The Quebec platelet disorder (QPD) is an autosomal dominant platelet disorder associated with delayed bleeding and α-granule protein degradation. The degradation of α-granule, but not plasma, fibrinogen in patients with the QPD led to the investigation of their platelets for a protease defect. Unlike normal platelets, QPD platelets contained large amounts of fibrinolytic serine proteases that had properties of plasminogen activators. Western blot analysis, zymography, and immunodepletion experiments indicated this was because QPD platelets contained large amounts of urokinase-type plasminogen activator (u-PA) within a secretory compartment. u-PA antigen was not increased in all QPD plasmas, whereas it was increased more than 100-fold in QPD platelets (P < .00009), which contained increased u-PA messenger RNA. Although QPD platelets contained 2-fold more plasminogen activator inhibitor 1 (PAI-1) (P < .0008) and 100-fold greater u-PA–PAI-1 complexes (P < .0002) than normal platelets, they contained excess u-PA activity, predominantly in the form of two chain (tcu-PA), which required additional PAI-1 for full inhibition. There was associated proteolysis of plasminogen in QPD platelets, to forms that comigrated with plasmin. When similar amounts of tcu-PA were incubated with normal platelet secretory proteins, many α-granule proteins were proteolyzed to forms that resembled degraded QPD platelet proteins. These data implicate u-PA in the pathogenesis of α-granule protein degradation in the QPD. Although patients with the QPD have normal to increased u-PA levels in their plasma, without evidence of systemic fibrinogenolysis, their increased platelet u-PA could contribute to bleeding by accelerating fibrinolysis within the hemostatic plug. QPD is the only inherited bleeding disorder in humans known to be associated with increased u-PA.
Introduction
Congenital platelet disorders are usually associated with defective primary hemostasis.1-3 The Quebec platelet disorder (QPD) is an autosomal dominant platelet disorder that has unusual clinical features: it is associated with moderate to severe delayed bleeding, that typically begins 12 to 24 hours after surgery or trauma, and its hemorrhagic manifestations can be controlled with fibrinolytic inhibitors but not with platelet transfusions.1,4-6 This disorder was initially designated as factor V Quebec because of the abnormalities found in platelet factor V of these patients.7 Two families from Quebec have been identified with this condition, which is now known to be associated with other platelet abnormalities that include reduced to low-normal platelet counts, proteolytic degradation of soluble and membrane proteins stored in platelet α-granules, an apparent quantitative deficiency of the α-granule protein multimerin, and defective aggregation with epinephrine.1,4-6,8 Although patients with the QPD have elevated levels of fibrinogen degradation products (FDPs) in their serum (because of platelet fibrinogen degradation), their plasma contains normal amounts of FDPs and D-dimers.6 Complex platelet abnormalities in these patients led us to redesignate their bleeding disorder as the Quebec platelet disorder.5
The cause of the QPD has been uncertain. Affected patients of both families share a characteristic pattern of platelet α-granule protein degradation that is not evident in unaffected family members or in patients with other congenital and acquired platelet disorders1,4-6,9 This degradation affects both plasma-derived and megakaryocyte-synthesized proteins stored in QPD α-granules, but it spares external membrane, dense-granular, and cytosolic platelet proteins.1,4,5 Moreover, some proteins (eg, fibrinogen, von Willebrand factor, and factor V) are degraded in QPD platelets but not in QPD plasma.1,4-6,8 The observation that endogenously synthesized and plasma-derived α-granule proteins were degraded in QPD platelets, despite their normal storage within α-granules,5 suggests some proteolysis occurs late, after megakaryocyte-synthesized and plasma-derived α-granule proteins enter the same compartment. This possibility led us to investigate QPD platelets for the presence of abnormal protease activity. We report that QPD platelets contain abnormal fibrinolytic activity, attributable to their stores of large amounts of urokinase-type plasminogen activator (u-PA). Furthermore, we observed that the consequence of adding similar amounts of exogenous u-PA to normal platelet proteins was the proteolysis of α-granule proteins to forms resembling the degraded proteins in QPD platelets.
Patients, materials, and methods
Patients
Blood was collected with informed consent and institutional, ethics board approval from unrelated healthy controls (n = 20) and 5 affected patients with QPD, representing both families with this disorder. Stored samples of washed platelet lysates, pelleted platelet lysates, and plasmas, collected from patients with QPD for previous studies,4-6 were included in some analyses.
Materials
The protease inhibitors aprotinin, E-64, AEBSF (4-(2-aminoethyl)-benzenesulfonyl fluoride), and leupeptin were from Boehringer Mannheim Canada (Laval, QC, Canada). All other protease inhibitors, PGE1, theophylline, ionophore A23187, adenosine 5′ diphosphate (ADP), and amiloride were from Sigma-Aldrich Canada (Oakville, ON, Canada). Fibrinogen (free of plasminogen and von Willebrand factor), Glu-plasminogen, plasmin, and bovine thrombin were from Enzyme Research (South Bend, IN). Polyclonal and monoclonal antibodies to u-PA were from Monosan (Uden, The Netherlands). Recombinant, active human plasminogen activator inhibitor 1 (PAI-1) was prepared and isolated as described.10 Polyclonal antibody against PAI-1 was raised in rabbits as previously described.11 Polyclonal anti–human plasminogen was from Biogenesis (Kingston, NH). Other antibodies and the procedures used for Western blotting of fibrinogen, fibronectin, von Willebrand factor, multimerin, factor V, thrombospondin-1, and osteonectin were as previously described.4 5 Purified recombinant single chain u-PA (scu-PA), high–molecular weight two-chain u-PA (tcu-PA), and low–molecular weight two-chain u-PA (LMW u-PA) were generously provided by Dr Jack Henkin (Abbott Laboratories, North Chicago, IL). Recombinant tissue-type plasminogen activator (t-PA) was from Hoffman-La Roche (Mississauga, ON, Canada). Supplies for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) were from Bio-Rad (Mississauga, ON, Canada). SeeBlue prestained SDS-PAGE protein standards were from Helixx Technologies (Scarborough, ON, Canada). Protein A Sepharose beads were from Amersham Pharmacia (Uppsala, Sweden). u-PA and t-PA enzyme-linked immunosorbent assays (ELISA) were purchased from American Diagnostica (AD; Greenwich, CT), and u-PA, PAI-1, and u-PA–PAI-1 complex ELISA were purchased from Oncogene Science (OS; Cambridge, MA). Results for u-PA ELISA were referred to by abbreviations of the assay manufacturer. 1 KB Plus DNA Ladder was from Gibco BRL (Burlington, ON, Canada).
Sample preparation
Double-centrifuged plasma (from blood collected in 9:1 vol/vol 3.2% buffered sodium citrate), washed platelet lysates, and platelet releasates were collected from patients and controls as previously described,4,5 with the following modifications. Resting platelets were prepared using anticoagulant and wash buffers supplemented with 2 μM PGE1 and 1 mM theophylline (final concentrations). Cell counts confirmed the washed platelets contained minimal leukocyte contamination. Washed platelets were solubilized (1 × 109 platelets/mL final)5 using buffer containing 0.5% Triton X-100 and multiple protease inhibitors (0.3 μM aprotinin, 2.8 μM E-64, 10 mM EDTA, 1 μM leupeptin, 5 mM N-ethyl-maleimide, 4 mM AEBSF, 1 μM pepstatin, 100 μM 1,10-phenanthrolene monohydrate, and 100 μg/mL soybean trypsin inhibitor [STI]). For some studies, platelet lysates were prepared without the serine protease inhibitors AEBSF, aprotinin, STI, and leupeptin. Platelet releasates4,5 were prepared from washed platelets, resuspended in albumin-free Tyrode buffer (pH 7.4 with 2 mM Ca++, 1 mM Mg++, and 5 mM HEPES; 1 × 109 platelets/mL) and activated (20 minutes, 37°C) using 50 μM ADP or 2 μM calcium ionophore A23187 (samples centrifuged 2000g for 10 minutes, followed by 14 000g for 15 minutes before freezing). All samples were frozen and stored at −70°C until analyzed. K562 cells stimulated with 12-O-tetradecanoyl-phorbol-13-acetate (TPA, 3 nM final; Sigma-Aldrich Canada) were used as a source of PAI-1 protein and u-PA messenger RNA (mRNA).12
Protein and protease analyses
Zymography was performed using 3% agarose substrate gels (SeaPlaque agarose; BioWhittaker Molecular Applications, Rockland, MD; in phosphate-buffered saline [PBS], pH 7.4) containing plasminogen-free fibrin or casein (1% wt/vol; Carnation Instant Skim Milk Powder, Nestle Canada, Toronto, ON), with or without added plasminogen (5 μg/mL final), similar to methods previously described.13,14 Protease activities were tested by spotting samples directly onto substrate gels, or after proteins were separated by nonreduced SDS-PAGE and renatured with 2% Triton X-100 in PBS, pH 7.4, for 1 hour. Casein gels with or without 1 mM amiloride were used for some determinations. Substrate gels were incubated with samples (37°C, 18 hours) and were photographed wet. Some samples were preincubated with protease inhibitors (same final concentrations as lysates; 20 minutes on ice) or recombinant PAI-1 (0-4000 ng/mL final after 1:1 dilution in a releasate pool, prepared from 5 QPD ionophore releasates; 1 hour, 22°C) before testing their proteolytic activity. Others were tested after immunodepletion with rabbit anti–human u-PA or control normal rabbit immunoglobulin G (IgG) bound to protein A Sepharose, similar to methods described.15
Plasma samples were assayed at 1:5 to 1:100 dilutions in the u-PA ELISA. Platelet lysates were tested at 1:2.5 and larger dilutions. Data for stored, washed, and pelleted platelet lysates were pooled because they contained similar amounts of u-PA at the dilutions tested. Data for new and stored lysates were analyzed separately because the stored samples were prepared with different protease inhibitors.4 5 All samples were tested undiluted in the t-PA ELISA, which was modified to include a lower concentration (2 ng/mL) standard. Some normal samples contained less u-PA or t-PA than the lowest standard of the AD ELISA when tested at recommended and lower dilutions. These amounts were reported as “less than” values when ranges for controls were determined, and they were rounded up to the nearest measurable value to calculate means and standard deviations for controls.
Active PAI-1 in platelet ionophore releasate and lysate (without added serine protease inhibitors) was assessed by measuring u-PA–PAI-1 complex generation, similar to methods previously described.16 Briefly, pooled samples of releasate and lysate, prepared from 5 control and 5 QPD donors, respectively, were incubated (30 minutes, 22°C) with or without added recombinant tcu-PA (200 ng/mL final in 20 μL sample) before measuring u-PA–PAI-1 complexes by ELISA (values expressed as an average of duplicate determinations).
For studies of α-granule protein degradation in vitro, recombinant tcu-PA (0-400 ng u-PA/mL) was incubated overnight (37°C) with releasate from control ionophore-stimulated platelets or with control platelet lysate, prepared without serine protease inhibitors (multimerin digests only) (0.4% Triton X-100, final; all reactions stopped with 4 mM AEBSF). Degraded proteins in these digests were compared to QPD platelet proteins by Western blotting after separation on SDS-PAGE or SDS-multimer gels.4-6 To determine whether u-PA formed high–molecular weight complexes when incubated with secreted platelet proteins, 10 ng recombinant u-PA was incubated with 10 to 60 μL control ionophore releasate for 1 to 18 hours.
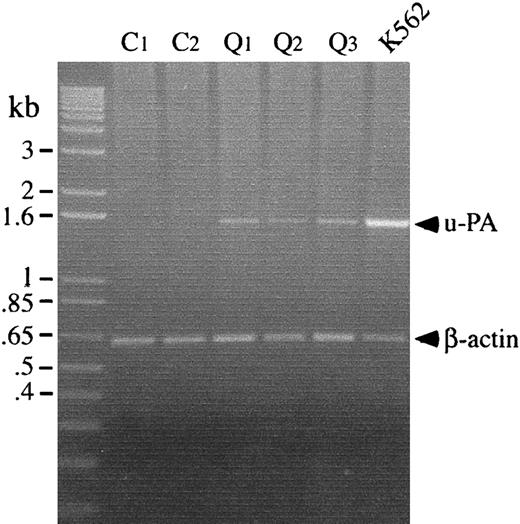
Analyses of platelet mRNA
Total RNA was extracted from platelets and from K562 cells, as previously described.17 Complementary DNA (cDNA) synthesis was carried out on 1 μg total RNA (20 μL final volume) using oligo dT as a primer and Thermoscript (Life Technologies, Burlington, ON, Canada) reverse transcriptase (RT), as recommended by the manufacturer. Polymerase chain reaction (PCR) was performed on 2 μL cDNA reaction in a final volume of 50 μL using Platinum Taq DNA polymerase (Life Technologies). Primers (synthesized by the Central Facility, McMaster University), expected products sizes, and cycle sequences for u-PA and β-actin reverse transcription–-PCR were u-PA forward, 5′-GGAATGGTCACTTTTACCG-3′, u-PA reverse, 5′-CTGCCCTGAAGTCGTTAG-3′, expected product 1.55 kb, 94°C at 30 seconds, 50°C at 30 seconds, 72°C at 2 minutes for 30 cycles; β-actin forward, 5′-CCTCGCCTTTGCCGATCC-3′, β-actin reverse, 5′-GGATCTTCATGAGGTAGTCAGTC-3′, expected product 620 bp, 94°C at 30 seconds, 55°C at 30 seconds, 72°C at 1 minute for 25 cycles. Products were analyzed on 1% agarose gels and visualized with ethidium bromide.
Results
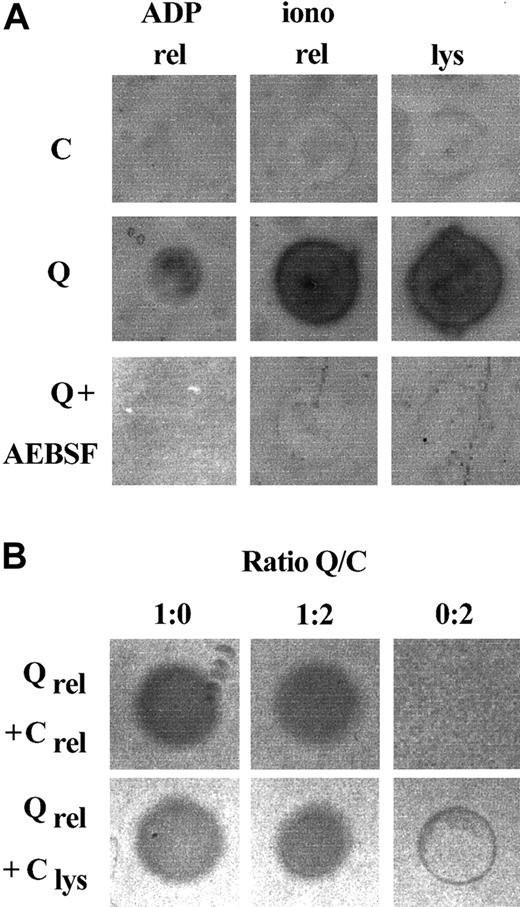
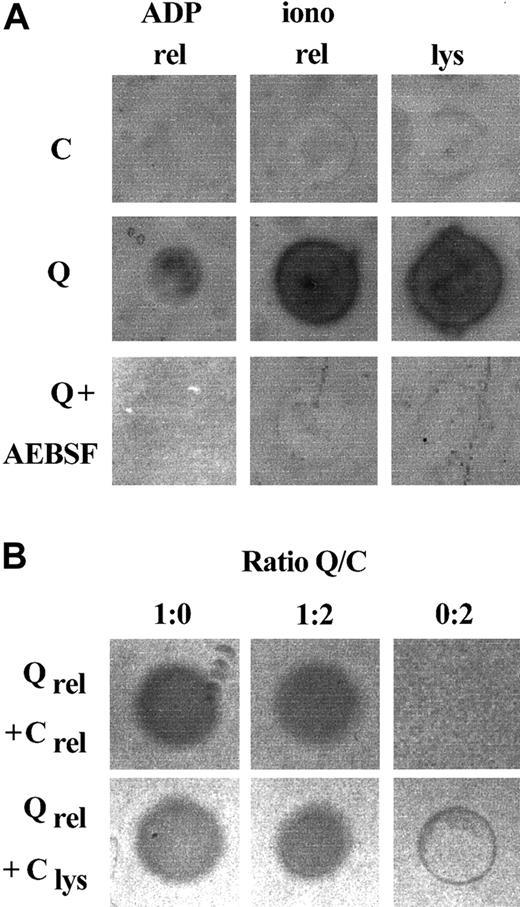
Because platelet fibrinogen was degraded in patients with the QPD, their platelet releasates and lysates were screened for proteolytic activity using fibrin substrate gels (Figure1). Fibrinolytic activity was evident in all QPD platelet releasates tested, but it was not detected in the same amounts of control releasates (Figure 1A; data representative of 5 patients and 12 controls). Fibrinolytic activity in QPD platelet releasates was inhibited by the serine protease inhibitor AEBSF (Figure1A), but it was not blocked by EDTA, leupeptin, the cysteine protease inhibitor E64, the aspartic protease inhibitor pepstatin, or the metalloproteinase inhibitor phenanthrolene (not shown). Similar fibrinolytic serine protease activity was present in lysates of QPD resting platelets, whereas it was undetectable in the same volume of control sample (Figure 1A). Fibrinolytic activity released by QPD platelets was not blocked in 1:2 mixtures with normal platelet releasate or lysate (Figure 1B), suggesting the defect was not due to an inhibitor deficiency.
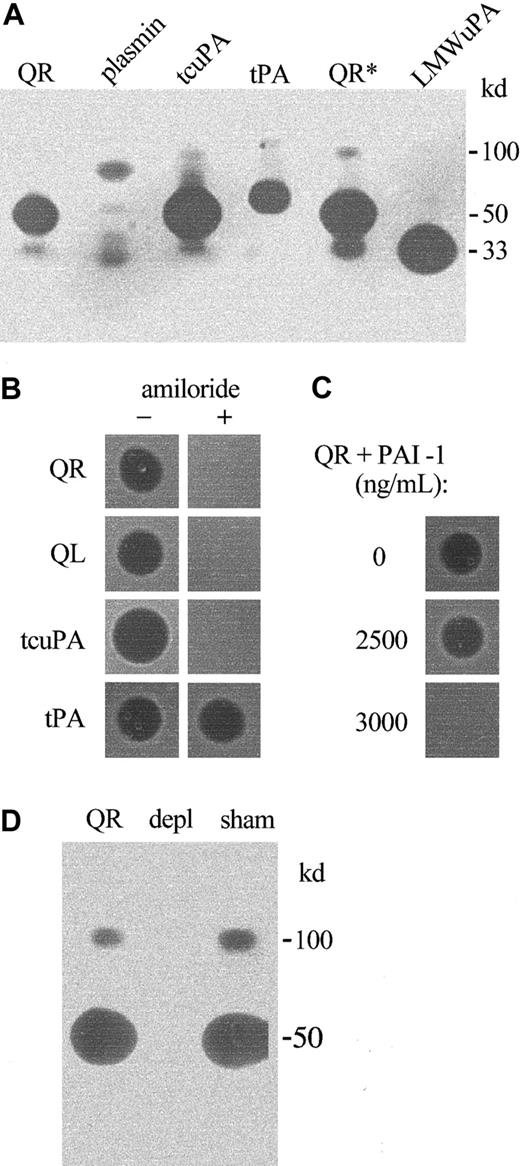
Fibrinolytic proteases in QPD platelets.
Platelet lysates (lys; 3 μL) and releasates (rel; 3 μL) from patients with QPD (Q) and control subjects (C) were spotted onto fibrin substrate gels. (A) There was abnormal fibrinolytic activity in QPD platelet lysates and releasates that was blocked by the serine protease inhibitor AEBSF. The secretagogue ionophore A23187 (iono) released more of this activity from QPD platelets than ADP. (B) The fibrinolytic enzymes released by QPD platelets were not inhibited in 1:2 mixtures with normal platelet lysates or releasates (tested ratios of patient ionophore releasate–control samples are shown). Protein rings, without evidence of fibrinolysis, were seen in the tests of control lysates.
Fibrinolytic proteases in QPD platelets.
Platelet lysates (lys; 3 μL) and releasates (rel; 3 μL) from patients with QPD (Q) and control subjects (C) were spotted onto fibrin substrate gels. (A) There was abnormal fibrinolytic activity in QPD platelet lysates and releasates that was blocked by the serine protease inhibitor AEBSF. The secretagogue ionophore A23187 (iono) released more of this activity from QPD platelets than ADP. (B) The fibrinolytic enzymes released by QPD platelets were not inhibited in 1:2 mixtures with normal platelet lysates or releasates (tested ratios of patient ionophore releasate–control samples are shown). Protein rings, without evidence of fibrinolysis, were seen in the tests of control lysates.
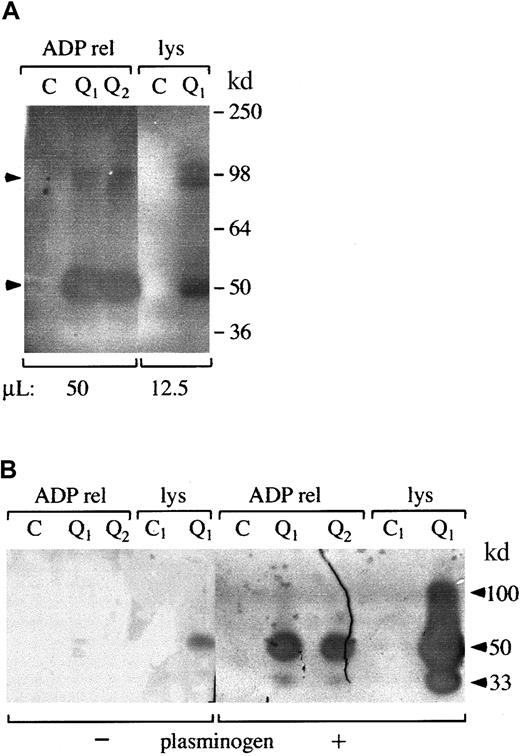
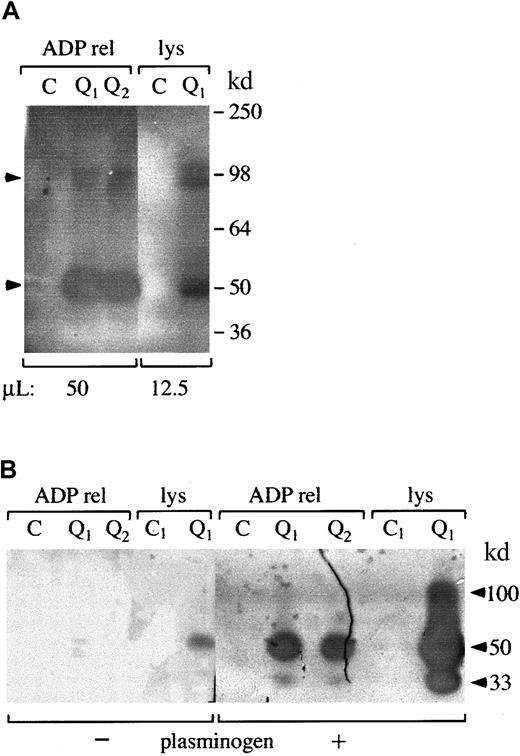
Zymograms indicated there were secretable 50-kd (major band) and 100-kd (minor band) (Mr nonreduced) fibrinolytic enzymes in QPD platelets that were not detectable in similar amounts of normal platelets (Figure 2A-B shows data representative of 5 patients and 12 controls). The activities of these fibrinolytic enzymes were destroyed by reduction (not shown). Comparisons of their activities on fibrin substrate gels, with and without added plasminogen (Figure 2B), indicated the 50- and 100-kd QPD platelet proteases had properties of plasminogen activators. QPD platelets also contained and secreted a 33-kd plasminogen activator that was not detected in the normal samples (Figure 2B).
Fibrin gel zymograms of proteases in QPD platelets.
Samples of QPD (Q1 and Q2 indicate patients from families 1 and 2, respectively) and control (C) platelet lysates (lys; A, 12.5 μL; B, 2 μL) and ADP releasates (ADP rel; A, 50 μL; B, 2 μL) were analyzed on fibrin substrate gels after 9% (A) or 5% to 15% (B) nonreduced SDS-PAGE. (A) Analyses using gels without added plasminogen indicated QPD platelets contained 50-kd (major band) and 100-kd (minor component) secretable, fibrinolytic proteases (arrows) that were not evident in the same amount of control samples. (B) Comparative analyses, using substrate gels with (+) or without (−) added plasminogen, indicated the 100- and 50-kd QPD platelet proteases had properties of plasminogen activators. A 33-kd plasminogen activator was also detected in QPD (but not in control) platelets and releasates (Mr based on 5%-15% nonreduced SDS-PAGE).
Fibrin gel zymograms of proteases in QPD platelets.
Samples of QPD (Q1 and Q2 indicate patients from families 1 and 2, respectively) and control (C) platelet lysates (lys; A, 12.5 μL; B, 2 μL) and ADP releasates (ADP rel; A, 50 μL; B, 2 μL) were analyzed on fibrin substrate gels after 9% (A) or 5% to 15% (B) nonreduced SDS-PAGE. (A) Analyses using gels without added plasminogen indicated QPD platelets contained 50-kd (major band) and 100-kd (minor component) secretable, fibrinolytic proteases (arrows) that were not evident in the same amount of control samples. (B) Comparative analyses, using substrate gels with (+) or without (−) added plasminogen, indicated the 100- and 50-kd QPD platelet proteases had properties of plasminogen activators. A 33-kd plasminogen activator was also detected in QPD (but not in control) platelets and releasates (Mr based on 5%-15% nonreduced SDS-PAGE).
ELISA and Western blots were used to determine whether the plasminogen activators in QPD platelets were either t-PA or u-PA. Although QPD and control plasmas contained similar amounts of t-PA, neither QPD nor control platelets contained detectable t-PA (Table1). Both the OS and AD u-PA ELISA indicated there was more than 100-fold more u-PA in QPD platelets than in normal platelets (Table 1). Furthermore, comparisons of platelet u-PA levels in unaffected family members and family members with the QPD indicated that only the affected patients had increased platelet u-PA levels (Table 1; data for stored platelet samples). The OS u-PA ELISA detected approximately 4-fold more u-PA in QPD platelets than the AD u-PA ELISA (Table 1), suggesting these assays differed in their ability to detect some forms of u-PA. The amounts of u-PA in normal plasma, measured by both OS and AD u-PA ELISA (Table 1), were similar to previously reported values.18-21 Each ELISA indicated patients with the QPD had larger increases in u-PA in their platelets than their plasmas because many patients had normal plasma u-PA levels (Table 1). u-PA ELISA confirmed QPD platelets released significant quantities of u-PA with secretagogue stimulation because their ADP releasates contained approximately 9% of their platelet u-PA, and their ionophore releasates contained approximately 48% of their platelet u-PA (averaged data, AD ELISA; n = 3 patients evaluated).
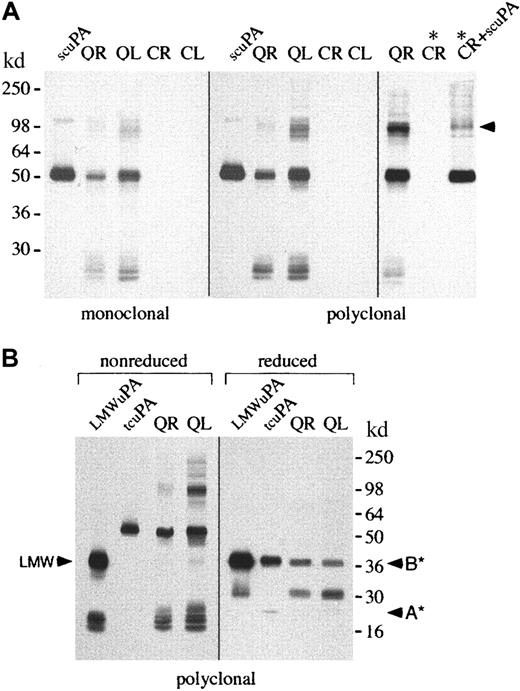
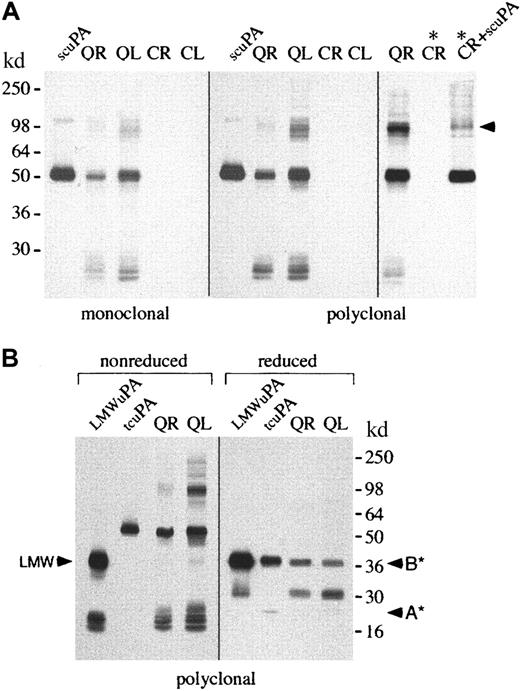
Western blots (probed with monoclonal and polyclonal u-PA antibodies) confirmed that QPD platelets and platelet releasates contained abnormally large amounts of u-PA (Figure3A shows data representative of 5 patients). Western blots of stored platelet lysates, from additional affected (n = 9) and unaffected (n = 5) members of both QPD families, confirmed this abnormality was present only in affected patients (not shown).
Western blots of u-PA in platelets.
u-PA in platelet lysates (L; 5 μL) and ionophore releasates (R; 5 μL) from patients with QPD (Q) and healthy control subjects (C) was visualized with monoclonal or polyclonal antisera after 10% SDS-PAGE. (A) Nonreduced analyses indicated QPD platelet lysates and releasates contained abnormally large amounts of u-PA. The predominant form of u-PA in QPD platelets comigrated, nonreduced, with recombinant scu-PA (scuPA, 10 ng), though larger and smaller forms were also detected. When control platelet releasate was incubated (lanes *) with recombinant scu-PA (lane CR + scuPA*), high–molecular weight u-PA complexes were generated that resembled large forms of u-PA in QPD releasates (arrow indicates the most abundant large form). (B) QPD platelet releasates and lysates (R and L, 5 μL nonreduced, 20 μL reduced) contained forms of u-PA with the characteristic nonreduced/reduced mobility of purified tcu-PA (tcuPA; 4.4 ng nonreduced, 17.6 ng reduced) and LMW u-PA (LMWuPA; 3.2 ng nonreduced, 12.8 ng reduced). Arrows indicate the A chain (A*) of reduced tcu-PA, the B chain (B*) common to reduced tcu-PA and LMW u-PA, and the nonreduced form of LMW u-PA (LMW). A reduced protein with the mobility of the A chain of tcu-PA was seen in prolonged exposures of the QR and QL lanes (not shown).
Western blots of u-PA in platelets.
u-PA in platelet lysates (L; 5 μL) and ionophore releasates (R; 5 μL) from patients with QPD (Q) and healthy control subjects (C) was visualized with monoclonal or polyclonal antisera after 10% SDS-PAGE. (A) Nonreduced analyses indicated QPD platelet lysates and releasates contained abnormally large amounts of u-PA. The predominant form of u-PA in QPD platelets comigrated, nonreduced, with recombinant scu-PA (scuPA, 10 ng), though larger and smaller forms were also detected. When control platelet releasate was incubated (lanes *) with recombinant scu-PA (lane CR + scuPA*), high–molecular weight u-PA complexes were generated that resembled large forms of u-PA in QPD releasates (arrow indicates the most abundant large form). (B) QPD platelet releasates and lysates (R and L, 5 μL nonreduced, 20 μL reduced) contained forms of u-PA with the characteristic nonreduced/reduced mobility of purified tcu-PA (tcuPA; 4.4 ng nonreduced, 17.6 ng reduced) and LMW u-PA (LMWuPA; 3.2 ng nonreduced, 12.8 ng reduced). Arrows indicate the A chain (A*) of reduced tcu-PA, the B chain (B*) common to reduced tcu-PA and LMW u-PA, and the nonreduced form of LMW u-PA (LMW). A reduced protein with the mobility of the A chain of tcu-PA was seen in prolonged exposures of the QR and QL lanes (not shown).
Western blots were used to determine whether the u-PA in QPD platelets comigrated, nonreduced and reduced, with purified scu-PA, tcu-PA, or LMW u-PA (Figure 3). There was considerable heterogeneity in the forms of u-PA found in QPD platelet lysates, and their releasates contained identical forms (Figure 3). On nonreduced gels (Figure 3A; Figure3B, left panel), the most abundant form of u-PA in QPD platelets comigrated with scu-PA and tcu-PA, whereas only a small proportion comigrated with LMW u-PA (Figure 3B). After reduction (Figure 3B, right panel), the most abundant form of u-PA in QPD platelets had the mobility of tcu-PA, indicating most u-PA in QPD platelets had been activated. Some of the less abundant forms of u-PA in QPD platelets were proteolyzed and did not comigrate with scu-PA, tcu-PA, or LMW u-PA (Figure 3A-B and longer exposures, not shown). A small proportion of their total u-PA was larger than scu-PA and tcu-PA and resembled high–molecular weight complexes generated by incubating exogenous scu-PA (Figure 3A, right panel) or tcu-PA (not shown) with normal platelet releasate proteins.
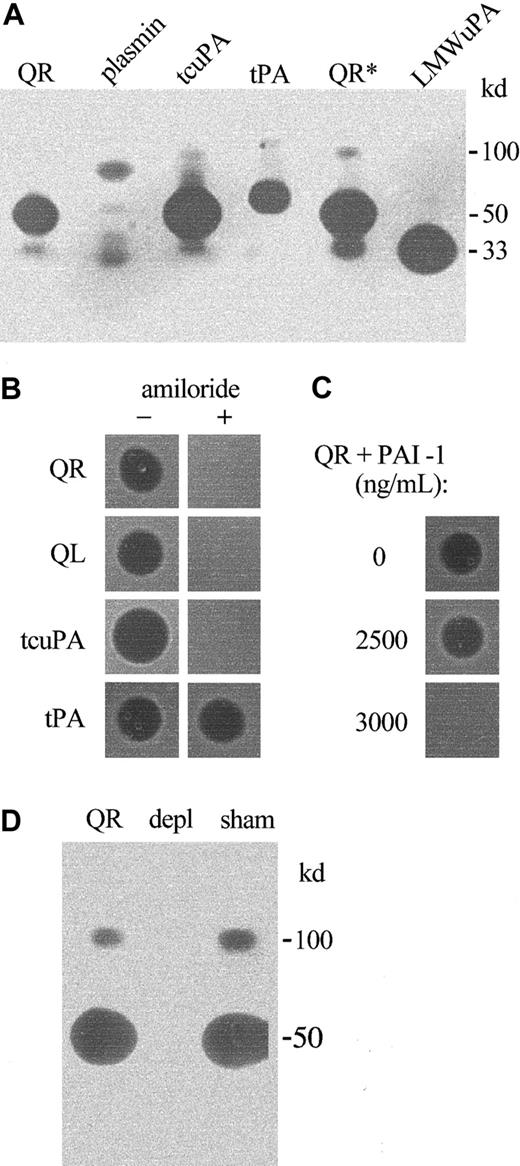
Zymograms indicated none of the QPD platelet plasminogen activators comigrated with t-PA or plasmin, and they confirmed the 50- and 33-kd plasminogen activators in QPD platelets comigrated with tcu-PA and LMW u-PA, respectively (Figure 4A). The activities of the 100-, 50-, and 33-kd QPD platelet plasminogen activators were blocked by 1 mM amiloride, which inhibited tcu-PA but not t-PA activity, as previously reported22 (Figure 4B). All the plasminogen activators in QPD releasates were neutralized when recombinant PAI-1 was added to final concentrations of 3000 ng/mL or more (Figure 4C), which was more than the concentration of PAI-1 in normal and QPD platelet lysates (Table 1). Furthermore, antibodies to u-PA selectively removed all detectable plasminogen activators (Figure4D) and fibrinolytic proteases (not shown) from QPD releasates. These observations indicated that the fibrinolytic, plasminogen-activating proteases detected in QPD platelets were different forms of the enzyme u-PA.
Plasminogen activators in QPD platelets.
Plasminogen activators were analyzed on casein substrate gels containing plasminogen. (A, D) Samples tested after 10% nonreduced SDS-PAGE. (B, C) Samples spotted directly onto substrate gels. (A) QPD releasate (QR, 6 μL; QR*, 10 μL) contained plasminogen activators that comigrated with tcu-PA (tcuPA, 8.8 ng) and LMW u-PA (LMWuPA, 3.2 ng), but not with purified plasmin (10 ng) or t-PA (tPA, 1 IU). (B) Unlike t-PA (0.5 IU), tcu-PA (5 ng) and the plasminogen activators in 1 μL QR and QL (QPD lysate) were inhibited on gels with added (+) 1 mM amiloride. (C) Large amounts of recombinant PAI-1 (final concentrations shown) were required to fully neutralize the plasminogen activators in pooled QR. (D) Zymograms indicated that the 100-, 50-, and 33-kd proteases in QR (5 μL/lane) were removed by rabbit antibodies to human u-PA (depl), but not by normal rabbit IgG (sham). The 33-kd protease in lanes sham and QR was evident on the original gel.
Plasminogen activators in QPD platelets.
Plasminogen activators were analyzed on casein substrate gels containing plasminogen. (A, D) Samples tested after 10% nonreduced SDS-PAGE. (B, C) Samples spotted directly onto substrate gels. (A) QPD releasate (QR, 6 μL; QR*, 10 μL) contained plasminogen activators that comigrated with tcu-PA (tcuPA, 8.8 ng) and LMW u-PA (LMWuPA, 3.2 ng), but not with purified plasmin (10 ng) or t-PA (tPA, 1 IU). (B) Unlike t-PA (0.5 IU), tcu-PA (5 ng) and the plasminogen activators in 1 μL QR and QL (QPD lysate) were inhibited on gels with added (+) 1 mM amiloride. (C) Large amounts of recombinant PAI-1 (final concentrations shown) were required to fully neutralize the plasminogen activators in pooled QR. (D) Zymograms indicated that the 100-, 50-, and 33-kd proteases in QR (5 μL/lane) were removed by rabbit antibodies to human u-PA (depl), but not by normal rabbit IgG (sham). The 33-kd protease in lanes sham and QR was evident on the original gel.
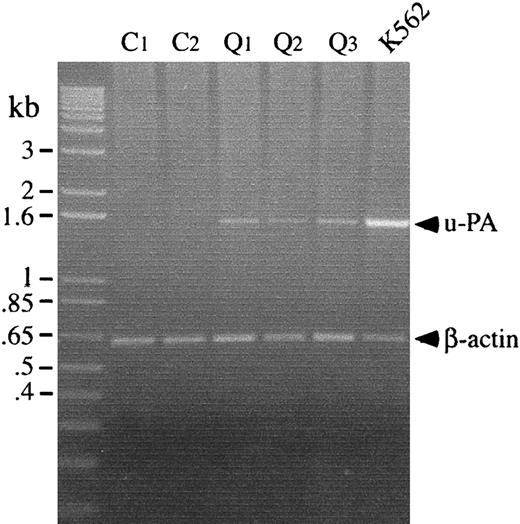
RT-PCR analyses were performed to determine whether the u-PA abnormalities in the QPD platelets were associated with increased u-PA mRNA levels in platelets. Although platelets from patients and controls contained similar amounts of β-actin mRNA, only QPD platelets contained detectable u-PA mRNA (Figure5).
Platelet u-PA and β-actin mRNA, analyzed by RT-PCR.
u-PA and β-actin transcripts were amplified separately before running the products in lanes of the gel. Lanes compare transcripts amplified from platelets of 2 controls (C) and 3 patients with the QPD (Q), with transcripts from TPA-stimulated K562 cells. QPD platelets contained normal amounts of β-actin mRNA and increased u-PA mRNA, which was not detectable in control platelets.
Platelet u-PA and β-actin mRNA, analyzed by RT-PCR.
u-PA and β-actin transcripts were amplified separately before running the products in lanes of the gel. Lanes compare transcripts amplified from platelets of 2 controls (C) and 3 patients with the QPD (Q), with transcripts from TPA-stimulated K562 cells. QPD platelets contained normal amounts of β-actin mRNA and increased u-PA mRNA, which was not detectable in control platelets.
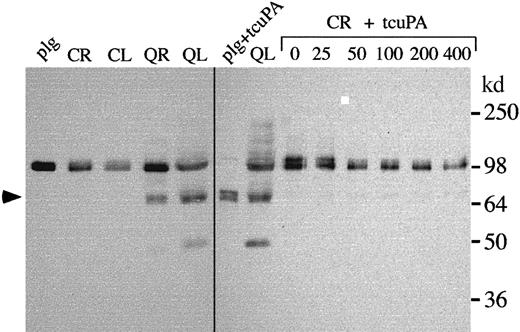
Unregulated u-PA activity in QPD platelets was further investigated by measuring platelet PAI-1 antigen and u-PA–PAI-1 complexes using ELISA. QPD platelets contained approximately 2-fold more PAI-1 antigen and more than 100-fold more u-PA–PAI-1 complexes than normal platelets (Table 1). Western blots confirmed some of the PAI-1 in QPD platelets had formed complexes with u-PA, though the proportions of complexed PAI-1 varied slightly between patients (Figure6A; Pt 3 indicates the patient with the highest concentrations of platelet u-PA–PAI-1 complexes by ELISA). Increased u-PA–PAI-1 complexes were also detected in QPD platelet releasates using ELISA (Figure 6B), but they were difficult to detect by Western blotting (Figure 6A and analyses of larger sample volumes, not shown). The high–molecular weight PAI-1 complexes stored in QPD platelets expressed epitopes recognized by u-PA antibodies (Figure 6A, lane *), and they comigrated with PAI-1 complexes generated in vitro by adding tcu-PA to normal platelet releasate (Figure 6A, right panel). All the QPD platelets tested contained proteolyzed forms of PAI-1 (Figure 6A, arrow) that were not evident in normal platelets, but only traces of similar proteolyzed forms were detected in control releasates incubated with tcu-PA (Figure 6A and longer exposures, not shown).
PAI-1 in control and QPD platelets.
(A) PAI-1 in control (C) and QPD (Q; 3 patients [Pt] are shown) samples was analyzed by Western blotting with rabbit anti–human PAI-1 after 10% nonreduced SDS-PAGE. Lanes compare 5 μL lysate (L) and ionophore releasates (R), 20 μL K562 media, and 5 μL control ionophore releasate, incubated with 0 to 400 ng/mL tcu-PA (CR + tcuPA), as indicated. Lane * shows the PAI-1 affinity purified from QL using monoclonal anti–u-PA. QPD platelet lysates contained PAI-1 in high–molecular weight complexes (bands near the 98-kd marker) that comigrated with the complexes generated by adding tcu-PA to control releasates. Proteolyzed PAI-1 (arrow) was detected in QPD platelet lysates and in long exposures (not shown) of control releasates incubated with tcu-PA. (B) u-PA–PAI-1 complex ELISA indicated that, unlike control samples, QPD platelet lysates and releasates were unable to generate additional u-PA–PAI-1 complexes in vitro with added (+) tcu-PA (data representative of 2 separate experiments with pooled samples).
PAI-1 in control and QPD platelets.
(A) PAI-1 in control (C) and QPD (Q; 3 patients [Pt] are shown) samples was analyzed by Western blotting with rabbit anti–human PAI-1 after 10% nonreduced SDS-PAGE. Lanes compare 5 μL lysate (L) and ionophore releasates (R), 20 μL K562 media, and 5 μL control ionophore releasate, incubated with 0 to 400 ng/mL tcu-PA (CR + tcuPA), as indicated. Lane * shows the PAI-1 affinity purified from QL using monoclonal anti–u-PA. QPD platelet lysates contained PAI-1 in high–molecular weight complexes (bands near the 98-kd marker) that comigrated with the complexes generated by adding tcu-PA to control releasates. Proteolyzed PAI-1 (arrow) was detected in QPD platelet lysates and in long exposures (not shown) of control releasates incubated with tcu-PA. (B) u-PA–PAI-1 complex ELISA indicated that, unlike control samples, QPD platelet lysates and releasates were unable to generate additional u-PA–PAI-1 complexes in vitro with added (+) tcu-PA (data representative of 2 separate experiments with pooled samples).
Assays of active PAI-1 indicated that although pooled QPD platelet lysates and releasates contained abnormally large amounts of u-PA–PAI-1 complexes before exogenous u-PA was added, they were unable to generate additional complexes with exogenous u-PA (Figure 6B). Furthermore, the amounts of u-PA–PAI-1 complexes generated when u-PA was added to pooled normal releasates and lysates were similar to the amounts contained in pooled QPD releasates and lysates (Figure 6B). These data indicated the active forms of PAI-1 had been depleted in QPD platelets, likely because they had formed complexes with u-PA in vivo.
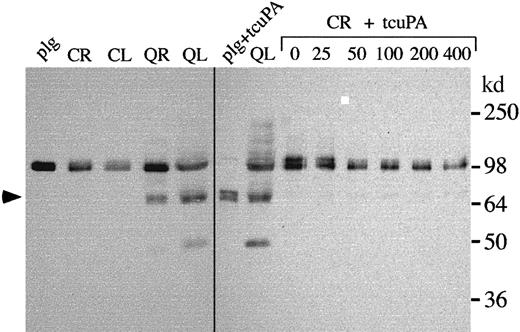
Western blots were used to determine whether the changes in u-PA in the QPD were associated with plasminogen proteolysis. QPD plasmas contained forms and amounts of plasminogen that were indistinguishable from normal controls (not shown). Although the plasminogen in normal, washed platelets comigrated with purified Glu-plasminogen, in QPD platelets much of the plasminogen was proteolyzed (Figure7), and there was a form that comigrated with plasmin on reduced (Figure 7) and nonreduced (not shown) gels. When normal platelet releasate was incubated with exogenous tcu-PA, there was loss of detectable intact plasminogen; however, the extent of plasminogen proteolysis was not as complete as in QPD platelets and the tcu-PA digests of purified plasminogen (Figure 7).
Plasminogen in control and QPD platelets.
Proteins were analyzed by Western blotting with rabbit anti–human plasminogen after 10% reduced SDS-PAGE. Lanes compare 12 μL control (C) and QPD (Q) lysates (L) and ionophore releasates (R) and control ionophore releasate, incubated overnight with 0 to 400 ng/mL tcu-PA (CR + tcuPA), as indicated. As references, Glu-plasminogen (plg; 20 ng/lane) and the plasmin generated by digesting Glu-plasminogen (60 ng) with 100 ng/mL tcu-PA (plg + tcuPA) are shown. QPD platelets contained proteolyzed plasminogen that comigrated with the heavy chain of plasmin (arrow; the light chain of plasmin was not visualized by the antisera). They also contained correspondingly reduced proportions of intact plasminogen and some smaller proteolyzed components, not evident in normal platelets. When control platelet releasate was incubated with exogenous tcu-PA, there was a similar loss of intact plasminogen, but the extent of plasminogen proteolysis was not as complete as in QPD platelets.
Plasminogen in control and QPD platelets.
Proteins were analyzed by Western blotting with rabbit anti–human plasminogen after 10% reduced SDS-PAGE. Lanes compare 12 μL control (C) and QPD (Q) lysates (L) and ionophore releasates (R) and control ionophore releasate, incubated overnight with 0 to 400 ng/mL tcu-PA (CR + tcuPA), as indicated. As references, Glu-plasminogen (plg; 20 ng/lane) and the plasmin generated by digesting Glu-plasminogen (60 ng) with 100 ng/mL tcu-PA (plg + tcuPA) are shown. QPD platelets contained proteolyzed plasminogen that comigrated with the heavy chain of plasmin (arrow; the light chain of plasmin was not visualized by the antisera). They also contained correspondingly reduced proportions of intact plasminogen and some smaller proteolyzed components, not evident in normal platelets. When control platelet releasate was incubated with exogenous tcu-PA, there was a similar loss of intact plasminogen, but the extent of plasminogen proteolysis was not as complete as in QPD platelets.
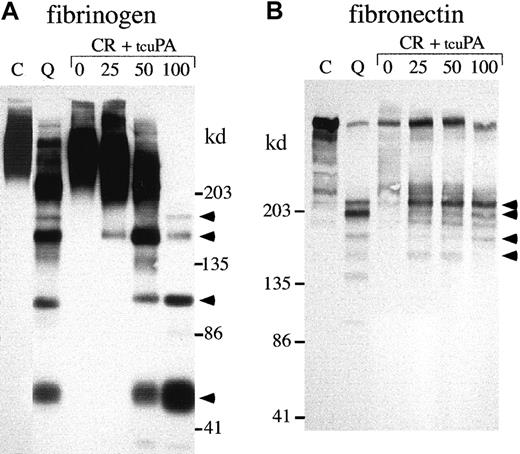
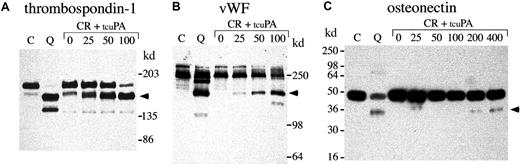
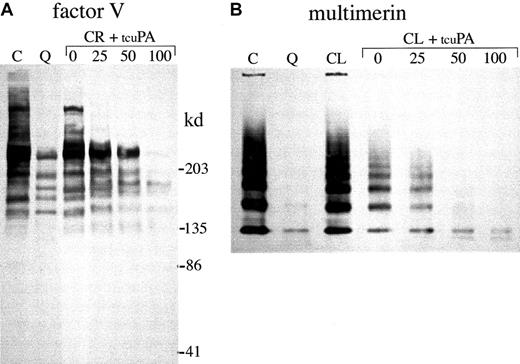
Next, we investigated whether exogenous tcu-PA (in concentrations similar to the increased u-PA in QPD platelets) could trigger the proteolysis of other stored platelet proteins to forms that comigrated with degraded proteins in QPD platelets (Figures8, 9,10). Adding tcu-PA to normal platelet releasate resulted in the degradation of α-granule fibrinogen to FDPs that comigrated with QPD platelet FDPs (Figure 8A). α-Granule fibronectin was also degraded when tcu-PA was added to normal platelet releasate (Figure 8B). Although the fibronectin degradation in vitro was not as extensive as in QPD platelets, there were many similarities in the sizes of degradation products (Figure 8B, arrows). When tcu-PA was incubated with normal platelet releasate, thrombospondin-1 was converted to a form that comigrated with the larger thrombospondin-1 degradation product in QPD platelets (Figure 9A, arrow). Osteonectin and von Willebrand factor were degraded when normal platelet releasate was incubated with tcu-PA to forms that comigrated with degraded osteonectin and von Willebrand factor in QPD platelets (Figure 9B-C, arrows). Platelet factor V was degraded when normal platelet releasate was incubated with tcu-PA (Figure 10A), resulting in a loss of factor V detectable by Western blotting, as in QPD platelets.4 5Because the amount of multimerin in platelet releasates was limiting, multimerin proteolysis was investigated by adding tcu-PA to normal platelet lysate, prepared without serine protease inhibitors (Figure10B). tcu-PA induced a striking loss of multimerin detected by Western blotting in these experiments, as in QPD platelets (Figure 10B shows multimerin multimers; findings in samples analyzed by reduced SDS-PAGE were similar [not shown]).
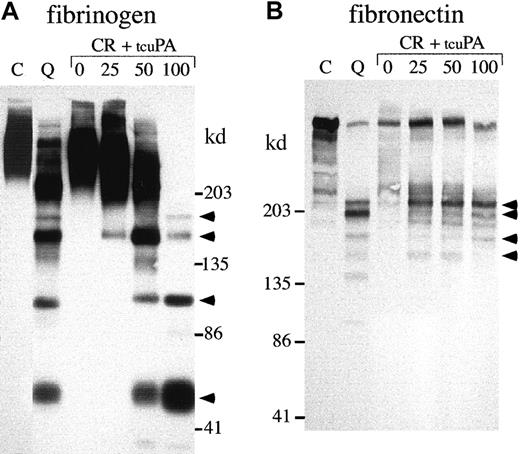
α-granule fibrinogen and fibronectin degradation.
Western blots compare nonreduced fibrinogen (A; 5%-8% SDS-PAGE) and fibronectin (B; 4%-8% SDS-PAGE) in QPD (Q) and control (C) platelet lysates with the degradation products generated by incubating normal platelet ionophore releasate (CR) with 0, 25, 50, or 100 ng/mL tcu-PA (tcuPA), as indicated (volumes of releasate and lysates: A, 3.6 μL/lane; B, 15 μL/lane). When tcu-PA was incubated with normal platelet secretory proteins, α-granule fibrinogen and fibronectin were proteolyzed to forms that comigrated (arrows) with their degraded forms in QPD platelets.
α-granule fibrinogen and fibronectin degradation.
Western blots compare nonreduced fibrinogen (A; 5%-8% SDS-PAGE) and fibronectin (B; 4%-8% SDS-PAGE) in QPD (Q) and control (C) platelet lysates with the degradation products generated by incubating normal platelet ionophore releasate (CR) with 0, 25, 50, or 100 ng/mL tcu-PA (tcuPA), as indicated (volumes of releasate and lysates: A, 3.6 μL/lane; B, 15 μL/lane). When tcu-PA was incubated with normal platelet secretory proteins, α-granule fibrinogen and fibronectin were proteolyzed to forms that comigrated (arrows) with their degraded forms in QPD platelets.
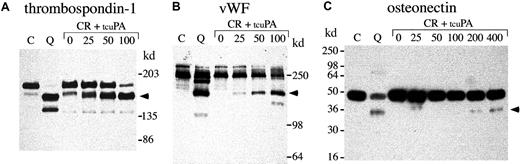
α-granule thrombospondin-1, von Willebrand factor, and osteonectin degradation.
Western blots compare thrombospondin-1 (A), von Willebrand factor (vWF) (B), and osteonectin (C) in QPD platelet lysates with degraded forms generated by incubating normal platelet ionophore releasate with tcu-PA (lanes and samples as in Figure 8; ng/mL tcu-PA are indicated). (A) When tcu-PA was incubated with normal platelet releasate, α-granule thrombospondin-1 was partially proteolyzed to a form that comigrated with the larger thrombospondin-1 degradation product in QPD platelets (arrow) (lanes compare 24 μL releasate and lysate, after reduced 4%-8% SDS-PAGE). (B) tcu-PA induced the degradation of von Willebrand factor to a form that comigrated with degraded von Willebrand factor in QPD platelets (arrow) (lanes compare 6 μL of releasate and lysate, after reduced 7% SDS-PAGE). (C) tcu-PA also induced the proteolysis of α-granule osteonectin, generating a form that comigrated with degraded osteonectin in QPD platelets (arrow) (lanes compare 9 μL releasate and lysate, after reduced 12% SDS-PAGE).
α-granule thrombospondin-1, von Willebrand factor, and osteonectin degradation.
Western blots compare thrombospondin-1 (A), von Willebrand factor (vWF) (B), and osteonectin (C) in QPD platelet lysates with degraded forms generated by incubating normal platelet ionophore releasate with tcu-PA (lanes and samples as in Figure 8; ng/mL tcu-PA are indicated). (A) When tcu-PA was incubated with normal platelet releasate, α-granule thrombospondin-1 was partially proteolyzed to a form that comigrated with the larger thrombospondin-1 degradation product in QPD platelets (arrow) (lanes compare 24 μL releasate and lysate, after reduced 4%-8% SDS-PAGE). (B) tcu-PA induced the degradation of von Willebrand factor to a form that comigrated with degraded von Willebrand factor in QPD platelets (arrow) (lanes compare 6 μL of releasate and lysate, after reduced 7% SDS-PAGE). (C) tcu-PA also induced the proteolysis of α-granule osteonectin, generating a form that comigrated with degraded osteonectin in QPD platelets (arrow) (lanes compare 9 μL releasate and lysate, after reduced 12% SDS-PAGE).
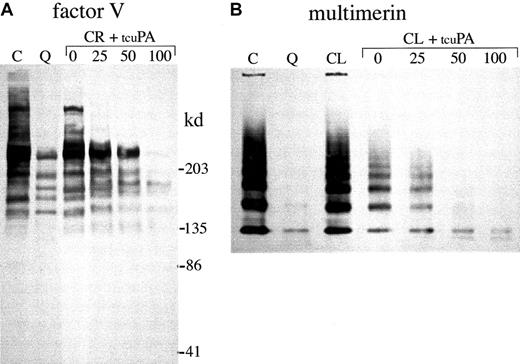
α-granule factor V and multimerin degradation.
Western blots compare proteins in QPD (Q) and control (C) platelet lysates (samples with all inhibitors) with the forms generated by incubating normal platelet ionophore releasate (CR; A) or lysate (CL; B, lysate without serine protease inhibitors) with 0, 25, 50, or 100 ng/mL tcu-PA (tcuPA), as indicated. (A) When u-PA was incubated with normal platelet secretory proteins, α-granule factor V was degraded, resulting in a loss of factor V detectable by Western blotting with polyclonal antisera, as in QPD platelets (lanes compare 18 μL lysate and releasate, after reduced 4%-8% SDS-PAGE). (B) Although there was some loss of detectable multimerin in lysates incubated without tcu-PA, tcu-PA reduced the detectable multimerin, as in QPD platelets (lanes compare 15 μL lysates, separated on nonreduced multimer gels and probed with a mixture of monoclonal and polyclonal antimultimerin). Traces of degraded multimerin (bands below the smallest multimerin polymer in normal platelets) were evident in the u-PA digests (B) and in QPD platelets, with longer exposures (not shown).
α-granule factor V and multimerin degradation.
Western blots compare proteins in QPD (Q) and control (C) platelet lysates (samples with all inhibitors) with the forms generated by incubating normal platelet ionophore releasate (CR; A) or lysate (CL; B, lysate without serine protease inhibitors) with 0, 25, 50, or 100 ng/mL tcu-PA (tcuPA), as indicated. (A) When u-PA was incubated with normal platelet secretory proteins, α-granule factor V was degraded, resulting in a loss of factor V detectable by Western blotting with polyclonal antisera, as in QPD platelets (lanes compare 18 μL lysate and releasate, after reduced 4%-8% SDS-PAGE). (B) Although there was some loss of detectable multimerin in lysates incubated without tcu-PA, tcu-PA reduced the detectable multimerin, as in QPD platelets (lanes compare 15 μL lysates, separated on nonreduced multimer gels and probed with a mixture of monoclonal and polyclonal antimultimerin). Traces of degraded multimerin (bands below the smallest multimerin polymer in normal platelets) were evident in the u-PA digests (B) and in QPD platelets, with longer exposures (not shown).
Discussion
Patients with Quebec platelet disorder have an unusual biochemical defect that causes their α-granule proteins to be degraded. Unlike patients with severe α-granule protein deficiencies, they suffer from bleeding that is paradoxically delayed and cannot be controlled with platelet transfusions.1 The purpose of our current study was to determine whether patients with QPD had a protease abnormality in their circulating platelets. We found that unlike normal platelets, QPD platelets contained large amounts of fibrinolytic, plasminogen-activating proteases. Moreover, we determined this was because QPD platelets contained markedly increased amounts of the enzyme u-PA, within a secretory compartment. These observations suggest u-PA could be involved in the pathogenesis of this unique storage pool disorder and its hemorrhagic complications.
Like normal platelets, QPD platelets store plasma-derived and megakaryocyte-synthesized proteins within their α-granules.5 Some u-PA has been reported to be associated with normal platelets and their external membranes when large quantities of platelet proteins have been analyzed.23-26 We observed that QPD platelets contained more than 100-fold more u-PA than normal platelets, which contained up to 1.3 ng u-PA/109 platelets. Furthermore, unlike normal platelets, QPD platelets released u-PA and high concentrations of u-PA–PAI-1 complexes in response to secretagogue stimulation. QPD platelet lysates, prepared with (Table 1) or without (Figure6B) high concentrations of serine protease inhibitors, also contained high concentrations of u-PA–PAI-1 complexes, suggesting QPD platelets costore u-PA with PAI-1 in α-granules.27 We suspect the increased u-PA in QPD platelets is synthesized by their megakaryocytes because only some patients had increased u-PA in plasma, and, unlike normal platelets, QPD platelets contained u-PA mRNA.
u-PA has a number of different forms, and its tcu-PA form has much greater plasminogen-activating activity than uncleaved scu-PA.28-31 These forms can be distinguished from each other and from LMW u-PA using nonreduced and reduced SDS-PAGE.28-31 Whereas normal platelets have been reported to contain mostly scu-PA,23,24 we found QPD platelets contained predominantly active tcu-PA, minimal scu-PA, some LMW u-PA, and a small amount of u-PA in high–molecular weight complexes. Moreover, unlike normal platelets, QPD platelets contained plasminogen that was proteolyzed and that comigrated with plasmin. The high–molecular weight u-PA complexes in QPD platelets resembled the complexes generated by incubating exogenous u-PA with normal releasate, and they included forms recognized by PAI-1 antibodies. These data suggest the very large forms of u-PA in QPD platelets, like the large forms in normal platelets,23,24 represent u-PA complexed to soluble platelet protease inhibitors, such as PAI-132-36 and protease nexin 1.37
The unregulated u-PA activity in QPD platelets indicates they do not contain sufficient protease inhibitors to fully neutralize their stored u-PA. Normal platelets contain large amounts of the u-PA inhibitor PAI-1 within their α-granules; however, most are functionally inactive, or latent, and incapable of neutralizing plasminogen activators.27,35,38-40 We observed that some of the u-PA in QPD platelets had been neutralized by PAI-1. However, the amount of exogenous, recombinant PAI-1 required to completely neutralize the u-PA secreted by QPD platelets exceeded the amount of PAI-1 in normal and QPD platelets. Because we found QPD platelets contained more than 100-fold more u-PA–PAI-1 complexes and 2-fold more total PAI-1 antigen than normal platelets, our data exclude a functional or quantitative PAI-1 deficiency as the cause of their unregulated u-PA activity. Although only a minority of the total PAI-1 in QPD platelets was contained in high–molecular weight complexes, we were unable to generate any further u-PA–PAI-1 complexes by adding u-PA to QPD platelet lysates or releasates in vitro. This suggests the increased u-PA in QPD platelets depletes their stores of active PAI-1. The relative proportions of total PAI-1, u-PA, and u-PA–PAI-1 complexes detected in QPD platelets by ELISA (954 ng PAI-1, 505 ng u-PA [OS assay], and 95 ng u-PA–PAI-1 complexes/109platelets) and the similar molecular masses of PAI-1 and u-PA41 infer that only a limited amount (approximately 5%) of the total PAI-1 in QPD platelets was active in neutralizing u-PA in vivo. Interestingly, this estimate corroborates previous reports that normal platelets contain less than 10% active PAI-1.27,35,38 39 The large amounts of u-PA in QPD platelets, and the relatively limited supply of u-PA inhibitors in platelets, could be part of the reason patients with QPD experience delayed bleeding that cannot be controlled with platelet transfusions.
The diversity of proteins degraded in QPD platelets has suggested that fairly broad-specificity protease(s) are involved. The FDPs secreted by QPD platelets are not recognized by a monoclonal antibody specific for plasmin-degraded fibrinogen.6 Using sensitive Western blots, we observed that QPD platelets contained proteolyzed forms of plasminogen with the mobility of plasmin, but we were unable to detect plasmin activity in QPD platelet releasates by zymography, even after u-PA was immunodepleted. tcu-PA and LMW u-PA are known to proteolyze fibrinogen in addition to plasminogen,42 but where they cleave fibrinogen has not been determined. Moreover, it is not yet known whether tcu-PA and LMW u-PA can cleave other potential substrates within platelets. We observed that the net effects of adding exogenous tcu-PA to normal platelet secretory proteins (in concentrations similar to the increased u-PA in QPD platelets) were a loss of intact plasminogen and the proteolysis of many α-granule proteins. There were phenotypic similarities in the sizes of the fibrinogen, fibronectin, von Willebrand factor, thrombospondin-1, and osteonectin degradation products generated to degraded proteins in QPD platelets. Furthermore, platelet multimerin and factor V were proteolyzed after adding tcu-PA, resulting in a loss of the forms detectable by Western blotting, as in QPD platelets. These observations provide indirect evidence that the changes to α-granule proteins, including multimerin, in the QPD likely reflect a complex process of proteolysis that may be initiated by increased platelet u-PA. The less extensive proteolysis of plasminogen—and some of the other α-granule proteins—in the in vitro digests compared to QPD platelets could reflect differences in the duration of substrate protein exposure to u-PA, or it could reflect contributions of factors, such as the environment within platelets, that enhance α-granule protein proteolysis in vivo. Although platelets have receptors on their external membranes for scu-PA and tcu-PA43 44 that could modulate some aspects of u-PA proteolysis in the QPD, we observed that membrane-tethered proteins were not required to degrade α-granule proteins to forms found in QPD platelets.
u-PA is normally expressed in many different tissues,45and it is thought to play a role in diverse physiological and pathological processes.31 In mice, u-PA deficiency causes problems with excess fibrin deposition, whereas its overexpression in the liver results in bleeding, marked hypofibrinogenemia, and systemic fibrinogenolysis.46,47 The QPD has biochemical abnormalities distinct from other platelet storage pool disorders1 and from congenital bleeding disorders associated with increased t-PA levels or t-PA–related proteins in plasma.48,49 Like α2-antiplasmin deficiency,50 the QPD is not associated with systemic fibrinogenolysis.6 This may be because plasma u-PA inhibitors effectively regulate the normal to increased u-PA levels in the plasma of patients with QPD. Our observations indicate patients with the QPD have an inherited, autosomal dominant defect that increases u-PA expression and storage in their megakaryocytes and platelets. Fibrinolytic inhibitors (such as tranexamic acid and epsilon amino caproic acid) rapidly and effectively control bleeding in patients with the QPD, yet they do not measurably improve QPD α-granule protein degradation, even when they are given for several weeks of therapy.6 This observation suggests that the moderate to severe bleeding in patients with the QPD could result from accelerated fibrinolysis within the hemostatic plug, where the concentrations of released u-PA may overwhelm protease inhibitors.
The QPD is the only inherited bleeding disorder in humans associated with increased levels of u-PA in blood. Unraveling its genetic cause is likely to provide further insights into this unusual and sometimes fatal bleeding disorder.
C.P.M.H. is the recipient of a Career Investigator Award from the Heart and Stroke Foundation of Ontario, and a Canada Research Chair in Molecular Hemostasis from the Government of Canada and a Premier's Research Excellence Award from the Ontario Government. We thank Dr Jack Henkin at Abbott Laboratories (North Chicago, IL) for the gift of recombinant u-PA.
Supported by grant NA 4379 from the Heart and Stroke Foundation of Ontario (C.P.M.H.) and a grant from Aventis Behring Canada (G.E.R.). W.H.A.K. is the recipient of a Medical Research Council of Canada/Heart and Stroke Foundation of Canada Post-Doctoral Fellowship Award.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Catherine P. M. Hayward, Department of Pathology and Molecular Medicine, McMaster University Medical Center, Rm 2N32, 1200 Main St West, Hamilton, Ontario, Canada L8N 3Z5; e-mail:haywrdc@mcmaster.ca.

![Fig. 6. PAI-1 in control and QPD platelets. / (A) PAI-1 in control (C) and QPD (Q; 3 patients [Pt] are shown) samples was analyzed by Western blotting with rabbit anti–human PAI-1 after 10% nonreduced SDS-PAGE. Lanes compare 5 μL lysate (L) and ionophore releasates (R), 20 μL K562 media, and 5 μL control ionophore releasate, incubated with 0 to 400 ng/mL tcu-PA (CR + tcuPA), as indicated. Lane * shows the PAI-1 affinity purified from QL using monoclonal anti–u-PA. QPD platelet lysates contained PAI-1 in high–molecular weight complexes (bands near the 98-kd marker) that comigrated with the complexes generated by adding tcu-PA to control releasates. Proteolyzed PAI-1 (arrow) was detected in QPD platelet lysates and in long exposures (not shown) of control releasates incubated with tcu-PA. (B) u-PA–PAI-1 complex ELISA indicated that, unlike control samples, QPD platelet lysates and releasates were unable to generate additional u-PA–PAI-1 complexes in vitro with added (+) tcu-PA (data representative of 2 separate experiments with pooled samples).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/2/10.1182_blood.v98.2.257/5/m_h81411272006.jpeg?Expires=1768413310&Signature=KGUai8EIInDEZVZP6mN5aIAeb9MNVjWjqiuH2TeDy2a1LBtX9h8fBNDLIe5gIuMTmMonLRYA5~fkWCW6rELF1jACiHhl50WmNsIK3MwDEiGFsQD~UBC3mWlsMag5Ra~Js~FxTcUi5yg1W5~MGBpR4NhleWIJPGjVcghcDidLZuDSwlmfLgqjJvqca2U7JsFYiKqpJmenR2PHRTFisxkZfAvQDJ~2Sj-FbZUUg3pL45BPnv9jU8E~E6rYNTpMjXb04b6tJcHvDw-jQ6TAipfsp8EU2WA6P9dTZO7g-ttsl2tluUGe9cbjp~EmkD2Gibk0Mpe5ThFrVq2UQyTgPAcfGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 6. PAI-1 in control and QPD platelets. / (A) PAI-1 in control (C) and QPD (Q; 3 patients [Pt] are shown) samples was analyzed by Western blotting with rabbit anti–human PAI-1 after 10% nonreduced SDS-PAGE. Lanes compare 5 μL lysate (L) and ionophore releasates (R), 20 μL K562 media, and 5 μL control ionophore releasate, incubated with 0 to 400 ng/mL tcu-PA (CR + tcuPA), as indicated. Lane * shows the PAI-1 affinity purified from QL using monoclonal anti–u-PA. QPD platelet lysates contained PAI-1 in high–molecular weight complexes (bands near the 98-kd marker) that comigrated with the complexes generated by adding tcu-PA to control releasates. Proteolyzed PAI-1 (arrow) was detected in QPD platelet lysates and in long exposures (not shown) of control releasates incubated with tcu-PA. (B) u-PA–PAI-1 complex ELISA indicated that, unlike control samples, QPD platelet lysates and releasates were unable to generate additional u-PA–PAI-1 complexes in vitro with added (+) tcu-PA (data representative of 2 separate experiments with pooled samples).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/2/10.1182_blood.v98.2.257/5/m_h81411272006.jpeg?Expires=1768413311&Signature=oMVyd4pKBwTmEDY~h46G4A12f5JFGIWX7b9OmRBq1F2uXXtUcdqu3p8YfIo0NJXrCp8iPtg8mig1dJtPcSngNQVSiNmkJINdhdXwrM8aNvT-OC7~7tkkZaxGKAlN7x~nKdSnS1Z2-ohFn7BXLhytabKbPJl8NbQ-tiMLSA9Pr26Y1EDSlTZw~xWnXUQsW6bddmUUgT6hQ2Kz6N7joQV9l21DGOihAAJ8DeBK3Fs-Z8hMnhopm2bFAM8cOBCsJTwVnUTo4RiLBa9P2NPQvDvq09DvS9E5rScpuvnQvmak~yTWptJGKUs2~LtFntS7r~Vu0R2hhWKemoITc44gDB0BwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)