Abstract
This report describes 2 patients with a clinical and hematologic diagnosis of chronic myeloid leukemia (CML) in chronic phase who had an acquired t(8;22)(p11;q11). Analysis by fluorescence in situ hybridization (FISH) and reverse transcription-polymerase chain reaction (RT-PCR) indicated that both patients were negative for the BCR-ABL fusion, but suggested that the BCR gene was disrupted. Further FISH indicated a breakpoint within fibroblast growth factor receptor 1 (FGFR1), the receptor tyrosine kinase that is known to be disrupted in a distinctive myeloproliferative disorder, most commonly by fusion to ZNF198. RT-PCR confirmed the presence in both cases of an in-frame messenger RNA fusion between BCR exon 4 and FGFR1 exon 9. Expression of BCR-FGFR1 in the factor-dependent cell line Ba/F3 resulted in interleukin 3-independent clones that grew at a comparable rate to cells transformed with ZNF198-FGFR1. The growth of transformed cells was inhibited by the phosphatidylinositol 3-kinase inhibitor LY294002, the farnesyltransferase inhibitors L744832 and manumycin A, the p38 inhibitors SB202190 and SB203580 but not by the MEK inhibitor PD98059. The growth of BaF3/BCR-FGFR1 and BaF3/ZNF198-FGFR1 was not significantly inhibited by treatment with STI571, but was inhibited by SU5402, a compound with inhibitory activity against FGFR1. Inhibition with this compound was associated with decreased phosphorylation of ERK1/2 and BCR-FGFR1 or ZNF198-FGFR1, and was dose dependent with an inhibitory concentration of 50% of approximately 5 μM. As expected, growth of BaF3/BCR-ABL was inhibited by STI571 but not by SU5402. The study demonstrates that the BCR-FGFR1 fusion may occur in patients with apparently typical CML. Patients with constitutively active FGFR1 fusion genes may be amenable to treatment with specific FGFR1 inhibitors.
Introduction
Chronic myelogenous leukemia (CML) is characterized in 85% to 90% of cases by the presence of the Philadelphia (Ph) chromosome and the BCR-ABL fusion gene. A further 5% to 10% of cases have other translocations, most commonly complex variants that involve one or more chromosomal regions in addition to bands 9q34 and 22q11, but also simple variants that typically involve 22q11 and a chromosome other than 9. In virtually all cases thus far reported these patients are found to be positive for BCR-ABL on molecular analysis.1 The status of the remaining 5% of patients, many of whom have distinctive clinical features, has been controversial. Until recently these patients were typically classified as either Ph negative, BCR-ABL negative CML, or as atypical CML, and many cases were considered eligible for entry into CML clinical trials.2-4 More recently BCR-ABL negative cases have been classified into a spectrum of myeloproliferative disorders/myelodysplastic syndromes (MPDs/MDSs) that includes chronic myelomonocytic leukemia (CMML) and other related diseases; the term “CML” is reserved for Ph postive and/or BCR-ABL positive cases only.5 The molecular pathogenesis of MPD/MDS is poorly understood, but a subset of patients presents with translocations that disrupt and constitutively activate protein tyrosine kinases, most commonly the platelet-derived growth factor β receptor or the fibroblast growth factor receptor 1 (FGFR1).6-13
With the development of targeted therapies such as STI571, it is clearly important that a diagnosis of BCR-ABL positive disease should be definitive. In combination with clinical and morphologic indications, the finding of the Ph chromosome or a variant translocation by cytogenetics is frequently considered to be sufficient evidence for the diagnosis of typical CML. Although this is undoubtedly true in the great majority of cases, we present data on 2 patients with clinical and morphologic indications of CML who presented with what appeared to be a simple variant of the Ph chromosome. We show that these individuals did not harbor the BCR-ABL chimera but instead had a novel fusion between BCR and FGFR1. In contrast to BCR-ABL, BCR-FGFR1 is not sensitive to STI571 but is inhibited by SU5402. These findings have important therapeutic implications for patients withFGFR1 fusion genes.
Materials and methods
Case reports
Case 1.
A 65-year-old woman presented with fever and sweats. Apart from mild hypertension, she had no relevant past history. Clinically, her spleen was enlarged 8 cm below left costal margin. Full blood examination revealed hemoglobin, 124 g/L; white blood cell (WBC) count, 132.4 × 109/L; and platelets, 334 × 109/L with the following differential: neutrophils 48%, lymphocytes 8%, monocytes 1%, eosinophils 1%, basophils 3%, metamyelocytes 13%, myelocytes 21%, promyelocytes 3%, and blast cells 3%. Biochemical analysis was unremarkable apart from a raised lactate dehydrogenase (LDH) of 849 U/L. Bone marrow examination morphologically resembled chronic phase CML. Cellularity was markedly increased with an M/E ratio of 19:1. Granulopoiesis was increased with a prominent left shift. Eosinophils were increased with an occasional abnormal form with basophilic granulation. The differential was as follows: neutrophils 26%, eosinophils 16%, metamyelocytes 13%, myelocytes 12%, promyelocytes 14%, myeloblasts 4%, basophils 6%, lymphocytes 1%, and erythroblasts 5%. There was a mild to moderate diffuse increase in reticulin. Bone marrow cytogenetics revealed a t(8;22)(p11.2;q11.2) in all metaphases analyzed. The patient received hydroxyurea, which effectively controlled the WBC count and interferon-α (IFN-α) therapy has recently been commenced.
Case 2.
A 51-year-old woman presented with fatigue and dyspnea on exertion. There was no palpable lymphadenopathy or hepatosplenomegaly. Full blood examination revealed hemoglobin, 81g/L; WBC count, 198 × 109/L; and platelets 101 × 109/L with the following differential: neutrophils 45%, lymphocytes 3%, monocytes 7%, eosinophils 2%, basophils 4%, metamyelocytes 20%, myelocytes 8%, promyelocytes 2%, bands 6%, and blast cells 3%. The leukocyte alkaline phosphatase score was 3 and LDH was 719 U/L. The bone marrow was hypercellular with myeloid hyperplasia (M/E ratio of 8:1), eosinophilia, decreased megakaryocytes, fibrosis, and focal interstitial lymphocytosis. The differential count was neutrophils 21%, eosinophils 5%, metamyelocytes 6%, myelocytes 15%, promyelocytes 20%, myeloblasts 4%, basophils 1%, lymphocytes 19%, and erythroblasts 9%. Reticulin stain showed a moderate increase. Flow cytometry revealed that 4% of nucleated cells were of immature B-cell phenotype and were positive for terminal deoxynucleotidyl transferase (TdT), CD34, CD19, CD10, and CD24. No clonal rearrangement of immunoglobulin heavy or light chains was detected by Southern analysis. Bone marrow cytogenetics revealed a t(8;22)(p11.2;q11.2) in 20 of 20 metaphases. The patient's blood counts were partially controlled with hydroxyurea and IFN-α therapy has recently been commenced.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) for BCR-ABL was performed with the Vysis BCR-ABL D-FISH or ES translocation probes (Vysis, Downers Grove, IL) according to the manufacturer's instructions. Bacterial artificial chromosomes (BACs) bk514C6 and bk143F12 contain sequences near BCR on chromosome 22 and were kindly provided by Dr M. Rocchi (University of Bari, Italy). Two-color FISH to detect FGFR1 rearrangements was performed with PACs 162-N14 and 224-C10.14 PAC DNA was isolated from 1.5-mL cultures using a standard miniprep procedure and FISH was performed as described previously.9
Reverse transcription–polymerase chain reaction
RNA was extracted, reverse transcribed, and tested for BCR-ABL transcripts as described.15 Primers used for the detection of BCR-FGFR1 and the reciprocal fusion were: E1+, 5′-AGATCTGGCCCAACGATGACGA-3′; E5/6+, 5′-GAAATCTCCGAGAACCTGAGAG-3′; NB1+, 5′-GAGCGTGCAGAGTGGAGGGAGAACA-3′; C3+, 5′-GAGGTCCAAGGTGCCCTACAT-3′; E5+R, 5′-CTGACAGCACTTCTCAGCCATTTC-3′; FGFR9−, 5′-GAGGGTCTTCGGGAAGCTCATA-3′; and FGFR8+, 5′-TGGTACCAAGAAGAGTGACTTCC-3′.
Constructs
BCR was amplified from plasmid pGD210 (kindly provided by Dr G. Daley, Whitehead Institute, Boston, MA) using primers BCR1+Bam (5′-GTGGATCCGGCCGCGCCATGGTGGAC-3′) and BCR-Rsa (5′-TTTCCGTACCCAGCAACGTCTGCAG-3′) and digested with BamHI and RsaI. The FGFR1 moiety was liberated from pcDNA3.1/ZNF198-FGFR1 by digestion with Ssp1 andBamH1. The BCR and FGFR1 fragments were coligated into theBamH1 site of pcDNA3.1 (Invitrogen, Leek, The Netherlands) to give plasmid pcDNA/BCR-FGFR1. The insert of this plasmid was completely sequenced to confirm an in-frame fusion and that no mutations had been inadvertently introduced. In vitro mutagenesis was accomplished by overlapping polymerase chain reaction (PCR) amplifications with 2 mutant primers and cloning the product into the unique NheI and EagI sites of pCDNA/ZNF198-FGFR1. The Nhe1-Eag1 region was sequenced to confirm the presence of the mutation and that no other changes had been inadvertently introduced.
Transformation assay
The Ba/F3 cells were maintained in interleukin 3 (IL-3) medium (RPMI 1640 medium with 10% fetal calf serum [FCS] and 10% conditioned medium from the IL-3–producing WEHI-3B cell line). For electroporation, 1 × 107 Ba/F3 cells were washed in cold phosphate-buffered saline (PBS), resuspended in 1 mL prewarmed RPMI 1640 and incubated for 10 minutes at room temperature with 16 to 20 μg plasmid DNA in PBS. Cells were electroporated at 250 V/960 μF in a Gene Pulser II apparatus (Bio-Rad, Hemel Hempsted, United Kingdom). After 10 minutes on ice the cells were plated in 10 mL IL-3–containing medium for 48 hours and then selected and subcloned by limiting dilution (5000 or 500 cells/well) in IL-3 medium plus 1 mg/mL G418. After 12 days, resistant subclones were washed 3 times in PBS and plated out in medium without IL-3. For growth curves, 5000 cells from individual clones were plated in duplicate in 100 μL medium with or without IL-3. Wells were aspirated daily and cell proliferation was estimated by the MTS assay (Promega, Southampton, United Kingdom). Results were expressed as an increase in the optical density at 490 nm relative to time zero.
Protein analysis
Western blotting was performed according to standard procedures16 using an enhanced chemiluminescence system (Amersham, Little Chalfont, United Kingdom). Antibodies used were 4G10 (phosphotyrosine; Upstate Biotechnology, Dundee, United Kingdom), FGFR1, CREB-1, ERK-1, phospho ERK (Santa Cruz Biotechnology, Santa Cruz, CA) and MEK1/2, AKT, phospho AKT, and phospho-CREB (New England Biolabs, Hitchin, United Kingdom).
Other reagents
BaF3/BCR-ABL was kindly provided by Dr F. X. Mahon (Université Victor Segalen, Bordeaux, France). Specific inhibitors were obtained as follows: SU5402, Sugen (South San Francisco, CA); STI571, Novartis Pharma (Basel, Switzerland); L-744832, Merck Research Laboratories (Rahway, NJ); and manumycin A, PD98059, LY294002, SB202190, and SB203580, Calbiochem (Nottingham, United Kingdom). Inhibitors were used at concentrations recommended by the manufacturers or other investigators. For analysis of effects on growth, compounds were added at day 0 and proliferation over 4 days measured by the MTS assay as above. To test that inhibitors were working as expected, cells were pretreated for 15 minutes to 3 hours, lysed, and Western blotted with appropriate antibodies to test the phosphorylation status of specific downstream substrates.
Results
Exclusion of BCR-ABL positivity
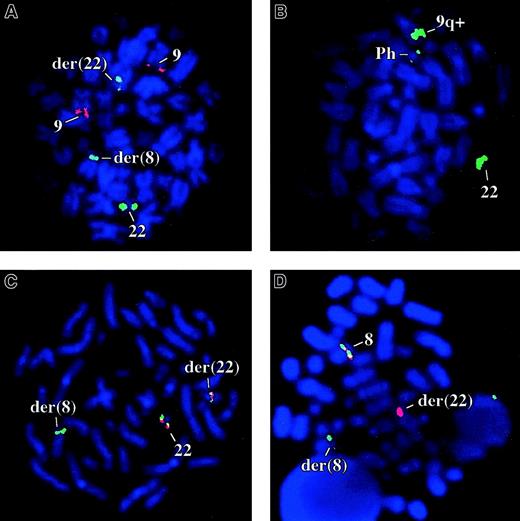
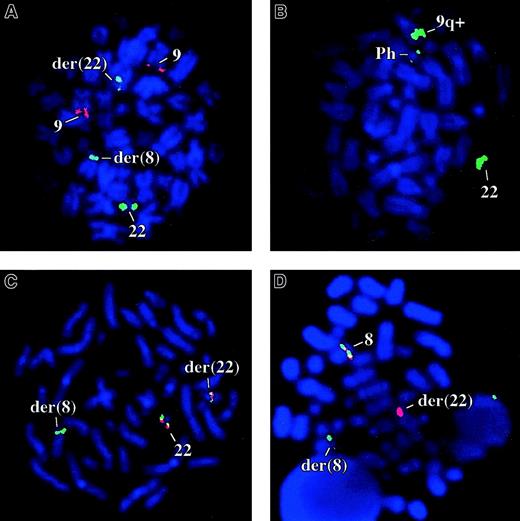
Bone marrow FISH studies using ES and D-FISH probes were negative for the BCR-ABL fusion in 10 of 10 metaphase and 100 interphase cells for patient 1 (Figure 1A). By D-FISH, the metaphases showed red (ABL) signals on both copies of chromosome 9; one large green (BCR) signal on the normal chromosome 22, with smaller green signals on the der(22) and on the der(8). These findings were consistent with the known karyotype and suggested that the chromosome 22 breakpoint must be close to, or within, BCR. Similarly patient 2 was negative for the BCR-ABL fusion by FISH (200 cells analyzed). Concordant with the FISH analysis, BCR-ABL transcripts were not detected in the peripheral blood or bone marrow of either patient by multiplex reverse transcription-PCR (RT-PCR; not shown).
FISH analysis.
(A) Patient 1 with the BCR-ABL D-FISH probes (ABL, red; BCR, green). (B) A Ph positive CML patient with bK143F12. (C) Patient 1 cohybridized with bK143F12 (green) and bK514C6 (red). (D) Patient 1 cohybridized with PAC 162N14 (green) and PAC 224C10 (red), clones that flank FGFR1.
FISH analysis.
(A) Patient 1 with the BCR-ABL D-FISH probes (ABL, red; BCR, green). (B) A Ph positive CML patient with bK143F12. (C) Patient 1 cohybridized with bK143F12 (green) and bK514C6 (red). (D) Patient 1 cohybridized with PAC 162N14 (green) and PAC 224C10 (red), clones that flank FGFR1.
The t(8;22) disrupts the BCR and FGFR1genes
To determine if the chromosome 22 breakpoint targetedBCR, we performed FISH analysis on cells from patient 1 with genomic clones that contained markers close to this gene. BAC bK143F12 contains D22S301, a marker distal to BCR, and BAC bK514C6 contains GNAZ, a gene proximal to BCR(www.biologia.uniba.it/rmc/). On Ph+ CML metaphases, bK143F12 hybridized strongly to the normal 22 and the 9q+ chromosomes but only weakly to the Ph chromosome and thus must encompass D22S301 and a point slightly proximal to the major breakpoint cluster region (M-bcr; Figure 1B). In contrast, bK514C6 hybridized to the normal 22 and Ph chromosomes only and therefore is entirely proximal to the M-Bcr (not shown). On t(8;22) metaphases, bK143F12 hybridized to both derivative chromosomes, whereas bK514C6 hybridized only to the der(22). These results suggested that BCR was disrupted (Figure1C).
Chromosome band 8p11 contains FGFR1, the receptor tyrosine kinase that is disrupted in an aggressive MPD that we have termed the 8p11 myeloproliferative syndrome (EMS).17 To determine ifFGFR1 was disrupted by the t(8;22) we performed 2-color FISH with PACs 162-N14 and 224-C10, clones that flankFGFR1.14 As shown in Figure 1D, both clones hybridized to the normal chromosome 8 but PAC162-N14 hybridized to the der(22), whereas PAC 224-C10 hybridized to the der(8). This indicated that the chromosome 8 breakpoint was close to or withinFGFR1.
Identification of the BCR-FGFR1 fusion
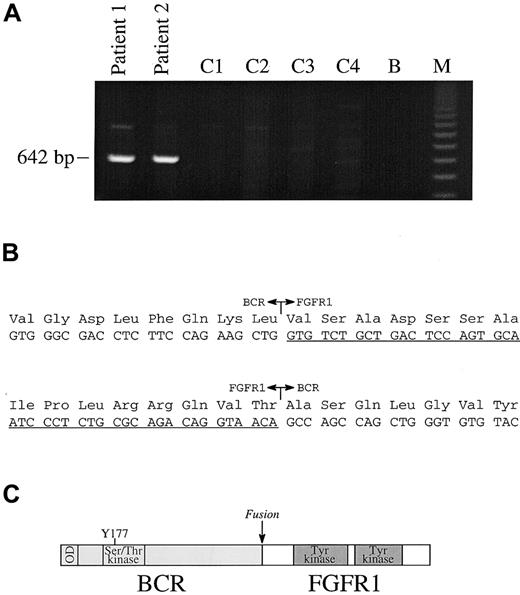
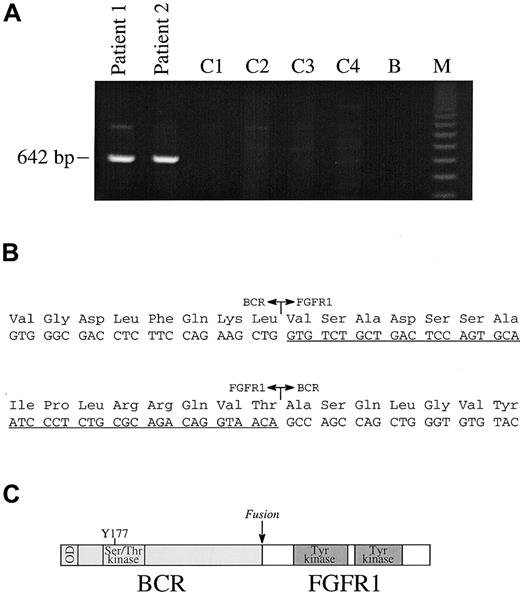
For all messenger RNA (mRNA) fusions thus far reported in EMS, the partner gene is fused to FGFR1 exon 9; however, a wide range of BCR breakpoints have been reported forBCR-ABL.1 We therefore used a multiplex PCR strategy with 4 forward BCR primers (E1+, exon 1; E5/6+, exon 5/6 junction; NB1+, exon 12 and C3+, exon 19) and a single reverse (FGFR9−) to look for a possible BCR-FGFR1 fusion. A 642-base pair (bp) product was obtained from both our patients, but not from control patients with Ph positive CML. PCR with single pairs of primers gave rise to the same size product only in the E1+/FGFR9− combination (Figure 2A). Sequencing of these products revealed an in-frame fusion between BCR exon 4 and FGFR1 exon 9 in both cases (Figure 2B). The BCR-FGFR1 fusion mRNA is predicted to be translated into a protein of 109 kd. PCR with primers to FGFR1 exon 8 (FGFR8+) and BCR exon 5 (E5+R) amplified a product specifically from the t(8;22) patients (not shown), which was confirmed to be the reciprocal FGFR1-BCR fusion by sequence analysis.
RT-PCR analysis.
(A) RT-PCR analysis with primers E1+ and FGFR9− show the presence of a specific band in patients 1 and 2, but not in 4 control patients with CML (C1-C4); B indicates blank; M, 123-bp ladder. (B) In-frame fusions of BCR-FGFR1 and FGFR1-BCR. FGFR1 sequences are underlined. (C) Structure of BCR-FGFR1 in which BCR amino acid 584 is fused to FGFR1 amino acid 429. The chimeric protein is predicted to contain the BCR oligomerization domain (OD), the BCR Ser/Thr kinase domains, and Y177 Grb2-binding site joined to part of the FGFR1 juxtamembrane domain and the entire FGFR1 tyrosine kinase and C-terminal domains.
RT-PCR analysis.
(A) RT-PCR analysis with primers E1+ and FGFR9− show the presence of a specific band in patients 1 and 2, but not in 4 control patients with CML (C1-C4); B indicates blank; M, 123-bp ladder. (B) In-frame fusions of BCR-FGFR1 and FGFR1-BCR. FGFR1 sequences are underlined. (C) Structure of BCR-FGFR1 in which BCR amino acid 584 is fused to FGFR1 amino acid 429. The chimeric protein is predicted to contain the BCR oligomerization domain (OD), the BCR Ser/Thr kinase domains, and Y177 Grb2-binding site joined to part of the FGFR1 juxtamembrane domain and the entire FGFR1 tyrosine kinase and C-terminal domains.
Transformation of Ba/F3 cells by BCR-FGFR1
The BCR-FGFR1 fusion contains the oligomerization domain of BCR18 and the entire tyrosine kinase domain of FGFR1 (Figure 2C) and is therefore most likely to be the transforming product of the t(8;22). We and others have previously shown that the ZNF198-FGFR1 fusion is a constitutively active, 150-kd tyrosine kinase that can transform the IL-3–dependent cell line Ba/F3 to growth factor independence.16,19 To test if BCR-FGFR1 also has biologic activity, we constructed a fusion complementary DNA in the expression vector pCDNA3.1. Ba/F3 cells were electroporated with pCDNA/BCR-FGFR1, vector alone as a negative control, or pCDNA/ZNF198-FGFR116 as a positive control. Cells were selected initially at limiting dilution in G418 for the plasmid and then in the absence of IL-3 to test for growth factor independence. In the absence of IL-3, growth was observed for clones that had been transfected with BCR-FGFR1 or ZNF198-FGFR1, but not for clones transfected with vector alone. Western blotting with FGFR1 antibody detected proteins of the expected sizes in construct-transfected cells, but not in cells transfected with the vector only (Figure3A). Two BCR-FGFR1 and 3 ZNF198-FGFR1 clones that expressed comparable levels of fusion protein were selected for analysis. In the absence of IL-3, both BCR-FGFR1–expressing clones proliferated at a rate comparable to clones that expressed ZNF198-FGFR1, whereas clones transfected with the vector failed to proliferate (Figure 3B). As expected, transformation was kinase dependent because no proliferation was seen in the absence of IL-3 for clones that expressed a ZNF198-FGFR1 mutant in which the critical lysine residue in the active site (corresponding to FGFR1 residue 514) had been mutated to arginine (Figure 3B).
Kinase-dependent transformation.
(A) Western blot of one BCR-FGFR1 clone, 3 ZNF198-FGFR1 clones, and a clone that had been transfected with vector only probed with anti-FGFR1. (B) Proliferation of representative BCR-FGFR1, ZNF198-FGFR1, vector only, and ZNF198-FGFR1/K514R clones in the absence of IL-3.
Kinase-dependent transformation.
(A) Western blot of one BCR-FGFR1 clone, 3 ZNF198-FGFR1 clones, and a clone that had been transfected with vector only probed with anti-FGFR1. (B) Proliferation of representative BCR-FGFR1, ZNF198-FGFR1, vector only, and ZNF198-FGFR1/K514R clones in the absence of IL-3.
Specific growth inhibition with SU5402
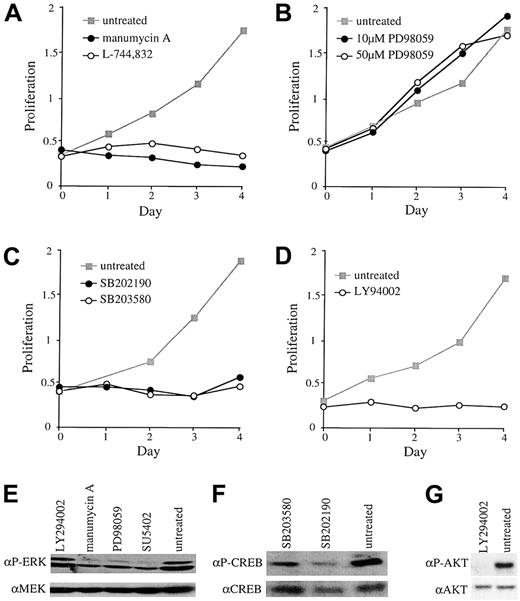
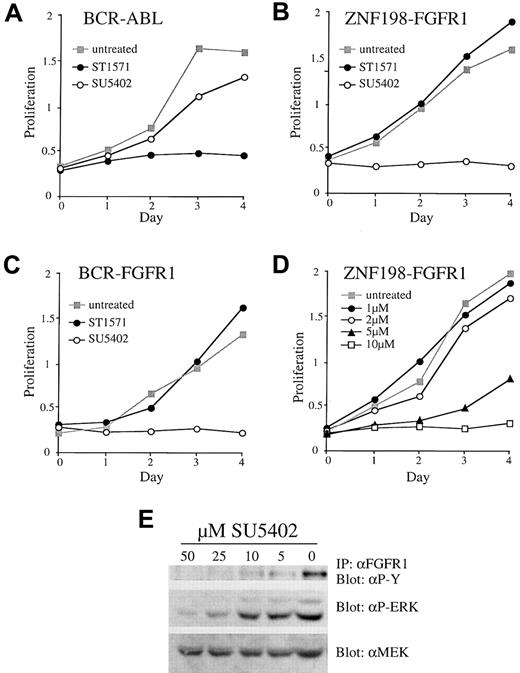
Cells expressing BCR-ABL are known to be sensitive to the tyrosine kinase inhibitor STI571.20 As expected from the known activities of this compound, we observed no growth inhibition of Ba/F3 cells expressing either BCR-FGFR1 or ZNF198-FGFR1 in the presence of 5 μM STI571, a concentration that completely inhibited the growth of Ba/F3 cells expressing BCR-ABL (Figure4A-C). We then tested SU5402, a compound known to have inhibitory activity against FGFR1.21 Clones expressing both BCR-FGFR1 and ZNF198-FGFR1 were inhibited by SU5402 in a dose-dependent fashion with an inhibitory concentration of 50% (IC50) of about 5 μM. Growth was completely inhibited at a concentration of 10 μM SU5402 (Figure 4B-D). Growth inhibition was specific because SU5402 had minimal effects on BaF3/BCR-ABL (Figure 4A) or BaF3/BCR-FGFR1 or BaF3/ZNF198-FGFR1 when grown in the presence of IL-3 (not shown). Increasing concentrations of SU5402 were associated with decreasing levels of tyrosine-phosphorylated ZNF198-FGFR1 or BCR-FGFR1 and decreasing phosphorylation of ERK1/2 (Figure4E).
Response to STI571 and SU5402.
Response of BaF3/BCR-ABL (A) and representative ZNF198-FGFR1 (B) and BCR-FGFR1 (C) clones to 5 μM STI571 or 10 μM SU5402. (D) Dose response of a representative ZNF198-FGFR1 clone to increasing amounts of SU5402. (E) Increasing concentrations of SU5402 added to a representative ZNF198-FGFR1 clone result in deceasing levels of tyrosine phosphorylated fusion protein and phospho ERK. Total protein levels are unchanged, as indicated by probing with anti-MEK.
Response to STI571 and SU5402.
Response of BaF3/BCR-ABL (A) and representative ZNF198-FGFR1 (B) and BCR-FGFR1 (C) clones to 5 μM STI571 or 10 μM SU5402. (D) Dose response of a representative ZNF198-FGFR1 clone to increasing amounts of SU5402. (E) Increasing concentrations of SU5402 added to a representative ZNF198-FGFR1 clone result in deceasing levels of tyrosine phosphorylated fusion protein and phospho ERK. Total protein levels are unchanged, as indicated by probing with anti-MEK.
Other signal transduction inhibitors
In a preliminary analysis to determine what signal transduction pathways might be important for transformation of Ba/F3 cells by FGFR1 fusion proteins, we tested the growth effects of a series of inhibitors. L-744832 and manumycin A are farnesyltransferase (FTase) inhibitors that can block the proliferation of Ras-transformed cells by mechanisms that are incompletely defined.22 23 Both of these compounds resulted in decreased levels of ERK 1/2 phosphorylation and completely blocked proliferation of Ba/F3 cells that had been transformed with BCR-FGFR1 or ZNF198-FGFR1 (Figure5A,E).
Growth effects of various inhibitors.
Response of representative clones expressing BCR-FGFR1 or ZNF198-FGFR1 to (A) 10 μM manumycin A or 10 μM L-744832, (B) 10 μM or 50 μM PD98059, (C) 40 μM SB202190 or SB203580, (D) 20 μM LY94002. (E) Treatment with SU5402, PD98059 (10 μM), and manumycin A, but not LY294002, results in reduced levels of ERK phosphorylation relative to untreated cells. Total protein was controlled for by probing with anti-MEK; results for L744832 are not shown. (F) Treatment with SB203580 or SB202190 results in reduced levels of phosphorylated CREB relative to untreated cells and total CREB. (G) LY294002 treatment results in reduced AKT phosphorylation relative to untreated cells and total AKT.
Growth effects of various inhibitors.
Response of representative clones expressing BCR-FGFR1 or ZNF198-FGFR1 to (A) 10 μM manumycin A or 10 μM L-744832, (B) 10 μM or 50 μM PD98059, (C) 40 μM SB202190 or SB203580, (D) 20 μM LY94002. (E) Treatment with SU5402, PD98059 (10 μM), and manumycin A, but not LY294002, results in reduced levels of ERK phosphorylation relative to untreated cells. Total protein was controlled for by probing with anti-MEK; results for L744832 are not shown. (F) Treatment with SB203580 or SB202190 results in reduced levels of phosphorylated CREB relative to untreated cells and total CREB. (G) LY294002 treatment results in reduced AKT phosphorylation relative to untreated cells and total AKT.
To test if the classical mitogen-activated protein kinase (MAPK) pathway (Raf-MEK-ERK), one of the principal mitogenic cascades downstream of Ras, might be required for transformation, we used the MEK inhibitor PD98059. This compound inhibited ERK1/2 phosphorylation at both 10 μM and 50 μM, as expected, but had no effect on the proliferation of transformed Ba/F3 cells (Figure 5B,E). In contrast 40 μM SB202190 or SB203580, inhibitors of p38 MAPK, nearly completely blocked the proliferation of both BCR-FGFR1– and ZNF198-FGFR1–transformed cells (Figure 5C). As expected, treatment with these compounds resulted in reduced levels of CREB phosphorylation (Figure 5F). Finally, the growth of transformed cells was also blocked by the phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 at 20 μM, a concentration that completely inhibited AKT phosphorylation (Figure 5D,G). No significant differences in the effect of any of these compounds were seen on comparison of individual clones that expressed BCR-FGFR1 with clones that expressed ZNF198-FGFR1.
Discussion
We have identified 2 patients who had a clinical and morphologic picture that was essentially indistinguishable from typical, BCR-ABL positive CML.24 Both patients had basophilia, a feature that is uncommon in BCR-ABL negative MPDs. Of note, both patients had eosinophilia, although the levels were within the range seen in CML, and one patient had an unusual marrow lymphocytosis. Instead of the t(9;22)(q34;q11), both patients had a t(8;22)(p11;q11), an abnormality that has not to our knowledge been previously described. Given the clinical and hematologic background, the t(8;22) might have been classified cytogenetically as merely a simple variant of the t(9;22). However, we found that both patients were negative for BCR-ABL and instead harbored a novel fusion between BCR and FGFR1.
Rearrangement and constitutive activation of FGFR1 is seen in patients with a distinct, aggressive biphenotypic hematologic malignancy that has been termed the “8p11 myeloproliferative syndrome” (EMS) or “stem cell leukemia-lymphoma syndrome” (SCLL).17,25The only curative treatment for this condition appears to be allogeneic bone marrow transplantation,25 although occasional patients may achieve complete cytogenetic remission with IFN-α based therapy (J.M.G., unpublished observations, June 2001). Three translocations associated with this disease, t(8;13)(p11;q12), t(8;9)(p11;q33), and t(6;8)(q27;p11), have been cloned and found to result in fusion of ZNF198, CEP110, and FOP, respectively, to FGFR1.7-13 Other variant translocations that involve FGFR1 have also been reported.14,26 EMS patients typically present with myeloid hyperplasia, eosinophilia but no basophilia and, unusually, an associated lymphoma that is generally of T-cell phenotype.17,25 Although it is difficult to generalize from just 2 cases, patients with BCR-FGFR1 appear to resemble classical CML more closely than individuals with other FGFR1 fusion proteins. One possible explanation for this is that the BCR moiety contributes to specific features of the patient phenotype. This could be by several mechanisms, for example, by providing a tyrosine residue that serves as a Grb2 binding site when phosphorylated, a function that is believed to be important for the action of BCR-ABL,27 by its intrinsic Ser/Thr kinase activity,28 by driving a particular level of transcription of the fusion gene, or as a consequence of a reduced level of normal BCR expression.
We have found that transformation of Ba/F3 cells by BCR-FGFR1 and ZNF198-FGFR1 is sensitive to FTase inhibitors, suggesting that Ras may be required. A more extensive analysis has indicated that Ras is required for transformation mediated by BCR-ABL,29 and recent preclinical data have suggested that FTase inhibitors may be useful agents to treat CML.30 We have also found that inhibition of the classical MAPK pathway has no effect on transformed cells, but that proliferation was blocked by inhibitors of p38 or PI3K. These findings are again similar to what has been described for CML in diverse experimental systems31 and suggest that several distinct proteins and pathways might be useful therapeutic targets in both diseases. However, because multiple pathways are activated by diverse signals, including IL-3, it is difficult to establish adequate controls for nonspecific toxicity and therefore we cannot conclude that any of the pathways we studied are definitely required for transformation mediated by BCR-FGFR1 or ZNF198-FGFR1.
Simple variants of the t(9;22) are seen in a small minority of patients with CML. For example, the Medical Research Council CML III trial included 9 such individuals, corresponding to 1.5% of all patients or 18% of those with variant translocations (Dr. P. Shepherd, personal communication, May 2001). Of those cases that have been reported, virtually all have been BCR-ABL positive by RT-PCR or FISH analysis, or have had a rearrangement within the M-Bcr as determined by Southern blotting.1,32 Thus, in common with individuals who are Ph positive, patients who present with apparently typical CML and a variant translocation are not always evaluated for BCR-ABL by molecular tests. Our findings indicate that occasional patients may therefore be misdiagnosed. This is not simply an academic point: BCR-ABL positive cases are amenable to treatment with STI571,33 but data from our cell line model indicate that FGFR1 fusion proteins are resistant to this compound. We found, however, that cells transformed with BCR-FGFR1 or ZNF198-FGFR1 are sensitive to SU5402, a compound known to have inhibitory activity against FGFR1.21 These data indicate the increasing importance of an accurate molecular diagnosis and suggest that patients with constitutively active FGFR1 fusion proteins may be amenable to treatment with specific FGFR1 inhibitors.
We would like to thank Drs B. Druker, F-X. Mahon, M. Rocchi, G. Daley, P. Shepherd, J. McMahon, B. Smolich, S. Kelsey, J. Gibbs, and E. Buchdunger for invaluable discussion and provision of reagents.
Supported by Leukaemia Research Fund Specialist Programme Grant 91/19 and the Turkish Ministry of Education.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Nicholas C. P. Cross, Wessex Regional Genetics Laboratory, Salisbury District Hospital, Salisbury, Wilts SP2 8BJ, United Kingdom; e-mail: n.c.p.cross@soton.ac.uk.