Abstract
Oxygen deprivation (hypoxia) is a consistent component of ischemia that induces an inflammatory and prothrombotic response in the endothelium. In this report, it is demonstrated that exposure of endothelial cells to hypoxia (1% O2) increases messenger RNA and protein levels of transforming growth factor-β2 (TGF-β2), a cytokine with potent regulatory effects on vascular inflammatory responses. Messenger RNA levels of the TGF-β2 type II membrane receptor, which is a serine threonine kinase, also increased. The stimulatory effect of hypoxia was found to occur at the level of transcription of the TGF-β2 gene and involves Smad proteins, a class of intracellular signaling proteins that mediates the downstream effects of TGF-β receptors. Transient transfection studies showed that the region spanning −77 and −40 base pairs within the TGF-β2 promoter (harboring a Smad-binding “CAGA box”) is activated in hypoxic cells compared with nonhypoxic controls (P < .01). Hypoxia also stimulated transcription from another promoter, 3TP-Lux, a reporter construct responsive to Smads and TGF-β. In addition, specific binding to a Smad-binding oligonucleotide was observed with nuclear extracts from hypoxic endothelial cells but not from nonhypoxic cells. It is concluded that Smad proteins, which can regulate endothelial responses to mechanical and inflammatory stress, also may play an important role in vascular responses to hypoxia and ischemia.
Introduction
One of the earliest responses of endothelium to hypoxia is inflammation, and the balance between positive and negative regulators of this inflammatory response is a determinant of endothelial adaptation to hypoxia. Proinflammatory endothelial responses induced by hypoxia involve production of cytokines, chemokines,1-3 tissue factor,4 and plasminogen activator inhibitor-1 (PAI-1).5 Recent studies indicate that activation of the transcription factor early growth response-1 (Egr-1) by hypoxia is responsible for the procoagulant profile and vascular remodeling that ultimately result in the tissue damage associated with ischemia.4 Adaptive responses to hypoxia are also regulated at a transcriptional level, which is best exemplified by activation of the transacting protein hypoxia-inducible factor-1 (HIF-1), which stimulates differential expression of specific genes and results in increases in glucose uptake, glycolytic enzyme production, and angiogenesis.6 7
The transforming growth factor-β (TGF-β) family of growth factors has an especially critical role in modulation of vascular inflammatory responses and remodeling,8-10 and up-regulation of TGF-β production is an invariant response to vascular injury, indicating that regulation of gene expression of the TGF-β proteins by quiescent and injured endothelium is likely to be a critical factor affecting the course of vascular inflammation. This hypothesis is supported by demonstration of defective vasculogenesis and increased and ultimately lethal vascular inflammation in mice null for TGF-βs,11,12 their membrane receptors,13,14or their downstream substrates, the Smad proteins.15 16
Three mammalian isoforms of TGF-β (TGF-β1, TGF-β2, TGF-β3)11,12,17,18 are secreted by endothelial cells in a latent form in which latency-associated peptide (LAP) and its 25 kd carboxy-terminal dimer, which is the mature TGF-β,19,20remain noncovalently bound. In vivo release of the mature dimer from LAP occurs via its degradation or conformational change19,21 and allows binding of the mature (bioactive) peptide to its specific membrane receptors TGF-βRI, TGF-βRII, and TGF-βRIII, of which types I and II are serine threonine kinases.22-24 Binding of the 25 kd TGF-β dimer to the high-affinity type II receptor results in recruitment and activation of the type I receptor by phosphorylation at its cytoplasmic domain by the type II receptor kinase. Activated TGF-βR1 phosphorylates intracellular Smad proteins, which propagate downstream signals of TGF-βs.25 Phosphorylation of Smad2 and Smad3 (receptor-activated Smads) causes their dissociation from TGF-βR1 and stimulates their assembly with Smad4 (co-Smad)22,26; this complex then translocates and accumulates in the nucleus. Heterodimeric Smad complexes can bind DNA in either orientation of a 5′-CAGA-′ consensus sequence (Smad-binding element [SBE]27-29) and can directly regulate transcription by recruiting coactivators or inhibitors of transcription.30
Recent studies of human endothelial cells and in vivo animal studies have shown that Smad proteins regulate endothelial responses to mechanical stress such as shear31,32 and to inflammatory stress such as exposure to bacterial lipopolysaccaride.33It seemed reasonable, therefore, to test the hypothesis that TGF-βs and their downstream Smad signaling pathway can be activated by hypoxic stress in vascular endothelium in humans. In this report, we show that hypoxia selectively up-regulates the TGF-β2 isoform by as much as 20-fold in endothelial cells in a time-dependent fashion. Furthermore, we show for the first time that hypoxia activates Smad-mediated gene transcription and Smad-DNA association in human endothelial cells. These data indicate a new function for Smads in endothelial cells as one of the signaling pathways activated by hypoxia, one that is capable of modulating inflammatory consequences of ischemia.
Materials and methods
Cell cultures
Primary human umbilical vein endothelial cells (HUVECs) were isolated and propagated from pooled primary cultures of umbilical veins by digestion with collagenase type II and were maintained as described by Gimbrone.34 Briefly, HUVECs were grown on gelatin (2%)-coated tissue culture flasks (Costar, Cambridge, MA) in endothelial cell growth medium that consisted of medium 199 (Gibco, Grand Island, NY) supplemented with endothelial cell growth supplement (50 μg/mL; Calbiochem, San Diego, CA), 20% fetal bovine serum, antibiotics (100 U/mL penicillin G, 100 μg/mL streptomycin), and heparin (50 μg/mL, porcine intestinal; Sigma, St Louis, MO). Cells were passaged by trypsin–ethylenediaminetetraacetic acid (EDTA) treatment, split 1:4, fed every 1 to 3 days, and used at second or third passage, when nearly confluent (70%). Cytoplasmic von Willebrand factor by immunocytochemistry and nonoverlapping cobblestone morphology verified endothelial cells. For exposure to hypoxia, cells were incubated under an atmosphere of 1% O2, 5% CO2, and 94% N2 at 37°C in a NAPCO 7301 incubator (Precision Scientific, Chicago, IL). Previous experiments in our laboratory have shown that during exposure to hypoxia, the pH of the medium remains unchanged, pO2 in the medium is 10 to 16 mm Hg, and viability is more than 95% as measured by trypan blue dye exclusion and determination of release of lactate dehydrogenase. Experiments were performed in parallel on normoxic and hypoxic cells that were exposed for the times indicated to hypoxia or recombinant TGF-β2 (R&D Systems, Minneapolis, MN). HUVEC viability assessed after each treatment was consistently more than 95%. HUVEC culture reagent α-amanitin was obtained from Sigma.
RNase protection assay
Total RNA was prepared from treated or control HUVECs using TRIzol (Gibco) and quantified as previously described.35Ribonuclease (RNase) protection analysis was done using the RiboQuant Multi-Probe RNase protection assay system (PharMingen, San Diego, CA). In this assay, high-specific–activity RNA probes are synthesized from a mixture of DNA templates of distinct lengths that correspond to a sequence in a distinct messenger RNA (mRNA) species. Template sets hCK-3 and hCR-4 (PharMingen) were used; hCK-3 contains templates for tumor necrosis factor-β (TNF-β), TNF-α, lymphotoxin-β, interferon-β, interferon-γ, TGF-β1, TGF-β2, TGF-β3, and the ubiquitously expressed genes for ribosomal protein L32 and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH); hCR-4 contains templates for type I interleukin-1 receptor (IL-1RI), type II IL-1 receptor (IL-1RII), p55 type I TNF-α receptor (TNF-αRI), p75 type II TNF-α receptor (TNF-αRII), IL-6 receptor α (IL-6Rα), glycoprotein 130, TGF-βRI, TGF-βRII, L32, and GAPDH. T7 polymerase–directed antisense RNA probe synthesis, hybridization to total RNA, RNase treatments, and gel resolution of protected probes were done exactly as described by the manufacturer. Briefly, probe was synthesized by labeling the template with [α-32P]UTP (New England Nuclear, Boston, MA) using T7 RNA polymerase. A total of 6 × 105 cpm of labeled probe in hybridization solution was added to 10 μg total RNA, and the samples were overlaid with mineral oil before brief heating to 90°C; hybridization continued for 16 hours at 56°C. After hybridization, the samples were digested for 45 minutes at 30°C with a mixture of RNase A and RNase T1 (as recommended in the kit), treated with proteinase K, extracted with phenol/chloroform/isoamyl alcohol (50:50:0.5), precipitated with ethanol, and resolved on 5% denaturing polyacrylamide sequencing gels. Radioactive fragments were detected by autoradiography and also by PhosphorImager (Molecular Dynamics, Sunnyvale, CA), from which quantification of mRNA was done by a volume integration protocol using ImageQuant software (Molecular Dynamics). To control for differences in sample processing, hybridization signals in each sample were divided by the signal for L32, which by independent analysis was found not to change with exposure to hypoxia (not shown). For determining fold increases in mRNA levels in hypoxic HUVECs compared with control, the cytokine/L32 signal ratio from hypoxic HUVECs was divided by the cytokine/L32 ratio obtained from normoxic HUVECs.
TGF-β assay
TGF-β–responsive mink lung epithelial cells (MLECs) stably transfected with the expression construct p800neoLUC containing a truncated PAI-1 promoter fused to the firefly luciferase reporter gene were kindly supplied by Dr Daniel B. Rifkin (New York University School of Medicine, New York, NY). Cells were maintained in Dulbecco modified Eagle medium containing 10% fetal calf serum, 2 mM L-glutamine, and antibiotics (100 U/mL penicillin G, 100 μg/mL streptomycin, 250 μg/mL geneticin; Gibco). To assay bioactive and total TGF-β2 levels in HUVEC supernatants, MLECs were detached with trypsin, washed, plated at 1.6 × 105 cells per well in a 2 mL volume in 24-well tissue culture plates (Costar), and were allowed to attach for 18 hours at 37°C in 5% CO2. Medium in the wells was replaced by 2 mL of the following, placed in triplicate wells: control medium; control medium containing increasing concentrations of recombinant TGF-β2 to generate a standard curve; 1:10 dilution of conditioned HUVEC medium to measure bioactive TGF-β; and 1:10 dilution of conditioned HUVEC medium activated for 10 minutes by heating at 80°C to measure total TGF-β. Incubations were continued for 18 hours at 37°C, and MLEC lysates from each well were prepared using reporter lysis buffer (Luciferase Assay System, Promega, Madison, WI). Luciferase activity was measured as relative light units (RLUs; model TD-20/20 luminometer, Promega), which were converted to TGF-β activity (picomoles) using the TGF-β2 standard curve.20Total protein content of supernatants was quantitated using Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA),36 and TGF-β2 levels obtained by the luciferase assay from each well were normalized to protein. Mean RLUs from triplicate wells (which were within 5%-7% of one another) were converted to picomoles of TGF-β2. This assay is sensitive to all 3 isoforms of TGF-β; therefore, to specifically quantitate total TGF-β2 in hypoxic and nonhypoxic supernatants, aliquots of the identical supernatants used in MLEC assay were acid-activated and subjected to enzyme-linked immunosorbent assay (ELISA) (Quantikine, R&D Systems) as previously described.37 Results (not shown) confirmed that TGF-β2 levels were increased by 2-fold in hypoxic supernatants. Because measurement of bioactive TGF-βs by ELISA is not yet reliably accomplished by existing commercial kits, this assay can only confirm the increase in total TGF-β2 determined by the MLEC assay.
Nuclear run-on assay for TGF-β2
The rate of transcription of TGF-β2 in HUVECs was determined as described by Greenberg and Ziff38 in nuclei prepared from HUVECs cultured in either normoxic or hypoxic culture conditions. Briefly, a suspension of 6 × 109 nuclei in 200 μL glycerol (40%) in 50 mM Tris-HCl buffer (pH 8.3, with 5 mM MgCl2 and 0.1 mM EDTA) was mixed with 2 × reaction buffer (10 mM Tris-HCl [pH 8]; 5 mM MgCl2; 0.3 M KCl; 5 mM dithiothreitol; 1 mM each ATP, CTP, GTP; and 100 μCi [3.7 × 106 Bq] [32P]UTP [1.1 × 1014 Bq, 3000 Ci/mM; PerkinElmer Life Sciences, Boston, MA]). After incubation for 30 minutes at 30°C, [32P]-labeled RNA was extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1) and 50 μg transfer RNA, and 10% trichloracetic acid (TCA) were added. The mixture was filtered through a 0.45-μm Millipore filter (Millipore, Bedford, MA). Nuclear RNA from the filter was released by treatment with 20 mM HEPES buffer (pH 7.5, with 5 mM MgCl2 and 1 mM CaCl2) and deoxyribonuclease for 30 minutes at 37°C, addition of 45 μL of 0.5 M EDTA and 68 μL of 20% SDS, and incubation at 65°C for 10 minutes. Nuclear RNA was digested with proteinase K, extracted with phenol/chloroform/isoamyl alcohol, and ethanol-precipitated at −20°C overnight. TGF-β2, Glut-1, and GAPDH complementary DNA probes have been described.39-41 RNA was hybridized for 36 hours at 65°C to linearized plasmid DNA (TGF-β2, Glut-1, GAPDH) immobilized on nitrocellulose membranes.41 Filters were washed in SSC (2 ×) at 65°C 3 times for 30 minutes each, incubated at 37°C with 80 μg ribonuclease A, washed again, air dried, and exposed to DuPont reflection film (DuPont Pharmaceuticals, Wilmington, DE), with an intensifying screen, at −80°C, and also by exposure to PhosphorImager. For quantification of TGF-β2 transcripts and determination of the fold increase in transcript levels in hypoxic HUVECs compared with control, the TGF-β2/GAPDH signal ratio from hypoxic HUVECs was divided by the TGF-β2/GAPDH signal obtained from normoxic HUVECs.
Plasmid constructs
Plasmid constructs containing deletion mutants of the human TGF-β2 gene promoter linked to a chloramphenicol acetyl transferase (CAT) gene were generously donated by Dr S.-J. Kim.42TGF-β2/CAT constructs pB2-778, pB2-257, and pB2-40 were generated by polymerase chain reaction using genomic DNA containing the 5′-flanking DNA and the first exon of the TGF-β2 gene as template with different 5′-specific oligonucleotide primers, spanning from −778 to −40, and a common 3′-specific oligonucleotide primer at +63 relative to the transcription initiation site; amplified DNAs were cloned into the promoterless pGEM4/SV0CAT vector as described.42 The p3TP-Lux construct (a generous gift from Dr Joan Massagué, Memorial Sloan-Kettering Cancer Center, New York, NY) is a well described and widely used artificial promoter construct that was designed to have maximal responsiveness to TGF-β.23 This plasmid has 3 AP-1 sites corresponding to the collagenase promoter, concatemerized 5′ to a 400-nucleotide region of the PAI-1 promoter, followed by 70 base pairs (bp) of the adenovirus E4 promoter.43 Activity of this construct is increased significantly by TGF-β as well as by combined overexpression of Smad3 and Smad4. Smad4 is required for this transcriptional activity because 3TP-Lux is inactive in Smad4-null cells and is rescued by Smad4 expression.23 Expression vectors for Smad3 and Smad4 that contain the FLAG epitope tag at the amino domain have been described previously25 and were generously supplied by Drs Rik Derynck and Ying Zhang (University of California, San Francisco, CA).
Transfection of recombinant plasmids and reporter gene assays
For transient transfections, plasmids containing 5′ TGF-β2 promoter sequences and p3TP-Lux containing the luciferase reporter gene, with indicated expression vectors or corresponding empty constructs, were introduced by electroporation into early passage (second or third) HUVECs that were 70% confluent, as previously described.35 Briefly, 6 × 106 HUVECs, HepG2 (HB-8065, American Type Culture Collection [ATCC], Manassas, VA), A549 (CCL-185, ATCC), and MDA-MB-468 (HTB-132, ATCC) were resuspended in 600 μL respective growth media and reacted with 30 μg reporter plasmid DNA along with the indicated Smad expression plasmids or corresponding empty constructs (14 μg) and 0.5 μg plasmid cytomegalovirus β-galactosidase (pCMVβ-gal; Promega) (included in each transfection as internal control) for 10 minutes at 4°C. When multiple plasmids were cotransfected (eg, reporter and expression plasmids), the total amount of DNA was kept constant by supplementing the samples with empty expression vectors. Electroporation was performed using 220 V and 960 microfarads in a Bio-Rad Gene Pulser Electroporation System. Transfected cells were divided into 2 equal aliquots and cultured in 60-mm plates in endothelial cell growth medium for 6 hours, after which one of each pair of plates was placed in hypoxic conditions while the duplicate plate remained in nonhypoxic culture conditions for indicated periods. Adherent cells were washed with phosphate-buffered saline 36 hours after transfection, dislodged from plates using a rubber policeman, and the CAT assay performed at assay conditions predetermined to be within the linear range for CAT activities of the samples (data not shown). Luciferase activity was determined using the Luciferase Assay System (Promega). To ensure that the same amount of protein was analyzed, the protein concentration of each sample was determined according to the Bradford assay,36 and transfection efficiency was determined by β-gal activity. CAT/β-gal/protein or RLU/β-gal/protein ratios for hypoxic and nonhypoxic transfectants were compared to determine the fold induction by hypoxia. The displayed error bars in Figures 5 and 6, which are each representative of at least 3 independent experiments, represent ± 1 SD of the mean.
Electrophoretic gel mobility shift assays
Fresh medium with or without TGF-β2 (12.5 ng/mL) was added to 70% confluent HUVECs, and half of the duplicate flasks were placed in hypoxic conditions. At the end of the indicated incubation periods, nuclear extracts were prepared from hypoxic and nonhypoxic HUVECs as described by Andrews and Faller.44 Protein concentrations were determined by the Bio-Rad assay with bovine serum albumin standards; final protein concentration of nuclear extracts was usually between 5 and 10 mg/mL. Aliquots were stored at −70°C until use. A double-stranded oligonucleotide corresponding to the 26-bp sequence29 5′-AGTATGTCT-AGACTGA-3′ harboring the underlined consensus palindromic SBE was used as probe. The SBE probe was end-labeled with [α-32P]dATP using the Klenow fragment of DNA polymerase I for gel shift studies, and 50 to 100 pg (1 × 104-2 × 104 cpm) was incubated with 7 μg nuclear extract and 2 to 8 μg double-stranded poly(dIdC) in 20 μL of 10 mM HEPES (pH 7.9), 50 mM KCl, 5 mM MgCl2,1 mM EDTA, 1 mM dithiothreitol, and 5% wt/vol glycerol for 30 minutes at 4°C after preincubation of all components except probe for 15 minutes. For competition experiments, 20 to 500 molar excess of the indicated unlabeled oligonucleotide was added to the binding reaction mixture 5 minutes prior to probe addition. The following cold oligonucleotides were used for competition for nuclear extract binding to the probe: an 18 bp sequence corresponding to nucleotides 1 to 18 of the erythropoietin gene hypoxia-responsive enhancer45 5′-GCCCTACGTGCTGTCTCA-3′ harboring the HIF-1–binding core sequence (in italics); a 39 bp sequence corresponding to −77 to −40 bp of the TGF-β2 promoter with respect to transcriptional start site 5′-CCTAGCACGTCACTTTGTTGAAGGCAGACACGTGGTTCA-3′ harboring an underlined CAGA box and an HIF-1–binding core sequence46; and SBE. For supershift assays, each antibody (rabbit antihuman Smad2, anti-Smad3 [Zymed, South San Francisco, CA], rabbit anti-Smad4 [Upstate Biotechnology, Lake Placid, NY], or rabbit immunoglobulin G) was added to the binding reactions 15 minutes prior to the probes and incubated for another 30 minutes. After preelectrophoresis for 1 hour, 4% polyacrylamide gel electrophoresis was performed in 0.5 × Tris-borate buffer/1% glycerol without EDTA at 180 V for 2 hours at 4°C. The gel was dried, autoradiographed, and exposed to a PhosphorImager screen for 18 hours and analyzed using ImageQuant software.
Results
Effect of hypoxia on TGF-β mRNA and protein levels in HUVECs
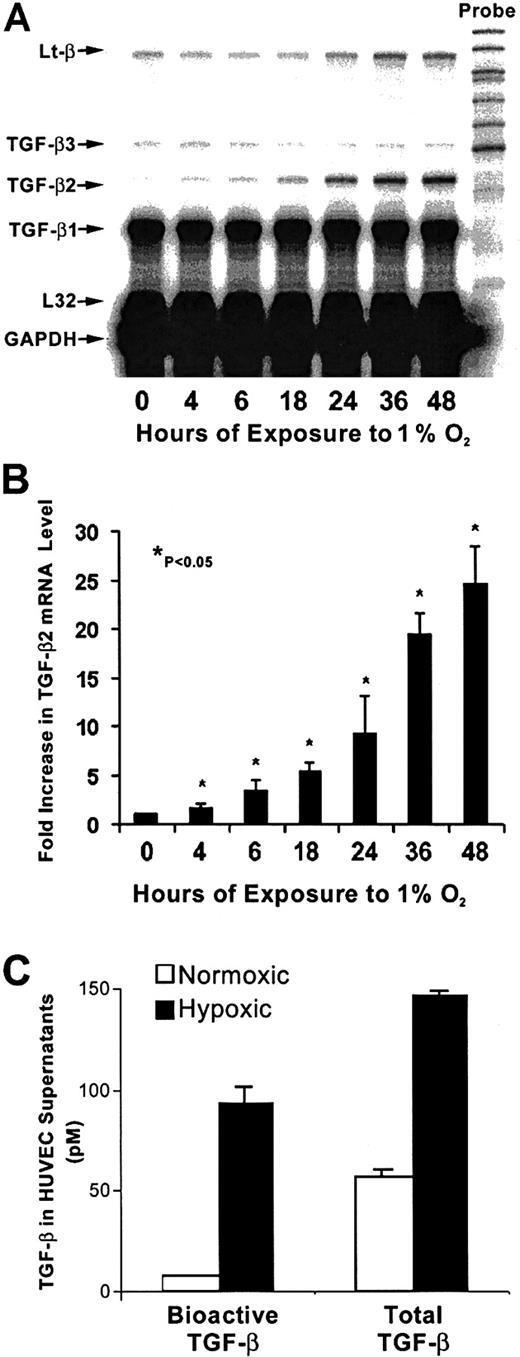
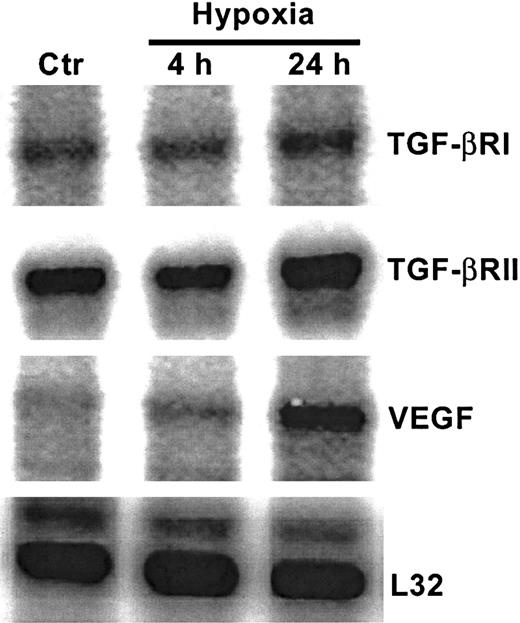
As shown in Figure 1A, in nonhypoxic HUVECs, TGF-β1 mRNA levels were greater than mRNA levels of TGF-β2 or TGF-β3. The strongest effect of hypoxia was on TGF-β2 mRNA levels, which increased by 4-fold after 4 hours of exposure to hypoxia compared with untreated HUVECs, followed by 10-fold and 25-fold increases at 24 hours and 48 hours, respectively (Figure 1A,B). At each time point, mRNA from nonhypoxic control HUVECs was also analyzed, and the TGF-β mRNA profile from these cultures remained unchanged (data not shown). Viability of endothelial cells at 24 hours and 48 hours, as disclosed by trypan blue and lactate dehydrogenase release, was more than 95%. A strong effect of hypoxia was observed on TGF-β bioactivation (Figure 1C). The level of bioactive TGF-β, which was assayed in supernatants prior to activation of latent TGF-β, was 12 times higher in supernatants from hypoxic HUVECs compared with normoxic control supernatants. Thus, 64% of TGF-β in supernatants from hypoxic HUVECs was bioactive, while only 14% of total TGF-β was bioactive in the nonhypoxic supernatants20 (Figure 1C). The level of total TGF-β (which is determined after activation of latent TGF-β) produced by HUVECs after 48 hours of exposure to hypoxia was increased by 2.6-fold compared with nonhypoxic cells (P < .05) both by the MLEC assay (Figure 1C) and by an ELISA that specifically measures TGF-β2 (data not shown). As seen in Figure 2, nonhypoxic HUVECs have readily discernable levels of both TGF-βRI and TGF-βRII mRNA. While hypoxia did not alter mRNA levels of TGF-βRI, there was a modest but consistent 1.8- to 2.5-fold increase in levels of the serine threonine kinase TGF-βRII in hypoxic HUVECs (P < .05) after correction for L32 expression (data not shown), suggesting that in these cells hypoxia could activate TGF-β signaling as well. Up-regulation of vascular endothelial growth factor in this experiment (Figure 2) confirms hypoxic conditions.47
Time course of hypoxia's effect on TGF-β mRNA levels in HUVECs.
(A) RNase protection analysis of HUVEC mRNA is shown. Total RNA was extracted from HUVECs after exposure to 20% O2 or 1% O2 at indicated times; 0 hours represents the normoxic sample. Each lane contained 10 μg total RNA hybridized to an antisense RNA probe cocktail that contained the templates for genes, protected fragments of which were separated on a 5% DNA sequencing gel and are indicated by arrows. (B) The graph represents the quantification results from 4 independent experiments. TGF-β2 signal was normalized to that from L32 (mRNA for ribosomal protein subunit); 0 hours is the mean of mRNA levels from control normoxic HUVECs. Each subsequent bar represents mRNA levels compared with its own normoxic control (not shown) at indicated hours. *P < .05. (C) TGF-β protein in supernatants from hypoxic and nonhypoxic HUVECs cultured for 48 hours was determined before and after heat activation to quantitate bioactive and total TGF-β levels, respectively, by MLEC bioassay. Results are from 4 independent experiments.
Time course of hypoxia's effect on TGF-β mRNA levels in HUVECs.
(A) RNase protection analysis of HUVEC mRNA is shown. Total RNA was extracted from HUVECs after exposure to 20% O2 or 1% O2 at indicated times; 0 hours represents the normoxic sample. Each lane contained 10 μg total RNA hybridized to an antisense RNA probe cocktail that contained the templates for genes, protected fragments of which were separated on a 5% DNA sequencing gel and are indicated by arrows. (B) The graph represents the quantification results from 4 independent experiments. TGF-β2 signal was normalized to that from L32 (mRNA for ribosomal protein subunit); 0 hours is the mean of mRNA levels from control normoxic HUVECs. Each subsequent bar represents mRNA levels compared with its own normoxic control (not shown) at indicated hours. *P < .05. (C) TGF-β protein in supernatants from hypoxic and nonhypoxic HUVECs cultured for 48 hours was determined before and after heat activation to quantitate bioactive and total TGF-β levels, respectively, by MLEC bioassay. Results are from 4 independent experiments.
Effect of hypoxia on serine threonine kinase TGF-β receptor mRNA levels.
RNase protection analysis of mRNA extracted from HUVECs after exposure to 20% O2 or 1% O2 at indicated times. Control represents mRNA from normoxic HUVECs. Each lane contained 10 μg total RNA hybridized to the antisense RNA probe cocktail. Results are from a single gel in which lanes are separated for display purposes.
Effect of hypoxia on serine threonine kinase TGF-β receptor mRNA levels.
RNase protection analysis of mRNA extracted from HUVECs after exposure to 20% O2 or 1% O2 at indicated times. Control represents mRNA from normoxic HUVECs. Each lane contained 10 μg total RNA hybridized to the antisense RNA probe cocktail. Results are from a single gel in which lanes are separated for display purposes.
Level of hypoxia-induced TGF-β2 gene expression in HUVECs
As suggested by Semenza,6,48 several adaptive responses to hypoxia involve changes in gene expression that occur at the level of transcription. To examine whether the hypoxia-induced stimulation of TGF-β2 mRNA resulted from an up-regulation of its transcriptional activation, we used in vitro transcription assays performed with nuclei isolated from HUVECs exposed to 20% or 1% O2 for 24 hours. The results from 1 of 2 experiments that yielded identical results are shown in Figure3 and demonstrate that hypoxia increased transcription of TGF-β2 by 15-fold after normalization to GAPDH (data not shown). Similarly, transcription of the hypoxia-responsive glucose transporter-1 (Glut-1)49 gene was also increased under hypoxic culture conditions, whereas transcription of the housekeeping gene GAPDH was largely unchanged. Furthermore, treatment with α-amanitin to block type II RNA polymerase activity exhibited very similar decay curves in hypoxic and normoxic HUVECs (Figure4B), indicating that hypoxia's effect on TGF-β2 gene expression occurred largely at the transcriptional level.
Effect of hypoxia on transcriptional activity of TGF-β2 gene in HUVECs.
Transcriptional analysis of TGF-β2, Glut-1, and GAPDH genes by nuclear run-on assay; 1 of 2 representative experiments is shown. (A) 5 μg of each of the plasmids indicated on the right bound to nylon membrane were hybridized with [32P]-labeled run-on transcripts from 6 × 109 nuclei isolated from HUVECs cultured for 24 hours in normoxia (20% O2) or hypoxia (1% O2). pUC19, the plasmid vector without insert, was used to estimate the background level. Radioactive bands were detected by autoradiography and also by PhosphorImager, from which transcripts were quantitated. (B) Fold increase in transcript levels in hypoxic HUVECs compared with normoxic controls was done by dividing the TGF-β2/GAPDH signal from hypoxic HUVECs by the TGF-β2/GAPDH signal obtained from normoxic HUVECs, which is shown as 1. The mean of results from 2 independent experiments is shown.
Effect of hypoxia on transcriptional activity of TGF-β2 gene in HUVECs.
Transcriptional analysis of TGF-β2, Glut-1, and GAPDH genes by nuclear run-on assay; 1 of 2 representative experiments is shown. (A) 5 μg of each of the plasmids indicated on the right bound to nylon membrane were hybridized with [32P]-labeled run-on transcripts from 6 × 109 nuclei isolated from HUVECs cultured for 24 hours in normoxia (20% O2) or hypoxia (1% O2). pUC19, the plasmid vector without insert, was used to estimate the background level. Radioactive bands were detected by autoradiography and also by PhosphorImager, from which transcripts were quantitated. (B) Fold increase in transcript levels in hypoxic HUVECs compared with normoxic controls was done by dividing the TGF-β2/GAPDH signal from hypoxic HUVECs by the TGF-β2/GAPDH signal obtained from normoxic HUVECs, which is shown as 1. The mean of results from 2 independent experiments is shown.
Effect of hypoxia on TGF-β2 message stability.
HUVECs subjected to normoxic conditions, C, or to 24 hours of hypoxia were treated with 1 μg/mL α-amanitin for the times indicated. (A) TGF-β2 message obtained with RNase protection analysis in HUVECs subjected to hypoxia for 24 hours and subsequently treated with α-amanitin for the times indicated. In parallel cultures, normoxic HUVECs were also cultured with and without α-amanitin (not shown except for the 0-hour control loaded in the first lane). (B) Semilog plot of TGF-β2 transcript half-life in the presence of hypoxic or normoxic conditions. Data were quantitated by PhosphorImager and the results plotted with linear regression; data from 1 of the 2 representative experiments are shown.
Effect of hypoxia on TGF-β2 message stability.
HUVECs subjected to normoxic conditions, C, or to 24 hours of hypoxia were treated with 1 μg/mL α-amanitin for the times indicated. (A) TGF-β2 message obtained with RNase protection analysis in HUVECs subjected to hypoxia for 24 hours and subsequently treated with α-amanitin for the times indicated. In parallel cultures, normoxic HUVECs were also cultured with and without α-amanitin (not shown except for the 0-hour control loaded in the first lane). (B) Semilog plot of TGF-β2 transcript half-life in the presence of hypoxic or normoxic conditions. Data were quantitated by PhosphorImager and the results plotted with linear regression; data from 1 of the 2 representative experiments are shown.
To further study hypoxia-induced TGF-β2 transcriptional activation, we performed transient transfection assays using CAT reporter constructs containing the 5′ TGF-β2 promoter sequences. First, activity of the CAT reporter construct pB2-778, which containscis-acting elements (Figure5A) that regulate transcription of the 5.8 kilobase TGF-β2 transcript,42 was determined. As shown in Figure 5B, activity of this construct was increased by 3-fold in response to hypoxia, and further studies using deletion mutants of this construct showed that the promoter sequence within pB2-77 is necessary and sufficient to mediate the 4.8 ± 1.2-fold increase in CAT activity obtained in hypoxic cells compared with nonhypoxic controls. A hypoxia-induced 4.8-fold increase in transcription from the TGF-β2 promoter region between −77 and +63 bp compared with nonhypoxic controls (P < .001) paralleled the increase seen in the mRNA levels after 24 hours of exposure to hypoxia, suggesting that the stimulatory mechanism involves changes associated with transcriptional events.
Effect of hypoxia on transcriptional activity of the TGF-β2 promoter in HUVECs.
(A) Recombinant constructs containing the human TGF-β2 gene promoter between −778 and +63 bp. The transcriptional start site (arrow) driving the CAT gene and selected protein-binding sites (Smad, HIF-1, AP-1) are indicated. (B) Transcriptional activity of the TGF-β2 promoter in response to hypoxia. HUVECs were transfected with pB2-778 or its deletion constructs pB2-77 or pB2-40; pB2-77 was also cotransfected with indicated Smad expression vectors and RSV-β-gal by electroporation. Each transfection was divided into 2 plates and exposed to either to 20% or 1% O2 for 36 hours. CAT activity was determined in whole-cell extracts, and its level, given as percent acetylation, was normalized to protein (Bradford assay, Bio-Rad) and to β-gal activity in each experiment. Results are expressed as fold increases in hypoxic CAT activity compared with normoxic CAT activity in each transfection. The mean ± SD of 4 independent experiments is shown.
Effect of hypoxia on transcriptional activity of the TGF-β2 promoter in HUVECs.
(A) Recombinant constructs containing the human TGF-β2 gene promoter between −778 and +63 bp. The transcriptional start site (arrow) driving the CAT gene and selected protein-binding sites (Smad, HIF-1, AP-1) are indicated. (B) Transcriptional activity of the TGF-β2 promoter in response to hypoxia. HUVECs were transfected with pB2-778 or its deletion constructs pB2-77 or pB2-40; pB2-77 was also cotransfected with indicated Smad expression vectors and RSV-β-gal by electroporation. Each transfection was divided into 2 plates and exposed to either to 20% or 1% O2 for 36 hours. CAT activity was determined in whole-cell extracts, and its level, given as percent acetylation, was normalized to protein (Bradford assay, Bio-Rad) and to β-gal activity in each experiment. Results are expressed as fold increases in hypoxic CAT activity compared with normoxic CAT activity in each transfection. The mean ± SD of 4 independent experiments is shown.
TGF-βs are known to be regulated by cis-acting Smad sites. Because the TGF-β promoter region within the hypoxia-responsive construct pB2-77 harbored a Smad-binding site (Figure 5A) and because endogenous Smad activity is known to be boosted by Smad overexpression,25 50 we tested the hypoxia response of the −77 to +63 bp CAT construct after Smad3 and Smad4 overexpression in HUVECs. Results show that overexpression of Smad3 and Smad4 resulted in a further 1.8-fold increase in construct pB2-77 activity in hypoxic HUVECs (Figure 5B) compared with nonhypoxic controls (P < .01). Baseline expression of constructs under normoxic conditions and response of pB2-778 or pB2-40 to overexpression of Smads did not have a significant effect on their CAT activities (data not shown).
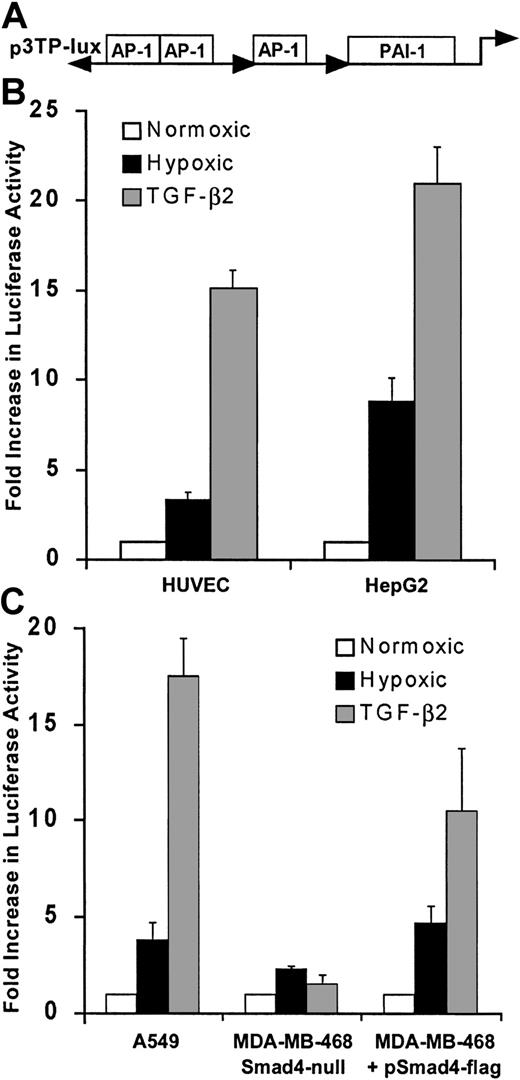
We next examined whether hypoxia stimulated activity of another promoter construct with known responsiveness to activation by Smad proteins using a well described and widely used artificial promoter construct, plasmid p3TP-Lux (Figure 6A), which is responsive to TGF-β as well as to Smad3 and Smad4 cooverexpression even in the absence of ligand.23 As seen in Figure 6B, hypoxia increased the luciferase activity of 3TP-Lux by 3.5-fold in HUVECs and by 10-fold in HepG2 cells, which are easier to transfect than HUVECs and thus are less subject to artifacts resulting from low transfection efficiencies. These results indicate that endogenous Smads are activated by hypoxia in both cell types. As expected, addition of TGF-β2 to HUVECs or HepG2 cells increased luciferase activity of 3TP-Lux (Figure 6A,B). To further determine the role of Smad proteins on hypoxia-induced activity of p3TP-Lux, we measured its response to hypoxia after transfection into the Smad4-null, epithelial breast adenocarcinoma cell line MDA-MB-468.16 In contrast to results obtained in HUVECs and HepG2, hypoxia-driven luciferase activity of p3TP-Lux was minimal in MDA-MB-468 (Figure 6C). When these cells were transiently transfected to express Smad4, there was a 4-fold increase in the p3TP-Lux activity, indicating that Smad4 is necessary for hypoxia-induced transcription in this cell line. Hypoxia also modulates expression of genes regulated by the transcription factor AP-1,51-53 and Smad3 and Smad4 potentiate transcription from AP-1,50 suggesting that Smad and AP-1 responses may both contribute to hypoxia-induced 3TP-Lux activity.
Hypoxia stimulates transcription from 3TP-Lux in HUVECs.
(A) Structural organization of the 3TP-Lux reporter plasmid, composed of 3 AP-1 binding sites and a TGF-β–responsive PAI-1 promoter fragment driving a luciferase reporter gene, is shown. (B) Response of 3TP-Lux is increased by 4-fold in HUVECs and 10-fold in HepG2 cells after exposure to 1% O2 or TGF-β2 (12.5 ng/mL) for 24 hours. Luciferase activity was determined and results corrected to CMV promoter-derived β-gal activity and protein content of the extracts. Results from 3 independent experiments are shown. (C) Response of p3TP-Lux to hypoxia or TGF-β2 (12.5 ng/mL) is absent in the Smad4-defective MDA-MB-468 cell line and can be reinstated by Smad4 expression. MDA-MB-468 in parallel with the lung carcinoma cell line A549 that expresses Smad4 was transfected with p3TP-Lux by electroporation either in the presence of Smad4 expression vector or empty vector. Transfected cells were divided into 3 identical culture dishes and were exposed to 20% O2 with or without TGF-β2 (12.5 ng/mL) or were exposed to 1% O2 for 24 hours. Luciferase activity from 3 independent experiments is shown.
Hypoxia stimulates transcription from 3TP-Lux in HUVECs.
(A) Structural organization of the 3TP-Lux reporter plasmid, composed of 3 AP-1 binding sites and a TGF-β–responsive PAI-1 promoter fragment driving a luciferase reporter gene, is shown. (B) Response of 3TP-Lux is increased by 4-fold in HUVECs and 10-fold in HepG2 cells after exposure to 1% O2 or TGF-β2 (12.5 ng/mL) for 24 hours. Luciferase activity was determined and results corrected to CMV promoter-derived β-gal activity and protein content of the extracts. Results from 3 independent experiments are shown. (C) Response of p3TP-Lux to hypoxia or TGF-β2 (12.5 ng/mL) is absent in the Smad4-defective MDA-MB-468 cell line and can be reinstated by Smad4 expression. MDA-MB-468 in parallel with the lung carcinoma cell line A549 that expresses Smad4 was transfected with p3TP-Lux by electroporation either in the presence of Smad4 expression vector or empty vector. Transfected cells were divided into 3 identical culture dishes and were exposed to 20% O2 with or without TGF-β2 (12.5 ng/mL) or were exposed to 1% O2 for 24 hours. Luciferase activity from 3 independent experiments is shown.
Nuclear extract binding to the Smad consensus sequence
Smad binding to DNA was studied by comparing gel retardation patterns of nuclear proteins from HUVECs exposed to hypoxia, normoxia, or TGF-β to DNA sequences that are specific for binding Smad proteins. As the probe in the electrophoretic gel mobility shift assay, we used the well-described oligonucleotide SBE which, as shown by x-ray crystallography, can associate with Smad3 by either or both of its palindromic Smad-binding sequences.29 As seen in Figure7A, after exposure to hypoxia a discrete gel-shifted band is observed that was absent from control nonhypoxic nuclear extracts. This band was identical in position to the one observed with nuclear extracts of TGF-β2–treated HUVECs and raised the possibility that hypoxia induced Smad binding to DNA. Binding to the SBE by nuclear extracts stimulated by hypoxia or by TGF-β2 was competed by an unlabeled 37 bp oligonucleotide corresponding to the hypoxia-responsive TGF-β2 promoter sequences between −77 and −40 bp and containing a Smad-binding site. However, unlabeled oligonucleotide corresponding to the HIF-1 binding site of the erythropoietin promoter did not compete binding of hypoxic nuclear extracts to the SBE. Results suggest that the shared Smad-binding site between the SBE and the hypoxia-responsive TGF-β2 promoter between −77 and −40 bp can bind nuclear proteins in hypoxic but not normoxic cells and that binding is independent of HIF-1.
Hypoxia induces sequence-specific DNA binding activity by Smads.
(A) Electrophoretic gel mobility shift assays were performed using a32P-labeled SBE probe as described in “Materials and methods.” Nuclear extracts were as follows: control normoxic HUVECs, C; HUVECs treated for 30 minutes with 12.5 ng/ml TGF-β2, T; or HUVECs exposed to hypoxia (1% O2) for 4 hours, H. Competition assays: no competition (−); 100-fold molar excess of unlabeled SBE (100 × SBE); 125-fold molar excess of erythropoietin (125 × EPO), an HIF-1–containing oligonucleotide; 125-fold molar excess of an oligonucleotide that corresponds to the −77 to −40 bp TGF-β2 promoter (125 × TGF-β2). (B) Antibody supershifting: nuclear extracts from HUVECs were preincubated with anti-Smad2, anti-Smad3, or anti-Smad4 antibodies for 30 minutes prior to the binding reaction. Constitutive binding is indicated by the unlabeled arrows.
Hypoxia induces sequence-specific DNA binding activity by Smads.
(A) Electrophoretic gel mobility shift assays were performed using a32P-labeled SBE probe as described in “Materials and methods.” Nuclear extracts were as follows: control normoxic HUVECs, C; HUVECs treated for 30 minutes with 12.5 ng/ml TGF-β2, T; or HUVECs exposed to hypoxia (1% O2) for 4 hours, H. Competition assays: no competition (−); 100-fold molar excess of unlabeled SBE (100 × SBE); 125-fold molar excess of erythropoietin (125 × EPO), an HIF-1–containing oligonucleotide; 125-fold molar excess of an oligonucleotide that corresponds to the −77 to −40 bp TGF-β2 promoter (125 × TGF-β2). (B) Antibody supershifting: nuclear extracts from HUVECs were preincubated with anti-Smad2, anti-Smad3, or anti-Smad4 antibodies for 30 minutes prior to the binding reaction. Constitutive binding is indicated by the unlabeled arrows.
To identify the transcription factors participating in the DNA association in nuclear extracts from TGF-β2 and hypoxia-treated HUVECs, supershift assays were performed with antibodies specific for Smad2, Smad3, and Smad4 and also with preimmune sera (not shown). As shown in Figure 7B, antibody to Smad3 and Smad4 supershifted the bands seen with nuclear extracts from TGF-β2– and hypoxia-treated HUVECs, while preimmune sera did not induce a supershift (data not shown). Furthermore, antibody to Smad2 did not induce a supershift but decreased the intensity of the specific association of hypoxia- and TGF-β–treated nuclear extracts with the SBE, indicating that Smad2 function may also be activated by hypoxia in endothelial cells. These results suggest that hypoxia induces Smad-DNA association similar to that induced by TGF-β2 and support the thesis that hypoxia results in Smad3 and Smad4 activation.
Discussion
In this report, we show that hypoxia increases TGF-β2 production and its bioactivation in human endothelial cells; furthermore, hypoxia effectively up-regulates TGF-β2 mRNA levels. This effect occurs at the transcriptional level, as evidenced by nuclear run-on analysis and reporter gene experiments using the TGF-β2 promoter, and is at least partly mediated by Smad proteins, because their overexpression further stimulates TGF-β2 promoter activation by hypoxia. It is possible that an autocrine mechanism in which bioactivation of TGF-β by hypoxia results in TGF-βR–mediated activation of Smads and results in TGF-β2 transcription; alternatively, Smad activation occurs by a pathway that is independent of TGF-βR. Activation of Smads by receptor and intracellular kinases other than those within TGF-β pathway have been described.54,55 For example, the intracellular kinase mitogen-activated protein kinase-1, an upstream activator of the stress-activated protein kinase/c-Jun N-terminal kinase, can activate Smads in endothelial cells.54 In view of the fact that hypoxia activates intracellular kinases p38, p38 mitogen-activated protein kinase, and stress-activated protein kinase/c-Jun N-terminal kinase,56 it is likely that Smad protein signaling in hypoxic endothelial cells is regulated by multiple mechanisms. Responses to hypoxia of negative regulators of the Smad pathway—for example, Smad7, which can regulate endothelial responses to inflammation and shear23 57—are unknown at this time.
Our results show that the HIF-1–binding oligonucleotide cannot compete association of hypoxic nuclear extracts with SBE. In addition, exposure to 200 μM hydrogen peroxide, which depletes HIF-1α accumulation in hypoxic cells and inhibits its ability to bind DNA,58 is associated with a 4-fold increase in TGF-β2 mRNA levels in hypoxic HUVECs and HepG2 (data not shown). Existing reports indicate a possible negative regulation of the Smad pathway by HIF-1, such as reduction of TGF-β1 gene expression in fetal skin by HIF-1 59 and inhibition of HIF-1 causing TGF-β3 gene expression and suppression of its downstream effects on trophoblasts.60 The potential interaction of Smad and HIF pathways in regulating gene expression in hypoxic endothelium is of interest. We are currently examining the effect of HIF-1α and its relative HIF-2α/endothelial PAS domain protein (EPAS)61 62 on activity of the TGF-β2 promoter by mutating its putative HIF-1-binding sites, and after HIF-1α overexpression.
Similar to our results, up-regulation of TGF-β bioactivation by hypoxia has been shown in perivascular glial cells of retina,63 but the mechanism is unclear. One possibility is that it occurs by hypoxic stimulation of endothelial expression of thrombospondin, which activates TGF-β by altering conformation of LAP.64 Thrombospondin transcript and protein are induced by hypoxia in a parallel time course to hypoxic activation of TGF-β2, with peak thrombospondin mRNA levels observed at 48 hours and protein levels at 72 hours.65 Interestingly, a hypoxia-induced increase in steady-state TSP-1 transcript levels is to some extent due to increases in mRNA stability.65 The possibility also exists that cellular redox states altered by hypoxia-induced gene activation and generation of reactive oxygen intermediaries, or changes in nitric oxide concentration, or inadvertent reoxygenation of HUVECs during our experiments contribute to bioactivation of TGF-β2 17,66; these possibilities are to be explored. Hypoxia causes an up-regulation of TGF-β2 expression in peritoneal mesothelial cells67 and down-regulates it in retinal cells68 and in dermal fibroblasts, where type II receptor mRNA was also found to be decreased.69 These results reflect the cell-specific nature of regulation of TGF-β genes within different contexts,17 including hypoxia. Recently, Smad2 binding to nuclear coactivator p300/CREB-binding protein was shown to inactivate nuclear factor–κB in endothelial cells in vitro and in vivo.33 It is possible that hypoxia-mediated changes in levels of transcriptional coactivators such as P300/CREB-binding protein70 may represent one mechanism responsible for cell- and context-specific actions of TGF-β2 in response to hypoxia.
Nonuniform responses in key endothelial functions such as endothelial cell growth, development, migration, and angiogenesis in response to TGF-β isoforms71,72 indicate that isoform-specific responses are biologically important. Prior to binding to TGF-βRII, TGF-β2 is anchored by membrane proteins73 or membrane or matrix proteoglycans, such as type III receptor (betaglycan),74,75 endoglin,23 biglycan, decorin, and fibromodulin.51,76 Therefore, hypoxia-induced regulation of these proteins can differentially modulate local concentrations of active TGF-β2 which, as suggested by Roberts,17 is pivotal in regulation of its actions.
Our studies do not address the consequences of Smad activation and TGF-β2 production on functions of hypoxic endothelial cells; these experiments are under way. A recent study has shown that TGF-β2 gene is specifically induced after infection of HUVECs with an adenovirus gene transfer vector that confers prolonged survival to endothelial cells even in the absence of serum.77 Furthermore, exposure of endothelial cells to TGF-β2 was shown to counteract the angiogenic effects of hepatocyte growth factor via inhibition of its signaling,78 and antibody to TGF-β2 was shown to stimulate DNA synthesis in pericytes.79 Based on these and previous findings regarding the antiinflammatory role of the TGF-β family of cytokines,75,80-82 we hypothesize that TGF-β2 induces an antiproliferative, antiapoptotic, and antiangiogenic signal to mitigate the inflammatory signals induced by hypoxia and augments an adaptive response in the endothelium. Similar to other adaptive responses, TGF-β2 response if prolonged or unopposed can result in progressive fibrosis and vascular thickening.10,83,84Understanding hypoxia-driven signaling by Smads can thus help elucidate the context and isoform-specificity of TGF-β2 actions in the vascular system and in the context of endothelial functions including tumor angiogenesis.15
We are thankful to Dr Isil Aksan, Molecular Biology and Genetics Department, Bogazici University, Istanbul, for helpful discussions.
Supported by the American Heart Association Heritage Affiliate Grant-in-Aid (O.A.B.); National Heart, Lung, and Blood Institute grant HL53573 (M.S.); and National Institute of Mental Health grant MH599990 (E.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Olcay Ayanlar Batuman, Div of Hematology/Oncology, Dept of Medicine, SUNY Downstate Medical Center, Box 20, 450 Clarkson Ave, Brooklyn, NY 11203; e-mail: obatuman@downstate.edu.

![Fig. 3. Effect of hypoxia on transcriptional activity of TGF-β2 gene in HUVECs. / Transcriptional analysis of TGF-β2, Glut-1, and GAPDH genes by nuclear run-on assay; 1 of 2 representative experiments is shown. (A) 5 μg of each of the plasmids indicated on the right bound to nylon membrane were hybridized with [32P]-labeled run-on transcripts from 6 × 109 nuclei isolated from HUVECs cultured for 24 hours in normoxia (20% O2) or hypoxia (1% O2). pUC19, the plasmid vector without insert, was used to estimate the background level. Radioactive bands were detected by autoradiography and also by PhosphorImager, from which transcripts were quantitated. (B) Fold increase in transcript levels in hypoxic HUVECs compared with normoxic controls was done by dividing the TGF-β2/GAPDH signal from hypoxic HUVECs by the TGF-β2/GAPDH signal obtained from normoxic HUVECs, which is shown as 1. The mean of results from 2 independent experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3324/6/m_h82311799003.jpeg?Expires=1767780042&Signature=xwaOcQ03yobgiRarIbc0-rh2ZfeWhZFBUD0pDqAhsW5YNVl6R4I0JEa5nM7-zzbRGGB7lU84eI0KWmlFSGQMIjwueGUXcejG8NYSvLltku5iS~kE-SEBKr4PAoYkEFMJSQ0gVwRQ2wtTgymMF0dJSlt1meCTZTHA2R4YsJmhWaeIi50yPI4BigrcX9~sf9W76B07ZtyOvE8aF1-T1GInDRgFr5CMYq9VI8FgLjnx-YVwjk60CQE0GsaGRcs5pJgrpIBOz~wd335VdsHcTW8N6KWJTyeeH55PITc~VqOYB39dIAUu9qZKyqAvZrQpvg5na0LZc-Ol-nJeVPU~dfpL3A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)