass filter.
Light microscopy immunocytochemistry
Immunolabeling was carried out on EA.hy926 cells grown on permanox tissue culture chambers or using cell suspensions. In both cases, the cells were fixed with periodate-lysine-paraformaldehyde25 containing 0.01% glutaraldehyde (PLPG) for 30 minutes. Cell suspensions were left to stand overnight with the fixative before dehydration and embedding in Histosec (Merck, Haar, Germany). For immunostaining, tissue culture chambers and dewaxed sections (5 μm thick) were washed 3 times with 0.01 M sodium phosphate, 0.15 M NaCl, pH 7.4, for 5 minutes and incubated at 22°C with goat anti-RRY18, goat anti-GPV20, or normal goat IgG (10 μg/mL). After 2 hours the cells were incubated with FITC-labeled rabbit antigoat IgG (1:30; Dako, Carpinteria, CA). Alternatively, when immunoperoxidase staining was performed, the slides were sequentially incubated with the same goat primary antibodies to human CK1 or goat IgG followed by a rabbit antigoat IgG (1:500; Dako) and processed according to an established peroxidase/antiperoxidase technique.26
Immunoelectron microscopy
The EA.hy926 endothelial cells were grown to be semiconfluent and cell suspensions were prepared by treating the cells with a solution containing 0.25% trypsin, 1 mM EDTA, 150 mM NaCl, 10 mM NaH2PO4, pH 7.8, for 20 to 60 seconds. The enzyme was inactivated by washing the cells with DMEM containing 10% fetal bovine serum. The cells were then washed several times with phosphate-buffered saline (PBS) to remove the serum and then fixed with PLPG for 30 minutes at room temperature. After fixation the cells were washed exhaustively with PBS, pH 7.4, and surplus fixative was quenched with PBS containing 50 mM NH4Cl, after which the cells were incubated with affinity purified anti-RRY18, anti-GPV20, or nonimmune goat IgG each at 5 to 10 μg/mL or mouse anti-gC1qR or mouse IgG each at 10 μg/mL for 4 hours at room temperature. Next, the cells were sequentially incubated with rabbit antigoat IgG (1:500; Dako) and a 10-nm gold-labeled antirabbit-IgG or goat antimouse IgG diluted 1:4 (Amersham Life Science). When the immunostaining procedure was completed the cells were postfixed with 3% glutaraldehyde and 1% osmium tetroxide for 1 hour each. The immunostained cells were then dehydrated and embedded according to standard procedures in a mixture of epon-araldite. Thin sections prepared from this material were contrasted with lead citrate and observed in a Hitachi H-700 electron microscope. In addition, aliquots of the immunostained cells were fixed with glutaraldehyde, washed with PBS and distilled water and then the gold particles were visualized, at the light microscopic level, using the Intense SE enhancement kit (Amersham Life Science).
In other experiments, EA.hy926 endothelial cells were grown and prepared for double immunolabeling immunoelectron microscopy. Double immunolabeling was carried out by incubating the cells simultaneously with goat anti-CK1 (anti-GPV20, 10 μg/mL) and anti-uPAR (4 μg/mL) for 1 hour under agitation. After incubation and washing, the cells were sequentially incubated with rabbit antigoat IgG (1:500; Dako), nonimmune goat serum to block unbound antigoat IgG, gold-labeled goat antirabbit IgG (Amersham Life Science, 15 nm gold, diluted 1:4) and gold-labeled goat antimouse IgG (Amersham Life Science, 5 nm gold, Auro-Probe EM-labeled diluted 1:4). Controls were prepared by replacement of both anti-CK1 and anti-uPAR by nonimmune goat and mouse IgG, respectively. After immunolabeling was completed, the cells were postfixed with 3% glutaraldehyde and 1% osmium tetroxide and embedded as previously described.
Laser scanning confocal microscopy
The HUVECs were grown on microscope slides for laser scanning confocal microscopy. Nonpermeabilized HUVECs were fixed with 2% paraformaldehyde, as reported perviously.27 After incubation for 15 minutes at 37°C, the cells were washed with 50 mM glycine in PBS for 5 minutes at room temperature to remove any trace amount of the paraformaldehyde followed by washing in binding buffer. All laser scanning confocal microscopy experiments had cells that were doubly labeled with 2 primary and secondary antibodies. The cells were incubated with either affinity purified goat anti-GPV20 (200 μg/mL) or goat IgG (200 μg/mL), mouse anti-uPAR (4 μg/mL) or mouse IgG (4 μg/mL), or mouse anti-gC1qR (4 μg/mL) or mouse IgG (4 μg/mL) for 1 hour at 37°C. At the end of the incubation, the cells were washed again and incubated simultaneously with a FITC-labeled rabbit antimouse (1:30; Calbiochem, San Diego, CA) or Alexa Fluor 594-labeled donkey antigoat IgG (H+L) conjugate (10 μg/mL) (Molecular Probes, Eugene, OR) for double labeling for 1 hour in the dark. The slides were then covered with a Vectorshield antifading mounting medium from Vector Laboratories (Burlingame, CA) and were visualized by using a confocal fluorescence microscope (Bio-Rad). Both projection-view and optical sections were restored electronically and were processed digitally. Optical scanning and digital processing of the images were performed to determine the topographic distribution of the FITC or Alexa Fluor/IgG associated with HUVECs as previously reported.27 In other experiments, the colocalization of HUVEC CK1 and von Willebrand factor (vWF) antigen on the surface of nonpermeabilized cells was determined. CK1 antigen was detected as above using a goat anti-GPV20 antibody followed by Alexa Fluor 594-labeled donkey antigoat IgG (H+L) conjugate (10 μg/mL) (Molecular Probes). The uPAR or gC1qR antigen also was detected as above using a mouse anti-uPAR antibody or mouse anti-gC1qR antibody followed by Alexa Fluor 594-labeled goat antimouse IgG conjugate (10 μg/mL) (Molecular Probes). Human endothelial cell vWF was detected by a rabbit antihuman vWF antisera at 1:100 (Dako) followed by a goat antirabbit IgG (H+L), F(ab′)2 FITC-labeled second antibody (Boehringer Mannheim, Indianapolis, IN).
Inhibition biotin-HK binding to endothelial cells by anti-GPV20, anti-uPAR, and anti-gC1qR antibodies
Confluent monolayers of HUVECs were washed 3 times with Hepes-carbonate buffer containing 2 mM CaCl2 and 1 mM MgCl2. Biotin-HK (7 nM) was incubated with the monolayers for 60 minutes at 37°C in the absence or the presence of increasing concentrations of affinity purified goat anti-GPV20, mouse anti-uPAR, mouse anti-gC1qR antibodies, normal goat antibodies, or normal mouse antibody, respectively. The relative binding of biotin-HK binding to the cells was determined using Immunopure streptavidin horseradish peroxidase conjugate (Pierce) and peroxidase-specific fast reacting substrate, 3,3′, 5,5′-tetramethylbenzidine dihydrochloride (turbo-TMP, Pierce), as previously described.12 Bound biotin-HK was quantified by measuring the absorbance of the reaction mixture at 450 nm using a microplate autoreader EL 311 from Bio-Tek Instrument (Winooski, VT).
Inhibitionof PK activation on endothelial cells by anti-GPV20 or anti-uPAR antibodies
Confluent monolayers of HUVECs were washed 3 times with Hepes-carbonate buffer containing 2 mM CaCl2 and 1 mM MgCl2 and 8 μM Zn2+. The endothelial cells were incubated with 20 nM HK for 1 hour at 37°C. After incubation, the cells were washed and incubated with PK (20 nM) for an additional hour. At the conclusion of the second incubation, the cells were washed and 0.8 mM H-D-Pro-Phe-Arg-pNA (S2302) (Diapharma, Franklin, OH) was added in the same buffer and hydrolysis proceeded for an additional hour.28 The generated kallikrein was quantified by measuring the absorbance of the reaction mixture at 405 nm using a microplate autoreader EL 311 from Bio-Tek Instrument.
Inhibition of ProUK-mediated plasminogen activation by antibodies to kininogen's binding proteins
Inhibition of ProUK (single-chain urokinase)-mediated plasminogen activation was performed similarly as previously reported.28 29 Confluent HUVECs in microtiter plate wells were incubated with plasminogen (1 μM) in Hepes-carbonate buffer for 1 hour at 37°C. At the end of the incubation, the cells were washed 3 times with buffer and the initial rate of plasmin formed was determined after the addition of 2 nM ProUK and 0.3 mM Val-Leu-Lys-pNA.HCl (S2251, Diapharma). In other experiments, HUVECs were incubated with HK (20 nM) and PK (20 nM) simultaneously for 1 hour at 37°C in the absence or presence of 350 nM anti-GPV20, 350 nM goat IgG, 60 nM anti-uPAR, 100 nM anti-gC1qR, or 100 nM mouse IgG. These concentrations of antibodies were those that produced 50% inhibition of HK binding to endothelial cells. At the end of the incubation, the HUVECs were washed and incubated with 1 μM plasminogen at 37°C for 1 hour. After washing again, the cells were incubated with 2 nM ProUK and 0.3 mM S2251 and the initial rate of plasmin formation was measured. The values presented represent the percent plasmin formed after the assembly of HK, PK, plasminogen, and ProUK.
Results
Characterization of CK1 on the membrane of cells in the intravascular compartment
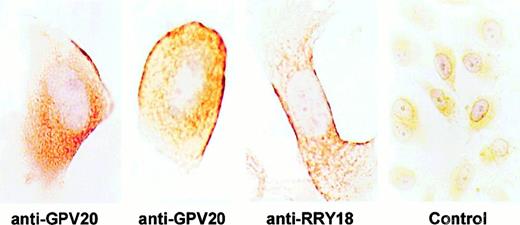
Investigations characterized the membrane expression of CK1 on EA.hy926 cells that were derived from HUVECs and HUVECs themselves. Anti-GPV20 antibody, which was reared to a 20–amino acid peptide of the HK-binding region on CK1,5 detected pure CK1 antigen on immunoblot as a doublet at 72 and 68 kd (data not shown). Immunostaining of nonpermeabilized EA.hy926 cells, grown on permanox tissue culture chambers, revealed the presence of immunoreactive CK1 on the endothelial cell border (Figure1). Using 2 antipeptide antibodies (anti-GPV20, anti-RRY18) to regions on CK1 that are expressed on the membrane of HUVEC,4 5 intense immunoperoxidase staining was detected on the external membrane of these cells (Figure1). When immunocytochemistry was carried out on 5-μm sections of these cells, some of the immunolabeling with anti-GPV20 (Figure2A) or anti-RRY18 (Figure 2B) appeared restricted to fluorescent dots sitting on the cell surface. No immunostaining was observed when the CK1 antipeptide antibodies were omitted (data not shown) or replaced by nonimmune goat IgG (Figure 2C).
Visualization of CK1 on nonpermeabilized EA.hy926 endothelial cells.
EA.hy926 cells were grown on permanox tissue culture chambers, fixed with PLPG, and then treated with anti-GPV20, anti-RRY18, or goat IgG (control). After incubation, the slides were treated with a rabbit antigoat antibody and immunostained by the peroxidase/antiperoxidase method as described in “Materials and methods.” The figure is a representative experiment of 2.
Visualization of CK1 on nonpermeabilized EA.hy926 endothelial cells.
EA.hy926 cells were grown on permanox tissue culture chambers, fixed with PLPG, and then treated with anti-GPV20, anti-RRY18, or goat IgG (control). After incubation, the slides were treated with a rabbit antigoat antibody and immunostained by the peroxidase/antiperoxidase method as described in “Materials and methods.” The figure is a representative experiment of 2.
Recognition of CK1 on sections of EA.hy926 endothelial cells.
Five- micrometer sections of a cell suspension fixed with PLPG were incubated with anti-GPV20 (A), anti-RRY18 (B), or goat IgG (C) as described in “Materials and methods.” The bound antibodies were visualized by an FITC-labeled rabbit antigoat antibody. The figure is a representative experiment of 2. Note the arrows in panels A and B point to the placement of membrane antigen.
Recognition of CK1 on sections of EA.hy926 endothelial cells.
Five- micrometer sections of a cell suspension fixed with PLPG were incubated with anti-GPV20 (A), anti-RRY18 (B), or goat IgG (C) as described in “Materials and methods.” The bound antibodies were visualized by an FITC-labeled rabbit antigoat antibody. The figure is a representative experiment of 2. Note the arrows in panels A and B point to the placement of membrane antigen.
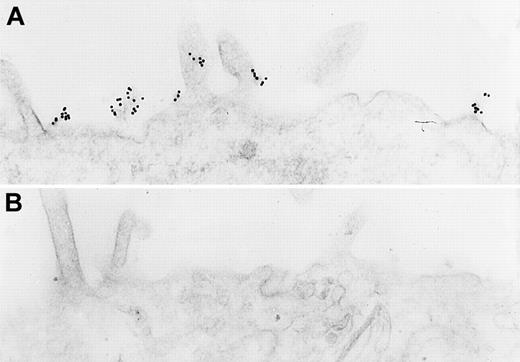
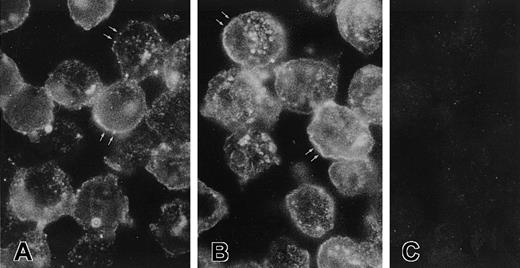
Additional studies confirmed the presence of CK1 antigen on the membrane of these cells. Immunoelectron microscopy of nonpermeabilized EA.hy926 cells showed that immunoreactive CK1 was on the endothelial cell membrane by immunocytochemical labeling with gold particles using anti-GPV20 (Figure 3A) and anti-RRY18 (Figure 3B). Visualization of the bound gold particles on light microscopy after silver enhancement revealed a concentration of the label on the cell periphery (Figure 3D,E). In fact, both anti-CK1 antibodies recognized the protein in clusters of 10-nm gold particles found in segments on the endothelial cell membrane (Figure 3A,B). In contrast, no gold particles were seen on the surface of cells incubated without the primary antibodies or with nonimmune goat IgG (Figure3C,F).
Immunoelectron microscopy for CK1 antigen on nonpermeabilized EA.hy926 endothelial cells.
Cell suspensions were fixed with PLPG and incubated with anti-GPV20 (A,D), anti-RRY18 (B,E), or nonimmune goat IgG (C,F). The bound cytokeratin antipeptide antibodies were visualized by an immunogold procedure. Clusters of 10-nm gold particles, indicating the presence of bound anti-GPV20 and RRY18, are seen segmentally on the endothelial plasma membrane (arrows) (A,B). Insets show immunostained cells at the light microscopic level after silver enhancement (D,E). The figure is a representative experiment of 2.
Immunoelectron microscopy for CK1 antigen on nonpermeabilized EA.hy926 endothelial cells.
Cell suspensions were fixed with PLPG and incubated with anti-GPV20 (A,D), anti-RRY18 (B,E), or nonimmune goat IgG (C,F). The bound cytokeratin antipeptide antibodies were visualized by an immunogold procedure. Clusters of 10-nm gold particles, indicating the presence of bound anti-GPV20 and RRY18, are seen segmentally on the endothelial plasma membrane (arrows) (A,B). Insets show immunostained cells at the light microscopic level after silver enhancement (D,E). The figure is a representative experiment of 2.
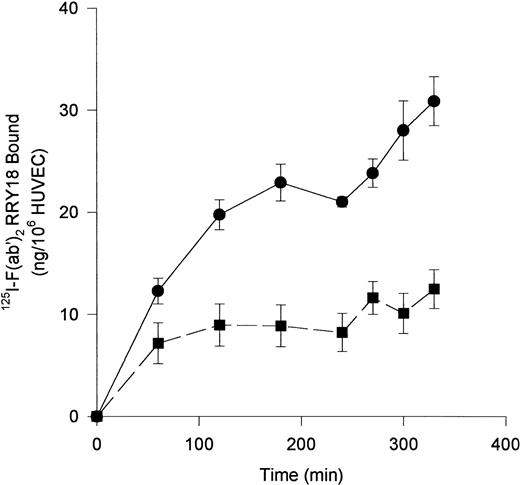
Investigations next proceeded to determine the amount of CK1 antigen expressed on HUVECs. An iodinated anti-RRY18 F(ab′)2was prepared for direct cell-binding studies (Figure4). When the iodinated F(ab′)2 was incubated with HUVECs, there was a plateau in binding at about 200 minutes (Figure 4). At that time, there was about 12 ng iodinated F(ab′)2 specifically bound per 106 HUVECs. This result calculated to be about 7.2 × 104 molecules of anti-RRY18 CK1 epitope per cell. Further investigations were performed to determine if CK1 antigen was on the membrane of HUVECs in suspension as determined by flow cytometry (Figure 5). Using flow cytometry, epitopes to CK1 were detected on HUVEC membranes using anti-GPV20 and anti-RRY184,5 (Figure 5). Additional studies were performed to determine if the other HK-binding proteins, uPAR and gC1qR, were present on HUVEC membranes. The uPAR was identified on HUVECs by flow cytometry (Figure 5). Moreover, unlike previous studies,6-8 gC1qR antigen was also expressed on the membrane of soluble HUVECs as seen on flow cytometry (Figure5). Confirmation that gC1qR antigen was expressed on the membrane of endothelial cells was performed by immunoelectron microscopy (Figure6). Using monoclonal antibody clone 74.5.2, gC1qR antigen was seen in clusters on the external membrane of EA.hy926 cells by immunoelectron microscopy using 10 nm immunogold (Figure 6).
Anti-RRY18 125I-F(ab′)2 binding to HUVEC.
HUVECs were incubated with 20 nM 125I-anti-RRY18 F(ab′)2 for the indicated time period in the absence (●) or presence (■) of a 50-fold molar excess of anti-RRY18 IgG. At each designated time point, samples were removed in triplicate and the amount of 125I-anti-RRY18 F(ab′)2 bound to HUVEC was determined as indicated in “Materials and methods.” The results presented are the means ± SEM of 3 to 5 independent experiments and expressed as nanograms 125I-anti-RRY18 F(ab′)2 bound per 106 HUVECs.
Anti-RRY18 125I-F(ab′)2 binding to HUVEC.
HUVECs were incubated with 20 nM 125I-anti-RRY18 F(ab′)2 for the indicated time period in the absence (●) or presence (■) of a 50-fold molar excess of anti-RRY18 IgG. At each designated time point, samples were removed in triplicate and the amount of 125I-anti-RRY18 F(ab′)2 bound to HUVEC was determined as indicated in “Materials and methods.” The results presented are the means ± SEM of 3 to 5 independent experiments and expressed as nanograms 125I-anti-RRY18 F(ab′)2 bound per 106 HUVECs.
Flow cytometry for HUVEC kininogen-binding proteins.
Suspensions of washed, unfixed, and nonpermeabilized HUVECs were incubated for 1 hour at 4°C with control IgG (open curves) (goat IgG [either 62 or 300 μg/mL] or mouse IgG [8 μg/mL]) or immune antibodies (shaded curves) (goat anti-GPV20 [300 μg/mL]), mouse anti-uPAR [8 μg/mlL] antibodies, affinity purified goat anti-RRY18 [62 μg/mL]) or mouse anti-gC1qR (4 μg/ml) antibodies. The binding of these antibodies to these cells was detected with secondary antibodies labeled with FITC. The figure is a representative experiment of 3 experiments.
Flow cytometry for HUVEC kininogen-binding proteins.
Suspensions of washed, unfixed, and nonpermeabilized HUVECs were incubated for 1 hour at 4°C with control IgG (open curves) (goat IgG [either 62 or 300 μg/mL] or mouse IgG [8 μg/mL]) or immune antibodies (shaded curves) (goat anti-GPV20 [300 μg/mL]), mouse anti-uPAR [8 μg/mlL] antibodies, affinity purified goat anti-RRY18 [62 μg/mL]) or mouse anti-gC1qR (4 μg/ml) antibodies. The binding of these antibodies to these cells was detected with secondary antibodies labeled with FITC. The figure is a representative experiment of 3 experiments.
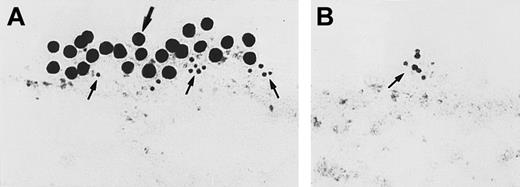
Immunoelectron microscopy for gC1qR antigen on nonpermeabilized EA.hy926 endothelial cells.
Cell suspensions were fixed with PLPG and incubated with anti-gC1qR or nonimmune mouse IgG. (A) The bound anti-gC1qR antibodies were visualized by an immunogold procedure as indicated in “Materials and methods.” Clusters of 10-nm gold particles, indicating the presence of bound anti-gC1qR, are seen segmentally on the endothelial plasma membrane. (B) Incubation of nonimmune IgG with the cells. The figure is a representative experiment of 2.
Immunoelectron microscopy for gC1qR antigen on nonpermeabilized EA.hy926 endothelial cells.
Cell suspensions were fixed with PLPG and incubated with anti-gC1qR or nonimmune mouse IgG. (A) The bound anti-gC1qR antibodies were visualized by an immunogold procedure as indicated in “Materials and methods.” Clusters of 10-nm gold particles, indicating the presence of bound anti-gC1qR, are seen segmentally on the endothelial plasma membrane. (B) Incubation of nonimmune IgG with the cells. The figure is a representative experiment of 2.
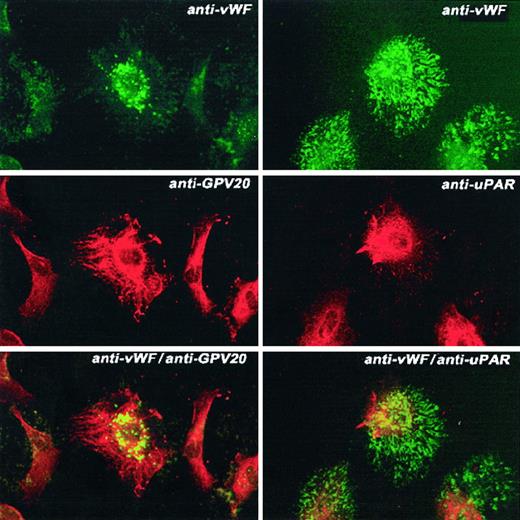
Colocalization of CK1 and uPAR on HUVECs
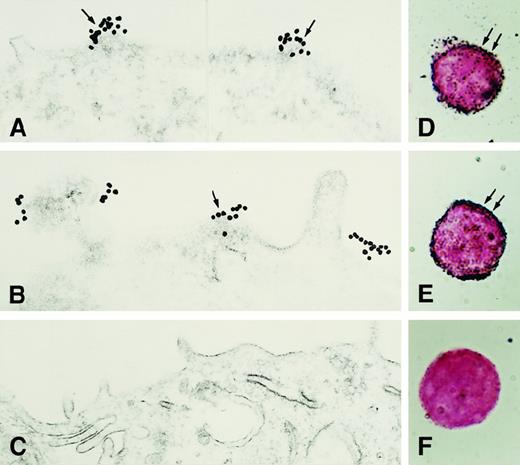
Next, studies were performed to determine how these 3 HK-binding proteins are associated on the membranes of HUVECs. Using the monoclonal antibody 3B10, FITC-labeled uPAR antigen was noted to be diffusely present over the cell surface sparring the nucleus (Figure7, upper left panel). Similarly, anti-CK1 antibody also demonstrated a complete cell surface distribution with nuclear sparring as detected by the Texas red label (Figure 7, middle left panel). When the double label of both the anti-CK1 and anti-uPAR antibodies was examined simultaneously, there was a blending of the color of the fluorescent labels attached to the second antibodies (Figure 7, bottom left panel). These latter data indicated that there was a colocalization of these proteins on the examined cells. This pattern of distribution was distinctly different from that seen with antibody to human vWF (Figure 8). Unlike antibody to CK1 or uPAR, antisera to vWF had a distinctly different distribution pattern on the surface of HUVECs in culture than that of CK1 or uPAR epitopes (Figure 8, bottom panels). To confirm the colocalization of CK1 and uPAR on the surface of endothelial cells as detected by laser scanning confocal microscopy, double immunogold labeling studies were performed (Figure9). Double immunogold labeling revealed a colocalization of CK1 and uPAR on the cell membrane of EA.hy926 endothelial cells. In fact, clusters of 15-nm gold particles corresponding with anti-GPV20 bound to immunoreactive CK1 coexisted on the cell surface of endothelial cells with the smaller size 5-nm gold particles representing immunoreactive uPAR (Figure 9A). These clusters of gold particles also were found in segments on the membrane of the cells (Figures 3 and 9). When the anti-CK1 antibody was replaced by normal goat IgG, only gold particles associated with the anti-uPAR antibody were found on the membrane of the cells (Figure 9B). Similarly, if the antibody to uPAR or both antibodies were replaced by nonimmune IgG, no gold particles were detected on the cell membrane (data not shown).
Colocalization of CK1 and uPAR on HUVECs.
HUVECs grown on microscope slides were fixed with 2% paraformaldehyde and not permeabilized. The panels to this figure are photomicrographs of the laser scanning confocal micrographs. The cells in this figure were doubly labeled with primary and secondary antibodies. Upper left panel represents cells that have been labeled with mouse anti-uPAR (8 μg/mL); upper right panel, mouse IgG. Their detection was performed with a second antibody conjugated with FITC. Middle left panel represents cells that have been labeled with goat anti-GPV20 (200 μg/mL); middle right panel, goat IgG (200 μg/mL). Their detection was performed with a second antibody conjugated with Alexa Fluor. Lower left panel represents detection of the 2 labels on these cells that were treated with both goat anti-GPV20 (200 μg/mL) and mouse anti-uPAR (8 μg/mL); lower right panel is a mixture of goat IgG (200 μg/mL), and mouse IgG (8 μg/mL). Recognition of the antibody binding was performed with the secondary antibodies labeled with Alexa Fluor and FITC simultaneously. The figure is a representative presentation of 3 independent experiments.
Colocalization of CK1 and uPAR on HUVECs.
HUVECs grown on microscope slides were fixed with 2% paraformaldehyde and not permeabilized. The panels to this figure are photomicrographs of the laser scanning confocal micrographs. The cells in this figure were doubly labeled with primary and secondary antibodies. Upper left panel represents cells that have been labeled with mouse anti-uPAR (8 μg/mL); upper right panel, mouse IgG. Their detection was performed with a second antibody conjugated with FITC. Middle left panel represents cells that have been labeled with goat anti-GPV20 (200 μg/mL); middle right panel, goat IgG (200 μg/mL). Their detection was performed with a second antibody conjugated with Alexa Fluor. Lower left panel represents detection of the 2 labels on these cells that were treated with both goat anti-GPV20 (200 μg/mL) and mouse anti-uPAR (8 μg/mL); lower right panel is a mixture of goat IgG (200 μg/mL), and mouse IgG (8 μg/mL). Recognition of the antibody binding was performed with the secondary antibodies labeled with Alexa Fluor and FITC simultaneously. The figure is a representative presentation of 3 independent experiments.
Colocalization of CK1 or uPAR and vWF antigen on HUVECs.
HUVECs grown on microscope slides were fixed with 2% paraformaldehyde and not permeabilized. The panels to this figure are photomicrographs of the laser scanning confocal micrographs. The cells in this experiment were doubly labeled with the primary and secondary antibodies. The upper left and right panels are nonpermeabilized endothelial cells treated with a rabbit antihuman vWF antibody followed by a second antibody conjugated with FITC. The middle left panel shows endothelial cells incubated with the anti-CK1 antibody anti-GPV20 and the middle right panel shows endothelial cells incubated with an anti-uPAR antibody. Both antibodies were detected with a second antibody conjugated with Alexa Fluor. The lower left panel is combined anti-vWF and anti-CK1 antibodies. The lower right panel is exposure of combined anti-vWF and anti-uPAR antibodies. Recognition of the antibody binding was performed with the secondary antibodies labeled with Alexa Fluor and FITC simultaneously. The figure is a representative presentation of 2 independent experiments.
Colocalization of CK1 or uPAR and vWF antigen on HUVECs.
HUVECs grown on microscope slides were fixed with 2% paraformaldehyde and not permeabilized. The panels to this figure are photomicrographs of the laser scanning confocal micrographs. The cells in this experiment were doubly labeled with the primary and secondary antibodies. The upper left and right panels are nonpermeabilized endothelial cells treated with a rabbit antihuman vWF antibody followed by a second antibody conjugated with FITC. The middle left panel shows endothelial cells incubated with the anti-CK1 antibody anti-GPV20 and the middle right panel shows endothelial cells incubated with an anti-uPAR antibody. Both antibodies were detected with a second antibody conjugated with Alexa Fluor. The lower left panel is combined anti-vWF and anti-CK1 antibodies. The lower right panel is exposure of combined anti-vWF and anti-uPAR antibodies. Recognition of the antibody binding was performed with the secondary antibodies labeled with Alexa Fluor and FITC simultaneously. The figure is a representative presentation of 2 independent experiments.
Colocalization of CK1 and uPAR on transmission electron microscopy.
Double immunogold labeling for CK1 and uPAR on nonpermeabilized EA.hy926 endothelial cells. (A) Cells incubated with goat anti-GPV20 antibody and mouse anti-uPAR antibody. Bound immunoglobulins were detected with gold-labeled antigoat IgG (15 nm) and gold-labeled antimouse IgG (5 nm), respectively. (B) Cells treated with the anti-uPAR antibody followed by gold-labeled antimouse IgG (5 nm). Goat anti-GPV20 was replaced by nonimmune goat IgG, which resulted in the absence of labeling with 15 nm gold-labeled antigoat IgG. The large arrow points to CK1 antigen; the small arrows point to uPAR antigen. The figure is a representative experiment of 2.
Colocalization of CK1 and uPAR on transmission electron microscopy.
Double immunogold labeling for CK1 and uPAR on nonpermeabilized EA.hy926 endothelial cells. (A) Cells incubated with goat anti-GPV20 antibody and mouse anti-uPAR antibody. Bound immunoglobulins were detected with gold-labeled antigoat IgG (15 nm) and gold-labeled antimouse IgG (5 nm), respectively. (B) Cells treated with the anti-uPAR antibody followed by gold-labeled antimouse IgG (5 nm). Goat anti-GPV20 was replaced by nonimmune goat IgG, which resulted in the absence of labeling with 15 nm gold-labeled antigoat IgG. The large arrow points to CK1 antigen; the small arrows point to uPAR antigen. The figure is a representative experiment of 2.
Additional investigations determined if the membrane expression of gC1qR colocalized with CK1 or vWF antigen on the membrane of endothelial cells (Figure 10). As seen in the bottom panel on the left, the color of the gC1qR antigen did not blend with the expressed CK1 antigen on the membrane of these endothelial cells, suggesting that these 2 epitopes were not colocalized (Figure 10). Further, the relative amount of gC1qR epitope on HUVEC (Figure 10, upper left panel) was greater than the relative amount of the CK1 epitope present on the HUVEC (Figure 10, middle left panel). Similarly, human vWF antigen did not colocalize with gC1qR (Figure 10, panels on the right).
Localization of CK1 and gC1qR and vWF and gC1qR on HUVECs.
HUVECs grown on microscope slides were fixed with 2% paraformaldehyde and not permeabilized. The panels to this figure are photomicrographs of the laser scanning confocal micrographs. The cells in this figure were doubly labeled with primary and secondary antibodies. The panels on the left are cells that were doubly labeled with anti-gC1qR and anti-CK1 (anti-GPV20). The panels on the right are cells that were doubly labeled with anti-vWF and anti-gC1qR. Upper left panel represents cells that have been labeled with mouse anti-gC1qR (4 μg/mL); upper right panel, rabbit antisera to human vWF. Their detection was performed with a second antibody conjugated with FITC. Middle left panel represents cells that have been labeled with goat anti-GPV20 (200 μg/mL) (anti-CK1 antibody); middle right panel, mouse anti-gC1qR (4 μg/mL). Their detection was performed with secondary antibodies conjugated with Alexa Fluor. Lower left panel represents detection of the 2 labels on these cells that were treated with both goat anti-GPV20 (200 μg/mL) and mouse anti-gC1qR (4 μg/mL); lower right panel is a mixture of rabbit antihuman vWF antisera and mouse anti-gC1qR (4 μg/mL). Recognition of the antibody binding was performed with the secondary antibodies labeled with Alexa Fluor and FITC simultaneously. The figure is a representative presentation of 3 independent experiments.
Localization of CK1 and gC1qR and vWF and gC1qR on HUVECs.
HUVECs grown on microscope slides were fixed with 2% paraformaldehyde and not permeabilized. The panels to this figure are photomicrographs of the laser scanning confocal micrographs. The cells in this figure were doubly labeled with primary and secondary antibodies. The panels on the left are cells that were doubly labeled with anti-gC1qR and anti-CK1 (anti-GPV20). The panels on the right are cells that were doubly labeled with anti-vWF and anti-gC1qR. Upper left panel represents cells that have been labeled with mouse anti-gC1qR (4 μg/mL); upper right panel, rabbit antisera to human vWF. Their detection was performed with a second antibody conjugated with FITC. Middle left panel represents cells that have been labeled with goat anti-GPV20 (200 μg/mL) (anti-CK1 antibody); middle right panel, mouse anti-gC1qR (4 μg/mL). Their detection was performed with secondary antibodies conjugated with Alexa Fluor. Lower left panel represents detection of the 2 labels on these cells that were treated with both goat anti-GPV20 (200 μg/mL) and mouse anti-gC1qR (4 μg/mL); lower right panel is a mixture of rabbit antihuman vWF antisera and mouse anti-gC1qR (4 μg/mL). Recognition of the antibody binding was performed with the secondary antibodies labeled with Alexa Fluor and FITC simultaneously. The figure is a representative presentation of 3 independent experiments.
Influence of antibody receptor blockade on kininogen-related functional activities
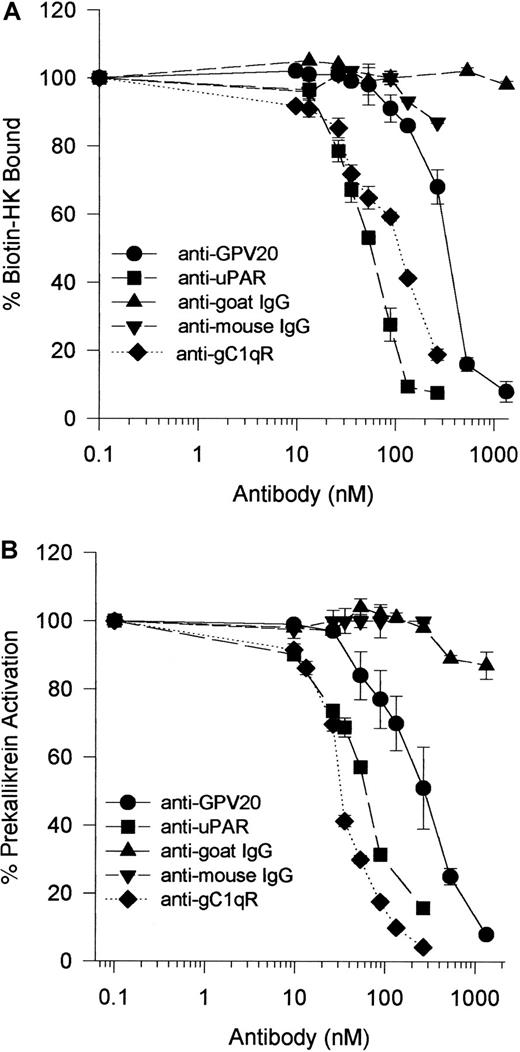
Because HK bound to CK1, uPAR, and gC1qR on HUVECs,1-5 investigations were performed to determine if antibodies to these receptors interfered with HK binding and, subsequently, PK activation on HUVECs.28,29 Anti-GPV20 antibody, which was made against a peptide of the HK cell-binding domain on CK1,5 blocked biotin-HK binding to HUVECs in a concentration-dependent fashion (IC50 = 350 nM) (Figure11A). Because PK must be bound to HK for it to be activated on HUVECs,28 30 studies determined that anti-GPV20 also blocked PK activation on HUVECs with an IC50 = 300 nM (Figure 11B). Accordingly, anti-uPAR antibody blocked biotin-HK binding and PK activation on HUVECs in a concentration-dependent fashion with an IC50 of 50 and 60 nM, respectively (Figure 11A,B). These combined data indicated that interference with HK binding to one binding protein blocked its interaction with the other membrane-binding protein as well and this effect interfered with PK activation. Although not colocalized to CK1 or uPAR, antibody to gC1qR also blocked HK binding (IC50 = 100 nM) and PK activation (IC50 = 35 nM) (Figure 11).
Inhibition of biotin-HK binding and PK activation on HUVECs by antibodies to kininogen HUVEC membrane-binding proteins.
(A) HUVECs were incubated with biotin-HK (7 nM) with increasing concentrations of goat anti-GPV20 (●) and goat IgG (s) for 1 hour at 37°C. HUVECs were also incubated with biotin-HK (7 nM) and various concentrations of mouse anti-uPAR (■), mouse anti-gC1qR (⧫), or mouse IgG (t) for 1 hour at 37°C. After incubation, the level of bound biotin-HK was determined. (B) HUVECs were incubated with HK (20 nM) and increasing concentrations of goat anti-GPV20 (●) or goat IgG (s) for 1 hour at 37°C. HUVECs were also incubated with HK (20 nM) and various concentrations of mouse anti-uPAR (■), mouse anti-gC1qR (⧫), or mouse IgG (t) for 1 hour at 37°C. At the conclusion of incubation, the cells were washed and incubated with PK (20 nM) for an additional hour at 37°C. The level of membrane-associated kallikrein was determined by the addition of the chromogenic substrate H-D-Pro-Phe-Arg-pNA (0.8 mM) as described in “Materials and methods.” The data are mean ± SEM of triplicate determinations from 3 different experiments.
Inhibition of biotin-HK binding and PK activation on HUVECs by antibodies to kininogen HUVEC membrane-binding proteins.
(A) HUVECs were incubated with biotin-HK (7 nM) with increasing concentrations of goat anti-GPV20 (●) and goat IgG (s) for 1 hour at 37°C. HUVECs were also incubated with biotin-HK (7 nM) and various concentrations of mouse anti-uPAR (■), mouse anti-gC1qR (⧫), or mouse IgG (t) for 1 hour at 37°C. After incubation, the level of bound biotin-HK was determined. (B) HUVECs were incubated with HK (20 nM) and increasing concentrations of goat anti-GPV20 (●) or goat IgG (s) for 1 hour at 37°C. HUVECs were also incubated with HK (20 nM) and various concentrations of mouse anti-uPAR (■), mouse anti-gC1qR (⧫), or mouse IgG (t) for 1 hour at 37°C. At the conclusion of incubation, the cells were washed and incubated with PK (20 nM) for an additional hour at 37°C. The level of membrane-associated kallikrein was determined by the addition of the chromogenic substrate H-D-Pro-Phe-Arg-pNA (0.8 mM) as described in “Materials and methods.” The data are mean ± SEM of triplicate determinations from 3 different experiments.
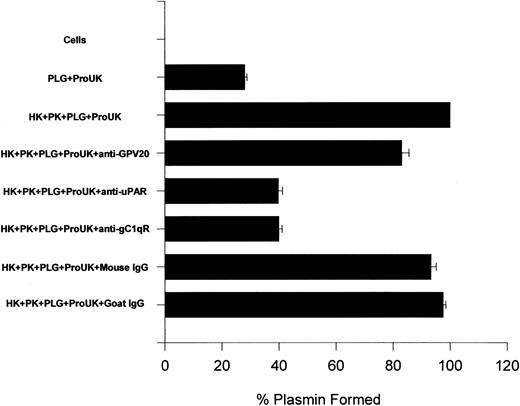
Further investigations were performed to ascertain the influence of these antibodies on ProUK-induced plasminogen activation (Figure12). The addition of HK and PK increased the extent of plasmin formation 2.6-fold over that induced by ProUK alone similar to what has been previously reported28 29 (Figure 12). In the presence of antibodies to CK1 that blocked HK binding to HUVEC 50%, the extent of plasmin formation was reduced by 24% (Figure 12). Anti-CK1 antibody concentrations at 1 μM completely blocked binding (data not shown). Alternatively, antibodies to uPAR or gC1qR at a concentration that would block HK binding by 50% reduced kallikrein-mediated ProUK activation and, subsequently, plasmin formation by 86% (Figure 12). These data indicated that interference with HK binding to uPAR and gC1qR had a greater influence on ProUK-mediated plasminogen activation than interference with binding to CK1.
The influence of antibodies to HK-binding proteins on HUVEC membranes on urokinase-induced plasminogen activation.
Microtiter plate wells with a confluent monolayer of HUVEC were incubated with plasminogen (PLG; 1 μM) for 1 hour at 37°C. After incubation, the cells were washed and the initial rate of formed plasmin was determined by the addition of 2 nM ProUK and 0.3 mM S2251. In other experiments, the HUVEC monolayer was incubated with 20 nM HK and PK for 1 hour at 37°C in the absence or presence of 350 nM anti-GPV20, 60 nM anti-uPAR, 100 nM anti-gC1qR, 350 nM goat IgG, or 100 nM mouse IgG. At the end of the incubation, the cells were washed and incubated with plasminogen for another hour at 37°C. After washing the cells again, the initial rate of formed plasmin was determined by the addition of 2 nM ProUK and 0.3 mM S2251. The data are expressed as the percent plasmin formed when compared to the assembly of HK, PK, plasminogen, and ProUK. The data shown are the mean ± SEM of 3 experiments.
The influence of antibodies to HK-binding proteins on HUVEC membranes on urokinase-induced plasminogen activation.
Microtiter plate wells with a confluent monolayer of HUVEC were incubated with plasminogen (PLG; 1 μM) for 1 hour at 37°C. After incubation, the cells were washed and the initial rate of formed plasmin was determined by the addition of 2 nM ProUK and 0.3 mM S2251. In other experiments, the HUVEC monolayer was incubated with 20 nM HK and PK for 1 hour at 37°C in the absence or presence of 350 nM anti-GPV20, 60 nM anti-uPAR, 100 nM anti-gC1qR, 350 nM goat IgG, or 100 nM mouse IgG. At the end of the incubation, the cells were washed and incubated with plasminogen for another hour at 37°C. After washing the cells again, the initial rate of formed plasmin was determined by the addition of 2 nM ProUK and 0.3 mM S2251. The data are expressed as the percent plasmin formed when compared to the assembly of HK, PK, plasminogen, and ProUK. The data shown are the mean ± SEM of 3 experiments.
Discussion
These investigations indicate a number of novel features. First, human endothelial cells have at least 2 epitopes of CK1 that are expressed on the external membrane of endothelial cells. Antipeptide antibodies to 2 different sequences unique to CK1 recognize antigen on the external membrane of HUVECs. The presence of cytokeratin, a member of the intermediate filament family of proteins, on the membrane of cells has only been recently appreciated.4,5,31-33 CK1 consists of a central helical rod domain flanked by 3 loops each consisting of 40 amino acids that protrude from its filament surface.34 These loops are glycine rich. CK1 does not have the structure of a transmembrane protein and does not have a phosphatidylinositol linkage as uPAR. However, the glycine-rich regions contain hydrophobic residues that may loop through cell membranes and interact with membrane proteins. The glycine loop region is separated from the rod region by the 35 amino acid H1 subdomain, which terminates the penetration of the N-terminal region into cell membranes.34 This region is the specific site where HK interacts with CK1 on protein coded by exon 1.5 Some individuals with mutations in the H1 domain have the dermatologic disorder, epidermolytic hyperkeratosis.35 36
The second finding is that CK1 and uPAR colocalize on the endothelial cell membrane. The finding that both CK1 and uPAR have the same surface distribution on laser scanning confocal microscopy and transmission electron microscopy on endothelial cells indicates that these proteins colocalize. The additional finding of a clustered, segmental distribution of these colocalized proteins on transmission electron microscopy is consistent with the finding of other GPI-anchored proteins and with caveolin being located in membrane rafts and adaptors.37-39 Further, the finding of the punctate antigen epitopes for CK1 on the membrane of endothelial cells also suggests a location in rafts. The uPAR is more highly expressed on HUVEC membranes than CK1 because there are about 250 000 sites/cell for uPAR versus 72 000 molecules/cell for CK1.40 The association of uPAR with CK1 is similar to the association of uPAR with Mac-1 (αMβ2integrin) on granulocytes. The ability of HK to interfere with fibrinogen binding to Mac-1 may be due to the colocalization of this integrin with uPAR, although recent data indicate the HK interacts directly with Mac-1.15,16,41 Although CK1 and uPAR are distinct proteins, together they must comprise a multiprotein receptor complex for HK on endothelial cells. Interference with the interaction of HK with either CK1 or uPAR blocks the interaction with the other protein. This notion suggests that HK must either interact with both proteins simultaneously or it preferentially binds to one protein blocking its interaction with the other. Cleaved forms of HK (HKa) have greater affinity for uPAR than intact HK.3 It is not known whether HKa preferentially binds to uPAR over CK1 on cell membranes. It is possible that intact HK may preferentially bind to one of these 2 proteins; when cleaved, an alternative binding pattern may occur. Further studies are needed to address these questions.
These investigations also unquestionably demonstrate that gC1qR is present on the membrane of cultured HUVECs and the endothelial cell line EA.hy926.22 Although some studies suggest that this antigen is not present on the membrane of endothelial cells,6-8 other investigators have demonstrated its presence.9,10 Further, recent investigations show that gC1qR is a receptor for the InIB invasion protein of Listeria monocytogenes, indicating that it must be a membrane protein.42 The differences in results among the various laboratories must be the result of using different antibodies directed to different epitopes of gC1qR. The membrane expression of gC1qR modifies the biologic activities of HK. Although gC1qR did not colocalize with CK1 and uPAR as described by laser scanning confocal microscopy, antibodies to this protein equally interfere with HK binding and subsequent PK activation on endothelial cells as antibodies to CK1 and uPAR. Furthermore, gC1qR appears to have greater influence on urokinase-mediated plasmin formation than CK1. These functional studies indicate that gC1qR also participates in the endothelial cell multiprotein receptor complex for HK like CK1 and uPAR.
Finally, the finding that CK1 and uPAR combined account for only 320 000 HK-binding sites on HUVECs is surprising. This value is 32% of the total number of HK-binding sites at 4°C and 3.2% of the total number of HK-binding sites at 37°C.20,23,43-45 The reason more HK binds to HUVECs at the higher temperature is not completely known. The possibility of the presence of other membrane-binding proteins for HK is also not completely known. gC1qR could be contributing to the total number of HK-binding sites as well. Qualitatively on immunofluoresence (data not shown) and laser scanning confocal microscopy, there are more gC1qR epitopes present on HUVEC than CK1 sites. Peerschke and colleagues suggest that the endothelial cell may have as many as 5 × 106 gC1qR molecules/cell, but when multimerization of the protein occurs, the number of sites reduces to 3.7 × 105 sites/cell.9 The HK bradykinin region binds to protease-activated receptor 1, but the addition of this protein cannot fully account for the total number of HK-binding sites demonstrated on HUVECs.22 Recent studies suggest that various glycosaminoglycans also are candidate HK-binding structures on the membrane of cells.46 The extent to which glycosaminoglycans contribute to HK binding to HUVECs is not known. What appears to be certain is that other candidate proteins or substances as yet need to be characterized as additional kininogen-binding protein, putative receptors, to fully understand the assembly of proteins that mediate activities related to the plasma kininogen/kallikrein system. How the present candidate kininogen receptors as well as other possible proteins interact with kininogen will help to more fully understand the assembly and activation of this proteolytic system.
The present studies indicate that CK1, uPAR, and gC1qR serve as a platform for PK activation when HK is bound. Because interference with cell binding blocks PK activation on HUVECs,28,30 the interaction of HK with its putative receptors directly modulates kallikrein-dependent activities such as factor XII and ProUK activation and bradykinin liberation. Our studies also indicate that both uPAR and gC1qR have a greater influence on urokinase-induced plasmin formation than CK1. These activities directly relate to the biologic activities of the plasma kininogen/kallikrein system to locally modulate vascular biology by regulating blood pressure, stimulating fibrinolysis, and influencing angiogenesis.47
We appreciate the efforts of Ikuko Mizukami, PhD, for the preparation of the purified 3B10 anti-uPAR monoclonal antibody, and Mr Chris Edwards for his assistance in laser scanning confocal microscopy. We also appreciate Dr Cora-Jean Edgell for providing the EA.hy926 cells.
Supported by National Institutes of Health grants HL52779 (A.H.S.), HL56415 (A.H.S.), CA39064 (R.F.T.), and CA42246 (R.F.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alvin H. Schmaier, University of Michigan, 1150 W Medical Center Dr, 5301 MSRB III, Ann Arbor, MI 48109-0640; e-mail:aschmaie@umich.edu.

![Fig. 5. Flow cytometry for HUVEC kininogen-binding proteins. / Suspensions of washed, unfixed, and nonpermeabilized HUVECs were incubated for 1 hour at 4°C with control IgG (open curves) (goat IgG [either 62 or 300 μg/mL] or mouse IgG [8 μg/mL]) or immune antibodies (shaded curves) (goat anti-GPV20 [300 μg/mL]), mouse anti-uPAR [8 μg/mlL] antibodies, affinity purified goat anti-RRY18 [62 μg/mL]) or mouse anti-gC1qR (4 μg/ml) antibodies. The binding of these antibodies to these cells was detected with secondary antibodies labeled with FITC. The figure is a representative experiment of 3 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/8/10.1182_blood.v97.8.2342/5/m_h80810927005.jpeg?Expires=1763881147&Signature=3O6Rshyc1jF4MucKunH8M-kOpyjzhPi~J27XmgWoDMG7Poq2tZ7dJXzRQnO7kstzBkI2XVA6k4qFcliqyRw07X0By6aQ1TI5ZOzY3aMFyxhMRZHO~aHLPMnXOqIYuny7JUZm1vRhiXNVPJIvybVkKU6-B50NX4jwW7h884EeU5gL~c3DDugdocnX9ziBC~EPpUqgy4cx9aidwgPsLhZdxYKnOCQVZWKPVyvPk0Vf-9FPymd538Ykq3-mS6sx7BRJJI8T~B~TkmDf-LR7Ynv~EmyJMOO5ROBPe4AAghbRLVF~NWA8vNTtctJygRmGoxuw7Eq1I2cEy3wrpJVS~OAYhg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)