Abstract
Two Japanese patients were newly diagnosed as having B subunit (XIIIB) deficiency of factor XIII (former type I deficiency). Both patients have a previously described one-base deletion at the boundary between intron A/exon II in the XIIIB gene, heterozygously or homozygously. A founder effect was proposed for this mutation because 3 unrelated patients with XIIIB deficiency also share 2 3′-polymorphisms. In one patient heterozygous for the above mutation, a novel mutation was also identified: a deletion of guanosine in exon IX (delG) of the XIIIB gene. To understand the molecular and cellular pathology of the delG mutation, expression studies were performed using a cultured mammalian cell line. Pulse-chase experiments showed that a resultant truncated XIIIB remained inside the cells and could not be secreted into the culture medium. Furthermore, immunocytochemical examinations by epifluorescence, confocal, and electron microscopes indicated impaired intracellular transportation of the truncated XIIIB from the endoplasmic reticulum to the Golgi apparatus. No mutations in the gene for the A subunit (XIIIA) were identified in this patient. Therefore, secretion of the truncated XIIIB must also be impaired in vivo, leading to a secondary XIIIA deficiency. These results support a previous conclusion that genetic defects of XIIIB are the basis for the former type I factor XIII deficiency.
Introduction
Factor XIII deficiency is an inherited hemorrhagic disease characterized by a lifelong bleeding tendency and abnormal wound healing in affected patients and spontaneous abortion in female patients.1-3 Coagulation factor XIII is a proenzyme of plasma transglutaminase and circulates in blood as a heterotetramer consisting of 2 catalytic A subunits (XIIIA) and 2 noncatalytic B subunits (XIIIB). Factor XIII catalyzes intermolecular cross-linking reactions between fibrin monomers themselves, fibrin and α2-plasmin inhibitor, and fibrin and fibronectin. These reactions increase the mechanical strength of the fibrin clot and its resistance to proteolytic degradation, and enhance the assembly of the extracellular matrix. Accordingly, deficiency of factor XIII results in defective cross-linking reactions.
More than 200 cases of factor XIII deficiency have been identified. In most cases, diagnosis of factor XIII deficiency is made by measuring the enzymatic activity of factor XIII, which represents the amount of functional XIIIA. Factor XIII deficiency had been classified by the presence or absence of antigens into 2 categories: type I, characterized by the lack of both XIIIA and XIIIB; and type II, characterized by the lack of XIIIA alone.4 Until now, most cases of factor XIII deficiency were diagnosed as type II and only 3 cases have been reported to be type I.4-7 The reliability of classification by immunologic assays depends on the specificity of antibodies against each subunit. From the results of genetic analyses on factor XIII deficiency, we previously concluded that type I deficiency results from genetic defects of XIIIB, and type II from genetic defects of XIIIA. We then proposed a new classification of factor XIII deficiency at the DNA level: XIIIA deficiency (former type II deficiency) and XIIIB deficiency (former type I deficiency), and a possible combined deficiency of XIIIA and XIIIB.8 This proposal was approved by the Scientific and Standardization Committee (SSC) in 1999 (http://www.med.unc.edu/isth/99FXIII.html).
We performed genetic analyses in all cases of XIIIB deficiency and identified 3 mutations in the XIIIB genes. Two mutations were found in one Japanese patient (patient 1): a deletion of adenosine, ag−(−)g, or IVS1-2delA, at the boundary of intron A/exon II, resulting in a loss of the obligatory AG splicing sequence; and a missense mutation in exon VIII, which substitutes Phe (TTC) for Cys430 (TGC), resulting in impaired transportation/secretion of the mutant protein.9,10 Another mutation, an AAC insertion in exon III, was identified in 2 Italian patients (patients 2 and 3), homozygously.11 This mutation creates a premature stop codon in the second Sushi domain of XIIIB. Patients 2 and 3 were unrelated and a founder effect was proposed for XIIIB deficiency among the Italians.12
In the present study, 2 Japanese patients were newly diagnosed as having XIIIB deficiency. Both were found to be heterozygous or homozygous for the known ag−(−)g mutation. A novel mutation (deletion of guanosine in exon IX, delG) was also identified in theXIIIB gene of one patient. This study represents the first time that a XIIIB protein, truncated due to the delG mutation, was successfully expressed in a mammalian cell line and examined for its properties to understand further the molecular and cellular pathology of XIIIB deficiency.
Patients, materials, and methods
Patients
Two Japanese patients (patients 4 and 5) with factor XIII deficiency were identified through postoperative bleeding and umbilical bleeding, respectively. They were unrelated and came from 2 separate areas of Japan (Fukushima in the northeast part of Japan and Fukuoka on Kyushu Island). The functional activity of factor XIII in the plasma of these 2 patients was less than 10% of normal individuals.
Venous blood was drawn from normal individuals and patients with XIII deficiency and their family members after informed consent had been obtained. Genomic DNA was isolated from peripheral blood cells by standard techniques.
Immunoblot analysis
Plasma samples were diluted with a solution (20 mM Tris, pH 7.5, 120 mM sodium chloride) at 1:5, 1:10, 1:20, and 1:40 for XIIIA and 1:20, 1:40, 1:80, and 1:160 for XIIIB. An equal volume of a sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris, pH 6.8, 2% SDS, 15% glycerol, and 0.05% bromphenol blue) was added to the samples and mixed. After heat denaturing, 20 μL of each sample was subjected to 8% SDS–polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to nitrocellulose membranes (0.45 mm, ADVANTEC, Tokyo, Japan) using a semidry blotter (ATTO AE 6677, Tokyo, Japan) and incubated with either anti-XIIIA or anti-XIIIB antibodies (Calbiochem, San Diego, CA) at a dilution of 1:1000. Detection was achieved using a horseradish peroxidase (HRP)-conjugated antirabbit IgG antibody and an enhanced chemiluminescent substrate. The signal was semiquantified by the densitometric method.
In vitro amplification of the XIIIB gene and nucleotide sequence analysis
The XIIIB gene of each patient was amplified from 0.5 μg genomic DNA using 3 pairs of specific primers by polymerase chain reaction (PCR) using a LA PCR kit (Takara, Kyoto, Japan). The sequences of specific primers for the XIIIB gene were as follow: for exons I to V (∼7 kb), 5′-AAT TCT AGA CTC GTT ACA AAA GGA CTT AGA CAG AGG-3′ (sense) and 5′-GAT GGA TCC ACA GGA ATT TTG TCA GAG CTA AT-3′ (antisense); for exons VI to X (∼7 kb), 5′-TAG CTG CAG TCT GCC ATT CTC TCT ATG ATG-3′ (sense) and 5′-CAT AAG CTT AGT GTT ATG TTA ACT CAA AAA TGA CAG C-3′ (antisense); for exons XI and XII (∼1.7 kb), 5′-CAA GAA TTC CTG TTC TTA CCA AGA TGT AAC AAG -3′ (sense) and 5′-TAT GAA TTC TAG TCA ATG GGC ATT AGG AAA TG-3′ (antisense). PCR was performed with denaturation at 98°C for 20 seconds, and annealing and extension at 68°C for 10 minutes, for 30 cycles. Nucleotide sequence analysis was carried out with a BigDye Terminator cycle sequencing kit (PE Biosystems, Foster City, CA) using a 310 Genetic Analyzer (PE Biosystems).
Screening for mutations in the XIIIB gene by PCR–restriction fragment length polymorphism analysis
Three known mutations of the XIIIB gene (ag−[-]g, Cys430-Phe, and AAC insertion) and 2 polymorphisms (one at the 3′-untranslated region [A/G27970] and another in the 3′-flanking region) were determined as described previously.9,11 12For detection of a novel mutation (delG), an oligonucleotide with one base mismatch (in italic) was designed as an antisense primer, 5′-CTC TGT TGC ACT GCA CAG AGA ATT-3′ (E9Gdel). A fragment containing exon IX was amplified by a combination of E9Gdel and a sense primer, 5′-ATA AAG CTT TTG AAA CTT GTC ATA ATT TTT GTT TTA-3′ (B4-5H). PCR was carried out with denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds, 35 times. An amplified product of 260 bp was treated withEcoRI endonuclease at 37°C for 90 minutes; then the sample was applied to a 2% agarose gel.
Construction of expression vectors for wild-type and mutant recombinant XIIIBs
A full-length complementary DNA (cDNA) of XIIIB was cleaved from an expression vector for wild-type recombinant XIIIB (rXIIIB), ZMB3/rXIIIb,13 and subcloned into the EcoRI site of pBluescript II (Stratagene, La Jolla, CA), to yield pBSII/WT rXIIIB. A region of exon IX containing the novel delG mutation was amplified by PCR using patients' genomic DNA as a template. The amplified fragment was treated with RsaI and BsgI endonucleases, and then a released fragment of 145 bp was inserted into the RsaI-BsgI site of pBSII/WT rXIIIB to yield pBSII/Mut rXIIIB. An EcoRI fragment of the mutant rXIIIB cDNA was then cleaved from the pBSII/Mut rXIIIB and ligated into theEcoRI site of the ZMB3 expression vector to generate ZMB3/Mut rXIIIB.
Cell culture and selection of stable transfectants for wild-type and mutant rXIIIBs
Baby hamster kidney (BHK) cells were cultured in Dulbecco modified Eagle medium (DMEM; Nikken Biomedical Laboratory, Tokyo, Japan) supplemented with 10% (vol/vol) fetal bovine serum (MBL, Nagoya, Japan), and antibiotics (Gibco BRL, Gaithersburg, MD) at 37°C in a 5% CO2 incubator. Transfection was performed using the modified calcium phosphate precipitation method of Chen-Okayama. Stably transfected cells that expressed the wild-type or mutant rXIIIB were selected as described previously.10
Pulse-chase experiments
The day before these experiments were performed, rXIIIB-transfected cells (4 × 105 cells) were plated onto a 3-cm dish and incubated in serum-free DMEM without methionine (Met; Sigma, St Louis, MO) for 1 hour at 37°C before radiolabeling. Newly synthesized proteins were labeled with 25 μCi/plate of [35S]-Met in Met-depleted serum-free DMEM for 20 minutes (pulse), and further incubated in serum-free DMEM supplemented with unlabeled Met (chase). Samples of culture media and cell lysates were harvested at varying time intervals and stored immediately at −80°C. To remove nonspecificically absorbed proteins, the samples were pretreated with PANSOLBIN cells (Wako Chemicals, Tokyo, Japan) alone, and only supernatants were used for immunoprecipitation. The samples were incubated with a rabbit antihuman XIIIB antibody for 1 hour at 4°C and incubated further with PANSOLBIN cells at room temperature for 30 minutes with shaking. The precipitates were washed 3 × with 2 × Tris-buffered saline (TBS) containing 0.2% Tween 20. Bound antigen was eluted from PANSOLBIN cells by heating at 100°C for 5 minutes and subjected to 8% SDS-PAGE. The PAGE gel was dried and exposed to an imaging plate (Fuji Film, Tokyo, Japan). The exposed imaging plate was analyzed by FLA2000 (Fuji Film) using MacBAS version 2.x software.
Immunocytochemistry
The rXIIIB-transfected cells were washed twice with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 15 minutes, and then permeabilized with 0.1% Triton-X100 for 10 minutes at room temperature. The permeabilized cells were washed twice with PBS, incubated with 2% bovine serum albumin (BSA) in PBS, and reacted with a rabbit antihuman XIIIB antibody at a dilution of 1:1000 in PBS containing 2% BSA for 4 hours at room temperature. The washed cells were incubated for another 2 hours at room temperature with a goat antirabbit IgG conjugated with fluorescein 5-isothiocyanate (FITC; Organon Teknika, Durham, NC) at a dilution of 1:1000 in PBS containing 2% BSA. Immunocytochemical staining was repeated using a mouse monoclonal antibody against Golgi 58K protein (Golgi58; Biomaker, Rehovot, Israel) as a Golgi marker or against protein disulfide isomerase (PDI; Stressgen, Victoria, BC, Canada) as an endoplasmic reticulum (ER) marker, and a rhodamin-conjugated antimouse IgG (ICN, Aurora, OH). The anti-Golgi 58K protein and anti-PDI antibodies were used at dilutions of 1:200 and 1:1000, respectively. After 3 washes with PBS, cover slides were mounted with 0.01% paraphenylendiamine in PBS containing 90% glycerol, and the samples were observed under an epifluorescence or confocal microscope (Olympus IX-70, Tokyo, Japan).
Immunoelectronmicroscopy
The rXIIIB-transfected cells were reacted with a rabbit antihuman XIIIB antibody and a goat antirabbit IgG conjugated with FITC by the same procedure as immunocytochemistry, except for the usage of suspended cells. After reacting with an HRP-conjugated rabbit anti-FITC IgG (Dako, Glostrup, Denmark), the cells were washed 3 × with 1% BSA in PBS and incubated with a DAB–hydrogen peroxide solution at room temperature for 10 minutes. After 3 washings with 1% BSA in PBS, the samples were immersed for 10 minutes in 2.5% buffered glutaraldehyde, washed in 0.1% cacodylate buffer (pH 7.4), refixed with 1% osmium tetroxide (pH 7.2) for 1 hour, and washed again in the cacodylate buffer. After dehydration in ethanol, the pieces were embedded in Epon 812 (Nisshin EM, Tokyo, Japan). Ultrathin sections were cut for electron microscopy (EM109, Zeiss, Esslingen, Germany).
Results
Immunoblot analysis of plasma samples
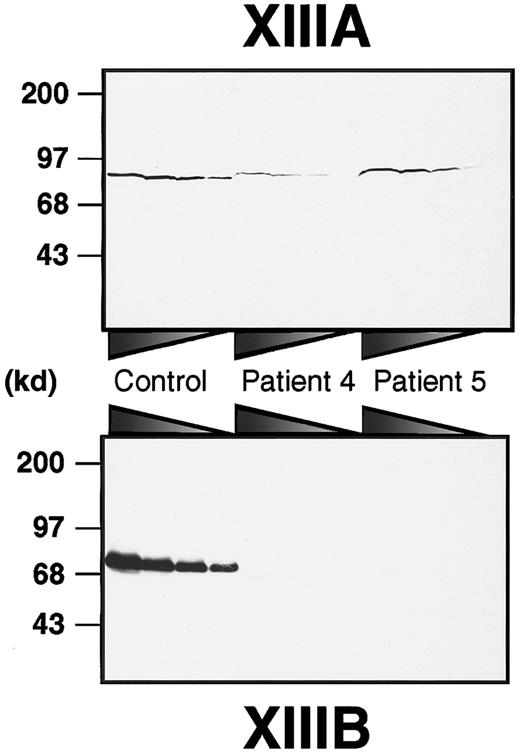
To obtain a rough estimate of the amounts of both subunits in each patient, immunoblot analyses of plasma samples were performed using anti–human XIIIA or XIIIB antibodies (Figure1). XIIIB was scarcely detectable in the patients, although it was clearly detected in a normal sample even at 160 × dilution. On the other hand, XIIIA was detectable in all plasma samples. In patient 4, the XIIIA was significantly decreased to about 12% of normal plasma. The plasma XIIIA level in patient 5 was approximately 40%, although it varied between 5% and 40% through his neonatal period. These results clearly indicate that both patients had complete XIIIB deficiency (former type I factor XIII deficiency).
Immunoblot analysis of XIIIA and XIIIB in patients' plasma.
Plasma samples were diluted at ratios of 1:5, 1:10, 1:20, and 1:40 for XIIIA, and 1:20, 1:40, 1:80, and 1:160 for XIIIB from the left to the right. Each sample was subjected to 8% SDS-PAGE under nonreducing conditions and immunoblot analysis using an anti-XIIIA or anti-XIIIB antibody.
Immunoblot analysis of XIIIA and XIIIB in patients' plasma.
Plasma samples were diluted at ratios of 1:5, 1:10, 1:20, and 1:40 for XIIIA, and 1:20, 1:40, 1:80, and 1:160 for XIIIB from the left to the right. Each sample was subjected to 8% SDS-PAGE under nonreducing conditions and immunoblot analysis using an anti-XIIIA or anti-XIIIB antibody.
Search for known mutations in the XIIIB gene
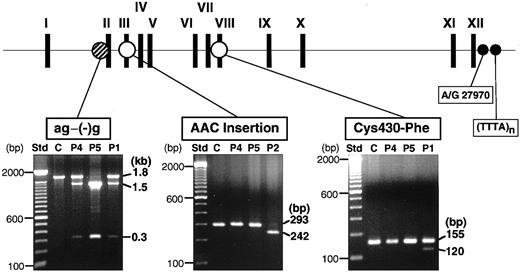
We had previously identified 3 mutations (ag−(−)g, Cys430-Phe, and AAC insertion) in the XIIIB genes of all the known patients with XIIIB deficiency worldwide (Figure2, top). To look for the presence of these mutations, genomic DNAs of these patients were tested by PCR–restriction fragment length polymorphism (RFLP) analyses. The ag−(−)g mutation, identified in patient 1 heterozygously, creates a recognition site for TaqI endonuclease,9 so that TaqI-treatment of a 1.8-kb fragment amplified from patient 1 yielded 3 fragments (Figure 2, bottom): an uncleaved fragment of 1.8 kb for the ag allele, and cleaved fragments of 1.5 and 0.3 kb for the (−)g allele. The same digestion pattern was observed in patient 4, indicating that patient 4 is a heterozygote for the ag−(−)g mutation. On the other hand, an amplified DNA from patient 5 was completely cleaved into 2 fragments of 1.5 and 0.3 kb, indicating that patient 5 was a homozygote for this mutation.
Screening of known mutations by PCR-RFLPs.
The locations of 3 known mutations in the XIIIB gene are indicated at the top. PCR-RFLP analyses were performed for 2 new patients (patient 4, P4; patient 5, P5), a normal control (C), and 2 previously identified patients (P1, P2): for the ag−(−)g mutation, a 1.8-kb amplified fragment was treated with TaqI; for the AAC insertion, a 430-bp amplified fragment withTru9I; for the Cys430-Phe mutation, a 155-bp amplified product with MboII. Each reaction mixture was electrophoresed in 1%, 2%, and 2% agarose gels, respectively. Patient 1 (P1) was a positive control for the ag−(−)g and Cys430-Phe mutations and patient 2 (P2) for the AAC insertion. Std indicates standard size markers (100-bp ladder). Two polymorphic sites (A/G27970 and TTTA repeats) at the 3′-flanking region of the XIIIBgene were also indicated by small filled circles.
Screening of known mutations by PCR-RFLPs.
The locations of 3 known mutations in the XIIIB gene are indicated at the top. PCR-RFLP analyses were performed for 2 new patients (patient 4, P4; patient 5, P5), a normal control (C), and 2 previously identified patients (P1, P2): for the ag−(−)g mutation, a 1.8-kb amplified fragment was treated with TaqI; for the AAC insertion, a 430-bp amplified fragment withTru9I; for the Cys430-Phe mutation, a 155-bp amplified product with MboII. Each reaction mixture was electrophoresed in 1%, 2%, and 2% agarose gels, respectively. Patient 1 (P1) was a positive control for the ag−(−)g and Cys430-Phe mutations and patient 2 (P2) for the AAC insertion. Std indicates standard size markers (100-bp ladder). Two polymorphic sites (A/G27970 and TTTA repeats) at the 3′-flanking region of the XIIIBgene were also indicated by small filled circles.
Neither of 2 other known mutations were detected in patient 4 or 5, although they were observed in the index cases (patients 1 and 2) in whom each mutation had been identified previously (Figure 2, bottom).
Genetic relationship among 3 Japanese patients
All 3 patients (patients 1, 4, and 5) with the ag−(−)g mutation are ethnic Japanese but unrelated. The ag−(−)g mutation was not found in either of 2 Italian patients (patients 2 and 3, data not shown). To search for the genetic relationship among the 3 Japanese patients, the ag−(−)g mutation and 2 known polymorphisms in the 3′-region of theXIIIB gene were determined among the patients and members of their families (Figure 3). The 2 known polymorphisms were nucleotide A/G27970 in the 3′-untranslated region, and a short tandem repeat of (TTTA)n (n = 7-10) in the 3′-flanking region (Figure 2, top). Gene frequencies of these polymorphisms among 90 normal Japanese individuals were as follow: G27970 and A27970, 0.813, and 0.187, respectively; and 10-, 9-, and 8-repeats of TTTA, 0.733, 0.217, and 0.050, respectively.
Family pedigrees and genotyping of Japanese cases with the ag−(−)g mutation.
Black and gray fill-ins indicate the ag−(−)g mutation and other mutations, respectively. Genotypes for the ag−(−)g mutation and 3′-polymorphisms (A/G27970 and TTTA repeats) were determined in the patients and their family members.
Family pedigrees and genotyping of Japanese cases with the ag−(−)g mutation.
Black and gray fill-ins indicate the ag−(−)g mutation and other mutations, respectively. Genotypes for the ag−(−)g mutation and 3′-polymorphisms (A/G27970 and TTTA repeats) were determined in the patients and their family members.
In a homozygote for the ag−(−)g mutation (patient 5), the polymorphism of G27970 and 10-repeat of TTTA were also found homozygously (Figure 3). Furthermore, both parents of patient 5 were heterozygous for the ag−(−)g mutation and possessed at least one allele of G27970 and a 10-repeat of TTTA. Therefore, this mutation should be traced to the same XIIIB allele containing the G27970 polymorphism and 10-repeat of TTTA. This conclusion is also in good agreement with the results for patients 1 and 4, where it was determined that the ag−(−)g mutation was likely to be linked with G27970 and the 10-repeat of TTTA.
Identification of a new mutation in patient 4
From the above results, it was predicted that patient 4 had another mutation(s) in the remaining XIIIB allele. Nucleotide sequence analysis of the XIIIB gene was then carried out for patient 4. First, PCR-amplified DNAs were sequenced directly. The ag−(−)g mutation was confirmed to be heterozygous, and a novel mutation was also found in exon IX. Doublet peaks of the nucleotide started to appear from the nucleotide position of 14522 in both sense and antisense directions (data not shown), suggesting the presence of a deletion/insertion. To confirm this mutation, PCR-amplified exon IX was subcloned, and 8 subclones were then sequenced. A novel mutation, the deletion of one guanosine at the nucleotide position of 14522 (delG), was identified in 3 out of the 8 subclones.
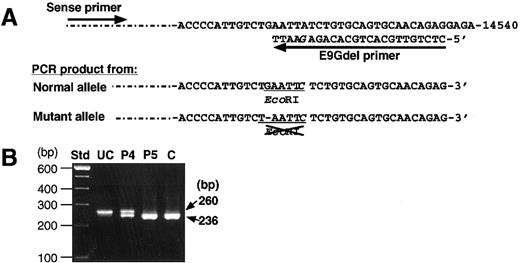
Analysis by PCR-RFLP was also performed to confirm the presence of the novel delG mutation. A PCR product amplified from the normal allele had a new EcoRI site, but one from the delG allele did not (Figure 4A). Therefore, the PCR product of 260 bp from a normal control would be completely cleaved withEcoRI endonuclease into 2 fragments of 236 bp and 24 bp, although the latter would scarcely be detectable because of its small size. The amplified products from patient 5 and a normal individual were completely cleaved by the EcoRI treatment (Figure 4B). However, the PCR product of patient 4 yielded 2 fragments of 236 bp and 24 bp, whereas an uncleaved fragment of 260 bp remained even after prolonged EcoRI digestion. These results indicate that the delG mutation was present heterozygously in patient 4. Because the delG mutation was not found among 90 normal Japanese individuals, it must be a second causative mutation in the remaining XIIIB allele of patient 4, which is responsible for XIIIB deficiency.
Detection of the delG mutation by PCR-RFLP.
(A) A mutagenic primer (E9Gdel) with one base mismatch (G in italics) was designed to create an EcoRI site only in the PCR product with a normal sequence. (B) Amplified products from patients (patient 4, P4; patient 5, P5), and a normal control (C) were treated withEcoRI and then electrophoresed in a 2% agarose gel. The product of 260 bp amplified from the normal allele is cleaved into 2 fragments of 236 bp and undetectable 24 bp. An uncleaved PCR product (UC) amplified from patient 4 was also applied as a negative control.
Detection of the delG mutation by PCR-RFLP.
(A) A mutagenic primer (E9Gdel) with one base mismatch (G in italics) was designed to create an EcoRI site only in the PCR product with a normal sequence. (B) Amplified products from patients (patient 4, P4; patient 5, P5), and a normal control (C) were treated withEcoRI and then electrophoresed in a 2% agarose gel. The product of 260 bp amplified from the normal allele is cleaved into 2 fragments of 236 bp and undetectable 24 bp. An uncleaved PCR product (UC) amplified from patient 4 was also applied as a negative control.
Sequencing analyses of all PCR products amplified from patient 5 showed the homozygous ag−(−)g mutation but no other mutations in theXIIIB gene, which is consistent with the results of the PCR-RFLP analyses.
Impaired secretion of truncated rXIIIB
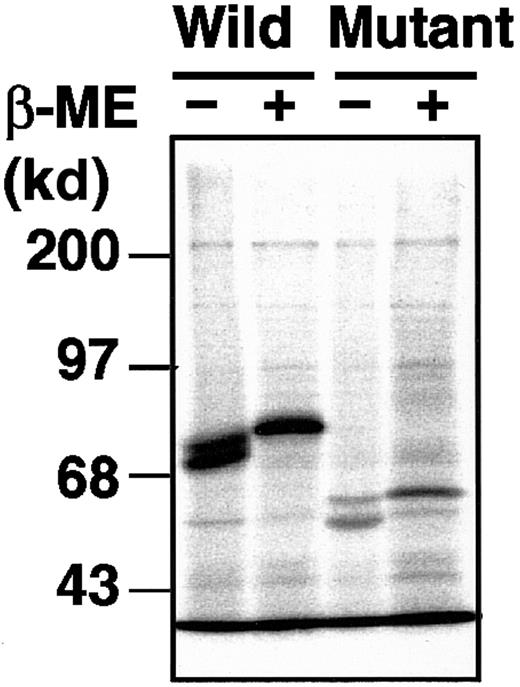
The delG mutation, newly identified in the XIIIB gene of patient 4, introduces a premature stop codon with a frameshift, resulting in the production of a truncated XIIIB. To understand the molecular and cellular pathology of this novel mutation, we performed expression studies using a mammalian cultured cell line (BHK) and expression vectors for wild-type and delG rXIIIBs. The rXIIIB-expressing cells were selected by expression levels determined by Northern blot and immunoblot analyses (data not shown). In metabolic labeling studies, a wild-type rXIIIB of about 80 kd and a truncated rXIIIB of about 60 kd were detected in the lysates of rXIIIB-transfected cells (Figure 5); thus, the actual size of each band corresponded well to its expected size. No dimer was observed for the mutant rXIIIB under nonreducing conditions, although the eighth Sushi domain of the mutant would contain an odd number of cysteine residues3 and thus one cysteine residue must be left unpaired. These results are in contrast to our previous report in which a Cys430-Phe mutant did form a dimer.10
Metabolic labeling of the wild-type and mutant rXIIIBs.
Radiolabeled cell lysate and culture medium, harvested at 0 hour after labeling, were immunoprecipitated with an anti-XIIIB antibody, followed by 10% SDS-PAGE under nonreducing or reducing conditions, with or without β-mercaptoethanol (β-ME).
Metabolic labeling of the wild-type and mutant rXIIIBs.
Radiolabeled cell lysate and culture medium, harvested at 0 hour after labeling, were immunoprecipitated with an anti-XIIIB antibody, followed by 10% SDS-PAGE under nonreducing or reducing conditions, with or without β-mercaptoethanol (β-ME).
In pulse-chase experiments, it was shown that the wild-type rXIIIB was exported from the cells to the medium immediately, and that nearly all labeled rXIIIB was found in the medium 2 hours after labeling (Figure6A). The exported rXIIIB was present at a fairly constant level in the medium for 24 hours (Figure 6B). The truncated rXIIIB, however, remained inside the cells and could not be secreted into the medium at all. After 2 hours, the truncated rXIIIB in the cells was decreased to 50% of that at 0 hour.
Pulse-chase experiments for rXIIIBs.
Experiments were carried out identically to those in Figure 5, except that radiolabeled samples were taken at various time intervals and electrophoresed under reducing conditions (A). The intensity of the band for cell lysates harvested at 0 hour was defined as 100%, and the relative intensity of each band was plotted (B). Open and closed circles show cell lysates and culture media of the wild type, respectively. Open and closed squares depict cell lysates and culture media of the delG mutant, respectively.
Pulse-chase experiments for rXIIIBs.
Experiments were carried out identically to those in Figure 5, except that radiolabeled samples were taken at various time intervals and electrophoresed under reducing conditions (A). The intensity of the band for cell lysates harvested at 0 hour was defined as 100%, and the relative intensity of each band was plotted (B). Open and closed circles show cell lysates and culture media of the wild type, respectively. Open and closed squares depict cell lysates and culture media of the delG mutant, respectively.
Intracellular localization of truncated rXIIIB
In general, a secretory protein goes through the ER to the Golgi apparatus before secretion. Therefore, we determined the intracellular localization of the wild-type and mutant rXIIIBs by an immunocytochemical staining method using a rabbit antihuman XIIIB antibody. Stably transfected cells of the wild-type rXIIIB showed a typical staining pattern of secretory proteins (Figure7A): no signal was detected in mock cells (data not shown). Intense signals for the wild-type rXIIIB were observed mainly near the nuclei, which likely corresponds to the Golgi, and were colocalized with the signals for Golgi 58K protein (Figure 7A, top). In contrast, cells of the mutant rXIIIB were stained in a reticular pattern throughout the cytoplasm (Figure 7A, bottom). The signals for the mutant rXIIIB were colocalized with those for an ER marker protein, PDI. The same patterns of distribution for the wild-type and mutant rXIIIB were also observed by confocal microscopy (data not shown). In addition, immunoelectron microscopic pictures clearly showed accumulation of rXIIIB in the Golgi of BHK cells expressing the wild-type (Figure 7B, top), whereas rXIIIB locates in the rough ER of cells expressing the mutant protein (Figure7B, bottom).
Intracellular localization of wild-type and mutant rXIIIBs.
Immunocytochemical staining with an anti-XIIIB antibody and an anti-Golgi 58K protein (Golgi58) or anti-PDI antibody was carried out using BHK cells expressing wild-type (WT) or mutant rXIIIB (delG). The samples were examined by an epifluorescence microscope × 400 (A). Electron microscopic immunolocalization of rXIIIB was also performed using an anti-XIIIB antibody and ultrathin sections of infected BHK cells (B). Boxed areas of pictures on the left (× 3000) were reexamined at a magnification of × 12 000 (right).
Intracellular localization of wild-type and mutant rXIIIBs.
Immunocytochemical staining with an anti-XIIIB antibody and an anti-Golgi 58K protein (Golgi58) or anti-PDI antibody was carried out using BHK cells expressing wild-type (WT) or mutant rXIIIB (delG). The samples were examined by an epifluorescence microscope × 400 (A). Electron microscopic immunolocalization of rXIIIB was also performed using an anti-XIIIB antibody and ultrathin sections of infected BHK cells (B). Boxed areas of pictures on the left (× 3000) were reexamined at a magnification of × 12 000 (right).
These results clearly indicate that intracellular transportation of the truncated rXIIIB to the Golgi was impaired and it was stacked within the ER.
Discussion
In the present study, 2 Japanese patients were newly diagnosed as having XIIIB deficiency. At this writing, as few as 5 cases of XIIIB deficiency have been identified in the world and these were found only in Italy and Japan. In contrast, more than 200 cases of XIIIA deficiency have been reported, and they were distributed worldwide. In general, XIIIA deficiency shows a nearly complete loss of the catalytic XIIIA protein in plasma, which is caused by defects in theXIIIA gene.14 On the other hand, the catalytic XIIIA in plasma significantly decreased but remained at about 5% to 40% of normal in some cases of XIIIB deficiency. Therefore, the clinical symptoms of XIIIB deficiency may be milder, and perhaps XIIIB deficiency is more difficult to discover than XIIIA deficiency. There may be many more undiagnosed cases of XIIIB deficiency in the world today; nevertheless, it appears to be an extremely rare condition.
The ag−(−)g mutation at the boundary of intron A/exon II, previously identified in a Japanese patient, was detected in 2 other Japanese patients (patients 4 and 5). PCR-RFLPs and nucleotide sequence analysis proved that patient 5, especially, was a homozygote for this mutation and had no other mutations in the XIIIB gene. It is thought that this defective XIIIB gene would lead to abnormal splicing of its messenger RNA,9 resulting in a complete loss of XIIIB in the patient's plasma (Figure 1). It is of significant interest that this same ag−(−)g mutation of the XIIIB gene was found among 3 Japanese patients (patients 1, 4, and 5) who live in 3 different parts of Japan: Kanazawa, Fukushima, and Fukuoka, respectively. Judging from the results of PCR-RFLPs for the ag−(−)g mutation and 3′-polymorphisms, it is very likely that the ag−(−)g mutation exists on a XIIIB allele containing G27970 and 10 repeats of TTTA, which is common among all 3 cases. Although these patients claim no immediate blood relation, they may share a common ancestor. We previously proposed a founder effect for XIIIB deficiency in Italians, which is caused by an AAC insertion.12 Similarly, a founder effect is now proposed for XIIIB deficiency in Japanese, which is caused by the ag−(−)g mutation.
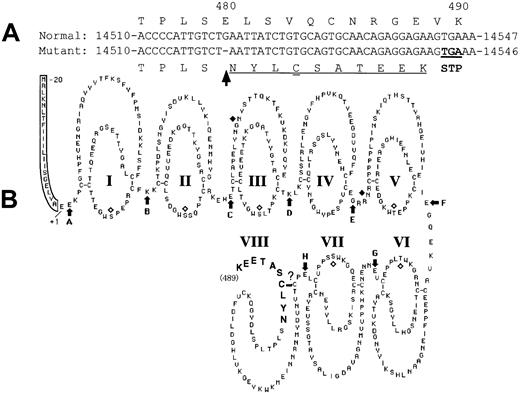
A novel mutation, a deletion of guanosine in exon IX (delG), was identified in one patient (patient 4) heterozygously as a second mutation. As a result of this delG mutation, a premature stop codon would be expected to be introduced in the XIIIB gene with a frameshift, resulting in the production of a truncated XIIIB (Figure8). The resulting truncated XIIIB will then consist of 489 amino acids containing 7 normal tandem repeats called “Sushi domains” and one partial Sushi domain to which 10 extra amino acids attached (Figure 8B, bold). A truncated rXIIIB of about 60 kd was actually synthesized in the mutant rXIIIB-transfected cells, as shown by the current expression studies. Pulse-chase experiments and immunocytochemical staining of the cells bearing the delG XIIIB mutant clearly demonstrated impaired intracellular transportation of the truncated rXIIIB from the ER to the Golgi, and the consequent impaired secretion. Thus, it is noteworthy that truncation as well as amino acid substitution of XIIIB results in the same severe disorder.10
Scheme of the mutant XIIIB.
The delG mutation (arrow) introduces a premature stop codon (double underlined) in the XIIIB gene with a frameshift (underlined) (A), resulting in the production of a truncated XIIIB (B) attached by 10 new amino acids (bold).
Scheme of the mutant XIIIB.
The delG mutation (arrow) introduces a premature stop codon (double underlined) in the XIIIB gene with a frameshift (underlined) (A), resulting in the production of a truncated XIIIB (B) attached by 10 new amino acids (bold).
There has been no information on the exact tertiary structure of XIIIB to date, because XIIIB is not readily crystallized mainly because of the heterogeneity in its carbohydrate chains. Therefore, it is not possible to predict likely effects of this delG mutation on the structure of the XIIIB molecule by molecular modeling, as was done for XIIIA.15,16 As discussed above, the mutant XIIIB has 7 intact Sushi domains. However, there must be some conformational change, at least in its eighth Sushi domain because it is truncated in the middle of the domain and extends the length of 10 entirely new amino acids. Moreover, an additional cysteine residue is introduced in this region of the mutant molecule and remains unpaired. These structural changes would lead to misfolding of the abnormal XIIIB molecule at the C-terminal region, and possibly affect the entire structure of XIIIB. The truncated and misfolded mutant XIIIB might then be trapped by some ER proteins like chaperones, which would prevent its transportation from the ER to the Golgi.17 This mechanism is called ER retention.18
In a cultured mammalian cell line, the delG mutation of theXIIIB gene resulted in impaired intracellular transportation of the truncated rXIIIB from the ER to the Golgi. Accordingly, the transportation of the truncated XIIIB must also be impaired in vivo, more specifically in the liver of patient 4 because XIIIB is synthesized by hepatocytes.19 20 Immunoblot analysis of the patient's plasma indicated that XIIIA was also significantly reduced (Figure 1). No mutations of the XIIIA gene, however, were identified in patient 4 by sequencing analysis. Accordingly, it is highly likely that the lack of plasma XIIIB resulted in a secondary loss of XIIIA in the patient's plasma because of the innate instability of the XIIIA molecule.
The results obtained by the current studies strengthen our previous conclusion that genetic defects of XIIIB are the basis for the former type I factor XIII deficiency.
The authors thank Mr. T. Tamura for sequencing these patients'XIIIA gene, Dr H. Kaetsu for providing purified plasma XIII and XIIIB, Drs T. Izumi and F. Tokunaga for helpful discussions, Dr T. Shiraishi for making a confocal microscope available, and Ms L. Boba for her help in preparation of the manuscript.
Supported in part by research grants from the Ministry of Education, Science and Culture, Japan (11470205,11770071), the Ministry of Health and Welfare, the Uehara Memorial Foundation (Japan), and the Ryoichi Naito Foundation for Medical Research (Japan).
The first and second authors contributed equally to the completion of this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Akitada Ichinose, Department of Molecular Patho-Biochemistry and Patho-Biology, Yamagata University School of Medicine, Iida-Nishi 2-2-2, Yamagata 990-9585 Japan; e-mail:aichinos@med.id.yamagata-u.ac.jp.