Abstract
Previously a novel gene was identified that encodes a glucocorticoid-induced leucine zipper (GILZ) whose expression is up-regulated by dexamethasone. This study analyzed the role of GILZ in the control of T-cell activation and its possible interaction with nuclear factor κB (NF-κB). Results indicate that GILZ inhibits both T-cell receptor (TCR)–induced interleukin-2/interleukin-2 receptor expression and NF-κB activity. In particular, GILZ inhibits NF-κB nuclear translocation and DNA binding due to a direct protein-to-protein interaction of GILZ with the NF-κB subunits. Moreover, GILZ-mediated modulation of TCR-induced responses is part of a circuit because TCR triggering down-regulates GILZ expression. These results identify a new molecular mechanism involved in the dexamethasone-induced regulation of NF-κB activity and T-cell activation.
Introduction
Glucocorticoid hormones (GCHs) are commonly used as therapeutic agents for many acute and chronic inflammatory and autoimmune diseases, in transplant patients, and in the treatment of leukemias and lymphomas.1-5 Their efficacy is in part due to the effect of GCHs on T-cell activity. T-cell activation, differentiation, and death are regulated by a number of stimuli that can contribute either to positive or negative selection.6-10 After antigen–T-cell receptor (TCR) interaction, clonal expansion and differentiation contribute to generate activated effectors and an immune response, followed by elimination of most of the activated cells to end the immune response and thus avoid a continuous clonal expansion and the generation of autoimmunity.11,12 TCR-driven activation involves the coordinated expression of a number of molecules involved in activation and clonal expansion, such as interleukin-2/interleukin-2 receptor (IL-2/IL-2R), as well as those involved in activation-induced cell death (AICD), such as Fas/Fas ligand (Fas/FasL).12-18Moreover, a number of genes coding for transcription factors are involved as regulators of T-cell activation and death. In particular, the role of nuclear factor κB (NF-κB) as a regulator of T-cell activation and apoptosis in thymocytes, peripheral T cells, and T-cell hybridomas has been described.19-24
Stimuli other than antigen-TCR interaction also regulate T-cell activation and death, including cytokines and coaccessory molecules.25-27 Among different stimuli, GCHs are critical regulators of T-cell development.28-32 In particular, GCHs inhibit T-lymphocyte activation and death and also counter activation-induced IL-2/IL-2R and Fas/FasL expression.31,33 The inhibitory effect on T-cell activation and death relates in part to the glucocorticoid-induced inhibition of NF-κB activity.24,34 In fact, it has been previously reported that GCHs antagonize NF-κB activity through augmentation of I-κBα expression through competition for the coactivator protein CBP/300 and through direct association of glucocorticoid receptor (GR) with NF-κB subunits.35-40It has also been reported that dexamethasone (DEX) treatment can down-modulate NF-κB/p65 transactivation potential in the absence of any I-κB up-regulation.40
Recently, DEX-induced genes able to inhibit the TCR-activated death have been described.41,42 In particular, we have previously identified a new gene, glucocorticoid-induced leucine zipper (GILZ), encoding a novel member of the leucine zipper family.42 GILZ is found to be expressed in normal T lymphocytes present in the thymus, spleen, and lymph nodes, while low or no expression was detected in other nonlymphoid tissues. Moreover, GILZ protein is detected in the nucleus of transfected clones.42 In thymocytes and peripheral T cells, GILZ gene expression is induced by DEX, a synthetic GCH with high affinity for the GR. Furthermore, GILZ expression protects T cells from TCR-activated apoptosis but not by treatment with other apoptotic stimuli. This antiapoptotic effect correlates with inhibition of activation-induced Fas/FasL up-regulation. Thus, GILZ is a candidate transcription factor involved in the regulation of T-cell activation.
In the present paper we have analyzed the role of GILZ in the control of IL-2/IL-2R expression induced by T-cell activation and the possible interaction with NF-κB molecules. Our results indicate that DEX-induced GILZ up-regulation inhibits TCR-induced NF-κB activation and nuclear translocation, IL-2 production, and IL-2R expression. This NF-κB inhibition is the result of a direct interaction between GILZ and p65/p52 molecules and constitutes a new molecular mechanism of GCH–NF-κB interaction. Furthermore, GILZ-mediated inhibition of TCR-induced activation appears to be part of a circuit because TCR down-regulates GILZ.
Materials and methods
Cell lines and animals
A spontaneously dividing CD3+, CD4+, CD2+, CD44+ subline of the ova-specific hybridoma T-cell line, 3DO,43 was used for the experiments. Cells were maintained in logarithmic growth in RPMI 1640 supplemented with 10% fetal calf serum, 10 nM HEPES, and antibiotics. Thymus cells were obtained from 4- to 6-week-old C3H/HeN mice (Charles River, Calco, Milan, Italy).
Human kidney epithelial carcinoma cell line 293 and embryonal carcinoma cell line NTera-2 were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and antibiotics.
Transfection of cultured cells and clone preparation
Transfected clones were prepared as previously described.44 Briefly, mouse GILZ complementary DNA (cDNA) coding sequence (414 base pairs) was cloned into a pcDNA3 plasmid (Invitrogen, San Diego, CA) for expression in 3DO cells. (Mouse GILZ GenBank accession number AF024519.) Cells were transfected by electroporation (300 mA, 960 μF) with 15 μg linearized pcDNA3 vector (control clones) or 15 μg linearized pcDNA3 vector expressing the GILZ cDNA. After 36 hours, cells were cultured in medium containing G418 0.8 mg/g active form per milliliter (Gibco, Life Technologies, Paisley, Scotland), and 100 μL/mL of cell suspension was plated in 96-well plates (4 for each transfection). Following 15 to 20 days, no more than 15% of the wells presented alive growing cells. These cells were considered clones and analyzed in ribonuclease protection for the expression of exogenous GILZ.
Luciferase assay
Eukaryotic expression plasmids pMT2T-p65 and pMT2T-p52 were obtained from Dr Dalla Favera (Columbia University, NY). The pMT2T-GILZ was constructed by inserting the human GILZ cDNA coding sequences (405 base pairs) into pMT2T expression vector at the EcoRI site. (Human GILZ GenBank accession number AF228 339.) Reporter plasmids containing tandem repeats of the murine IgK/HIV-κB (pBIIXLUC) site linked upstream to a minimal murine c-fos promoter and luciferase coding sequences were obtained from Dr Dalla Favera. The pEGFP-N1 N-terminal protein fusion vector was purchased from Clontech (Palo Alto, CA). Calcium phosphate–mediated transient DNA transfection and luciferase assays were performed as described.45NTera-2 or 293 cells were plated at 2 × 106/100-mm Petri dish 24 hours prior to an 8-hour transfection period. At 48 hours after transfection, cells were harvested and transcription activation was assayed as luciferase activity. The amounts of effector plasmids reported in the figures were cotransfected with 15 μg reporter plasmid (pBIIXLUC) and 3 μg pEGFP-N1. The pEGFP-N1 plasmid was used to normalize the transfection efficiency of each sample.
Similar assays were performed using a luciferase reporter gene (pGL3–IL-2) containing an IL-2 promoter region (1.9 kilobases of 5′-flanking sequence of IL-2 gene) obtained from Dr L. D'Adamio (Albert Einstein College of Medicine, NY).46 Control vector pRc/RSV-luc containing the coding region for luciferase gene was obtained from Dr M. Cippitelli (University “La Sapienza,” Rome). Each transfection was performed in triplicate. Cell lysis and luciferase quantification were performed using commercial reagents (Luciferase Reporter Gene Assay, Roche Diagnostics, Monza, Italy). The values are expressed as fold increases above the level of luciferase activity of cells transfected with the reporter plasmids. The values marked EC (endogenous control) represent the values obtained by transfection of the reporter plasmids along with the control pMT2T vector.
Antibody cross-linking and cell treatment
Hamster antimouse CD3ε (clone 145-2C11; Pharmingen, San Diego, CA) monoclonal antibody (mAb) at 1 μg/mL was allowed to adhere in flat-bottomed, high-binding 96-well plates (Costar, Cambridge, MA) at 4°C in 100 μL phosphate-buffered saline (PBS). After 20 hours, plates coated with mAb were washed and transfected clones were plated at 1 × 105 cells per well and incubated at 37°C for different times as indicated. Isotype-matched rat antimouse immunoglobulin G2b (IgG2b) mAb (clone R 35-38, Pharmingen) was used as control.47 48
IL-2 assay
Supernatants from clones untreated or treated with anti-CD3 mAb for 18 hours were tested for IL-2 concentration by 2-site enzyme-linked immunosorbent assay (ELISA) using mAb JES6-1A12 as the primary reagent and biotinylated monoclonal S4B6 as the secondary reagent. Both antibodies were purchased from Pharmingen. The IL-2 titer (means ± SD of replicate samples) was expressed as pg/mL, calculated by reference to standard curves constructed with known amounts of IL-2. The sensitivity limit was approximately 20 pg/mL.
Flow cytometry evaluation of IL-2R expression
A single suspension (1 × 106cells/sample) was incubated for 30 minutes on ice in 50 μL staining buffer (PBS plus 5% fetal calf serum) containing 10 μg/mL hamster antimouse IL-2R mAb directly conjugated to R-phycoerytrin or phycoerytrin-hamster IgG (isotype control). Both mAbs were purchased from Pharmingen. The percentage values of IL-2R histograms were calculated using lysis II research software (Becton Dickinson, Mountain View, CA).
Apoptosis evaluation by propidium iodide solution
Apoptosis was measured by flow cytometry as described elsewhere.47 Briefly, cells were centrifuged and the pellets resuspended in 1.5 mL hypotonic propidium iodide (PI) solution. The tubes were kept at 4°C in the dark overnight. The PI fluorescence of individual nuclei was measured by flow cytometry with standard FACScan equipment (Becton Dickinson).
Nuclear extracts and electrophoresis mobility shift assay
Cells (2 × 107/group) were washed with ice-cold PBS, and packed cells were resuspended in 1 mL hypotonic buffer (25 mM HEPES, 50 mM KCl, 0.5% Nonidet P-40 [NP-40], 0.1 mM dithiothreitol, 10 mg/mL leupeptin, 20 mg/mL aprotinin, and 1 mM phenylmethylsulfonyl fluoride [PMSF] solution in ethanol). Ten minutes after incubation on ice, the supernatants, containing cytoplasmic proteins, were separated from the nuclear pellets by centrifugation. Nuclear pellets were then washed with hypotonic buffer without NP-40 and resuspended in 10 mL lysis buffer (25 mM HEPES, 2 mM KCl, 0.1 mM dithiothreitol, 10 mg/mL leupeptin, 20 mg/mL aprotinin, and 1 mM PMSF). Fifteen minutes after incubation on ice, lysates were diluted with 10 vol dilution buffer (25 mM HEPES, 0.1 mM dithiothreitol, 10 mg/mL leupeptin, 20 mg/mL aprotinin, and 1 mM PMSF and 20% glycerol) and cleared in a precooled microfuge for 30 minutes at 14 000g. All DNA binding reactions were conducted for 20 minutes at room temperature in a final volume of 20 μL. The reactions were started by adding 10 μg nuclear protein extract to a reaction mix containing 1 μg poly [d(I-C)], 4 μL 5 × binding buffer (50 mM Tris, 250 mM NaCl, 5 mM ethyleneglycotetraacetic acid [EDTA], 25% glycerol, and 5 mM dithiothreitol), and approximately 20 000 cpm of the respective (γ32P)ATP-labeled double-stranded DNA (dsDNA) oligonucleotide. Cold competitor oligonucleotides were added to the reaction mix before the radiolabeled probe. The sample was then loaded on 5% native polyacrylamide gel in Tris-borate-EDTA buffer. After electrophoresis for 2.5 hours at room temperature and 10 V/cm, gels were dried, and separated protein-DNA complexes were visualized by autoradiography using Kodak XAR5 films. The following dsDNA oligonucleotides (Promega, Milan, Italy) were used in electrophoresis mobility shift assay (EMSA) analysis both as labeled or competitor cold probe: 5′-AGAGGGGACTTTCCGAGAGGC-3′ for NF-κB, 5′-TATGTGTAATATGTAAAA-3′ for OCT-1, and 5′-AAGAGGAAAATTTGTTTCATACAG-3′ for NF–activated T cell (NF-AT). For antibody-induced supershift assays 2 μL (0.2 μg/mL) antibody, anti-p65 or anti-p52 (Santa Cruz Biotechnology, Santa Cruz, CA), was incubated with 10 μg nuclear extract for 30 minutes at room temperature after the addition of radiolabeled probe.
Production of recombinant proteins
Human GILZ was in-frame cloned into the pGEX-4T2 plasmid (Pharmacia, Uppsala, Sweden). The pGEX-4T2 plasmid is a glutathione-S-transferase (GST) fusion vector carrying a tac promoter for chemically (isopropyl-β-D-thiogalactopyranoside [IPTG]) inducible high-level expression of the protein. GST fusion protein was expressed in Escherichia coli, grown at 30° C, and induced with 0.1 mM IPTG for 90 to 180 minutes. Following lysis by sonication, most of the induced protein was found in the soluble material, which was purified with Glutathione Sepharose 4B beads (Pharmacia) following the manufacturer's instructions.
In vitro interaction studies
Human GILZ, p65, p52, c-fos, and Fra-1 proteins were in vitro–translated with [35S]methionine by using the rabbit reticulocyte-coupled in vitro transcription translation system (Promega, Madison, WI) under the T7 promoter according to the manufacturer's instruction. In vitro–translated proteins were diluted with the binding buffer (final concentration: 25 mM HEPES, pH 7.5; 10% glycerol; 50 mM NaCl; 0.05% NP-40; 1 mM dithiothreitol) and precleared with glutathione beads for 45 minutes at 4°C. The labeled products were assayed for interaction with GST-GILZ fusion protein or GST protein, prepared by glutathione-coated bead purification from appropriate bacterial sonicates as previously reported.49 50 GST or GST protein bound to glutathione beads were incubated with in vitro–translated proteins for 20 minutes at 20°C. The beads were subsequently washed 5 times with 0.5 mL PBS, and bound proteins were recovered by boiling in SDS sample buffer and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). For I-κB competition experiments, GST was removed from GST-GILZ fusion protein following the manufacturer's instructions (Pharmacia) and GILZ (1 μg) was incubated overnight with in vitro–translated p65 (5 μL) diluted in the binding buffer described above, in the absence or in the presence of GST–I-κBα fusion protein (100 μg, Santa Cruz). Immunoprecipitation was performed with antibody anti-p65 (3 μL, Biomol Research Laboratories, Plymouth Meeting, PA), and Western blotting was performed with antibody anti-p65 (1:1000) or anti-GILZ (1:1000) or anti–I-κBα (1:1000) (Cell Signaling Technology, Beverly, MA).
Immunoprecipitation and coimmunoprecipitation
NTera-2 cells were transiently transfected as reported for the luciferase assay. Transfected cells were incubated for 30 minutes in medium lacking methionine and then metabolically labeled for 4 hours with [35S]methionine and immediately harvested. Whole-cell extracts were prepared and immunoprecipitations performed in radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 1% NP-40; 0.5% deoxycholate; 0.1% SDS; and 5 mM EDTA) supplemented with 1 mM PMSF. Antigen-antibody complexes were precipitated with protein A bound to Sepharose beads (Pharmacia) prior to SDS-PAGE. A rabbit polyclonal antiserum recognizing GILZ was prepared with the use of a fusion protein containing the full GILZ amino acid sequence as previously reported.42
Protein lysates (500 μg in radioimmunoprecipitation assay buffer) from untreated or DEX-treated thymocytes were immunoprecipiteted with 3 μL anti-p65 subunit rabbit polyclonal antibody. For coimmunoprecipitation experiments, Western blot was performed using both anti-p65 and anti-GILZ antibodies.
Immunofluorescence staining for nuclear translocation
To evaluate the nuclear translocation of NF-κB or NF-AT, clones untreated or treated with anti-CD3 mAb for 2 hours were processed for immunofluorescence by the paraformaldehyde-saponin procedure.51 After extensive washing in PBS with 1% HEPES, cells were fixed in 4% formaldehyde for 20 minutes on ice, washed again, and incubated at 4°C for 1 hour with blocking buffer (PBS with 3% bovine serum albumin and 1% glycine). For staining, cells were incubated for 45 minutes at 4°C with 100 ng polyclonal rabbit anti-p65 antibodies (Santa Cruz) or 100 ng polyclonal rabbit anti–NF-AT1 (Upstate Biotechnology, Lake Placid, NY) in buffer containing 0.1% saponin, washed, and incubated for 45 minutes at 4°C with Texas Red–conjugated goat antirabbit IgG in PBS-saponin. Cells were then washed, stuck on slides coated with poly-L-lysine, and mounted in buffered glycerol for fluorescence microscopic analysis. Photographs were taken on a Leitz Dialux 20 microscope.
Northern blot assay
Total RNA was extracted by the guanidium thiocyanate method and separated in 1.2% agarose gels containing 2.2 M formaldehyde and transferred to nitrocellulose filters (Scheicher and Schuell, Dassel, Germany). Nitrocellulose filters with 25 μg RNA were hybridized overnight with mouse GILZ cDNA 32P-labeled using the nick translation kit (Roche Diagnostics). Filters were washed 3 times in 1 × SSC with 0.05% SDS at room temperature, followed by 2 washes at 50°C in 0.1 × SSC with 0.1% SDS; β-actin was used as control.
Western blot analysis
Extracted proteins were separated on an SDS-polyacrylamide gel and studied by Western blotting as previously described.48Primary antibody was a rabbit polyclonal antiserum recognizing GILZ,42 and the secondary antibody was a horseradish peroxidase–labeled goat antirabbit IgG (Pierce, Rockford, IL). Anti–β-tubulin mAb (Calbiochem, San Diego, CA) was used as control. The antigen-antibody complexes were revealed by enhanced chemiluminescence following the manufacturer's instructions (SuperSignal, Pierce). The PhotoPlus I-κBα (Ser32) antibody kit for analysis of I-κBα phosphorylation was purchased from Cell Signalling Technology.
Statistical analysis
Each experiment was performed at least 3 times. Representative experiments are shown. Due to the nonnormal distribution of the data, nonparametric tests (Kruskall-Wallis' analysis of variance) were adopted for statistical evaluation.
Results
GILZ inhibits apoptosis and IL-2 and IL-2R expression
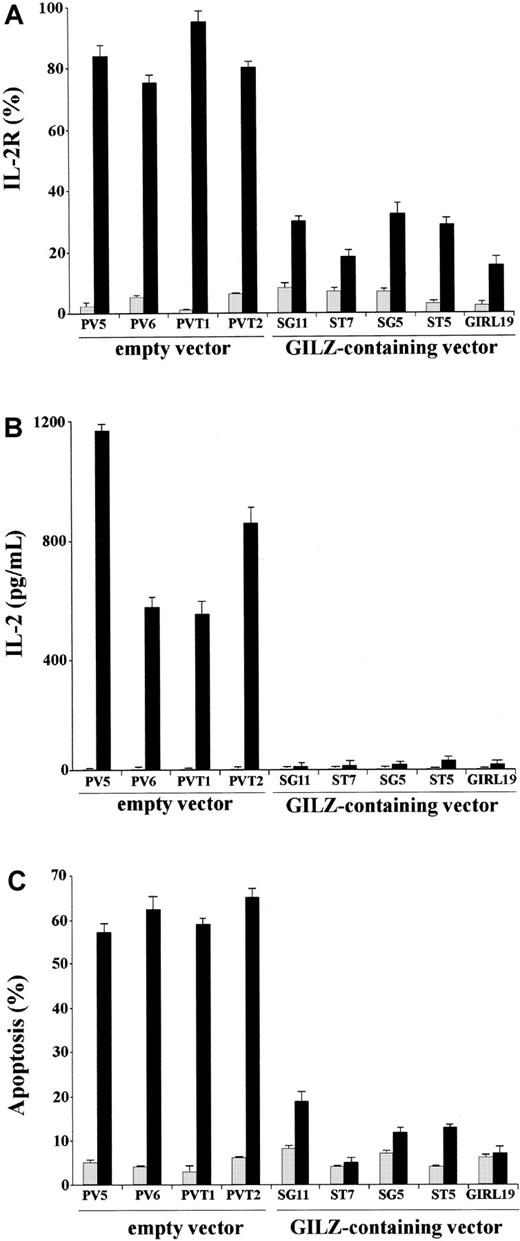
We performed experiments to evaluate whether GILZ could counter the TCR-induced increase of IL-2 and IL-2R. For that purpose we used empty vector– and GILZ-transfected clones nonstimulated or stimulated by treatment with anti-CD3 mAb. After stimulation with cross-linked anti-CD3 mAb, GILZ- and empty vector–transfected clones were stained with anti–IL-2R mAb or labeled for PI staining for flow cytometry apoptosis evaluation. Furthermore, the supernatants were used for IL-2 detection in an ELISA assay. Results show that, upon activation by anti-CD3 mAb, empty vector–transfected clones express high levels of IL-2R and produce IL-2 (Figure 1A-B). In contrast, anti-CD3–activated GILZ-transfected clones express low or undetectable IL-2 and IL-2R (Figure 1A-B). Moreover, GILZ-transfected clones are resistant to anti-CD3–activated apoptosis (Figure 1C). These results confirm previous data showing that GILZ inhibits TCR-induced apoptosis42 and further indicate that GILZ inhibits IL-2 and IL-2R expression.
GILZ inhibits anti-CD3–induced IL-2R expression, IL-2 production, and apoptosis.
(A) IL-2R expression in untreated (░) and anti-CD3–treated clones (▪) for 18 hours on plates coated with anti-CD3 (1 μg/mL), empty vector–transfected (PV5, PV6, PVT1, PVT2) clones, and GILZ-transfected (SG11, ST7, SG5, ST5, GIRL19) clones as evaluated by fluorescent anti–IL-2 mAb and flow cytometry analysis (see “Material and methods”). (B) IL-2 production (pg/mL) in untreated (░) and anti-CD3–treated (▪), empty vector–transfected (PV5, PV6, PVT1, PVT2), and GILZ-transfected (SG11, ST7, SG5, ST5, GIRL19) clones as evaluated by ELISA assay. (C) Percentage apoptosis in untreated (░) and anti-CD3–treated (▪), empty vector–transfected (PV5, PV6, PVT1, PVT2), and GILZ-transfected (SG11, ST7, SG5, ST5, GIRL19) clones as evaluated by PI assay.
GILZ inhibits anti-CD3–induced IL-2R expression, IL-2 production, and apoptosis.
(A) IL-2R expression in untreated (░) and anti-CD3–treated clones (▪) for 18 hours on plates coated with anti-CD3 (1 μg/mL), empty vector–transfected (PV5, PV6, PVT1, PVT2) clones, and GILZ-transfected (SG11, ST7, SG5, ST5, GIRL19) clones as evaluated by fluorescent anti–IL-2 mAb and flow cytometry analysis (see “Material and methods”). (B) IL-2 production (pg/mL) in untreated (░) and anti-CD3–treated (▪), empty vector–transfected (PV5, PV6, PVT1, PVT2), and GILZ-transfected (SG11, ST7, SG5, ST5, GIRL19) clones as evaluated by ELISA assay. (C) Percentage apoptosis in untreated (░) and anti-CD3–treated (▪), empty vector–transfected (PV5, PV6, PVT1, PVT2), and GILZ-transfected (SG11, ST7, SG5, ST5, GIRL19) clones as evaluated by PI assay.
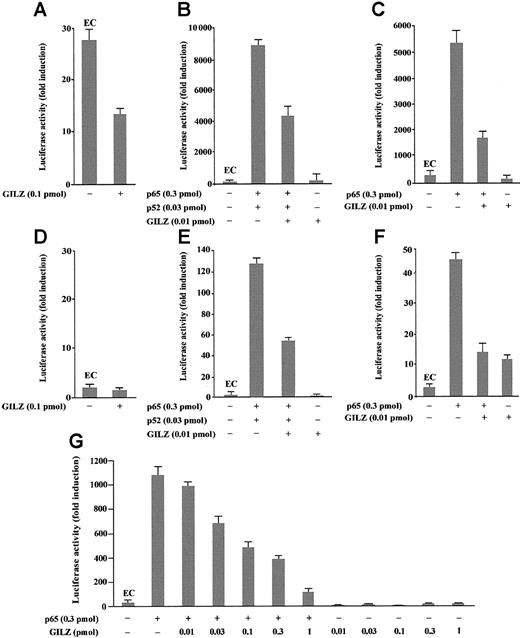
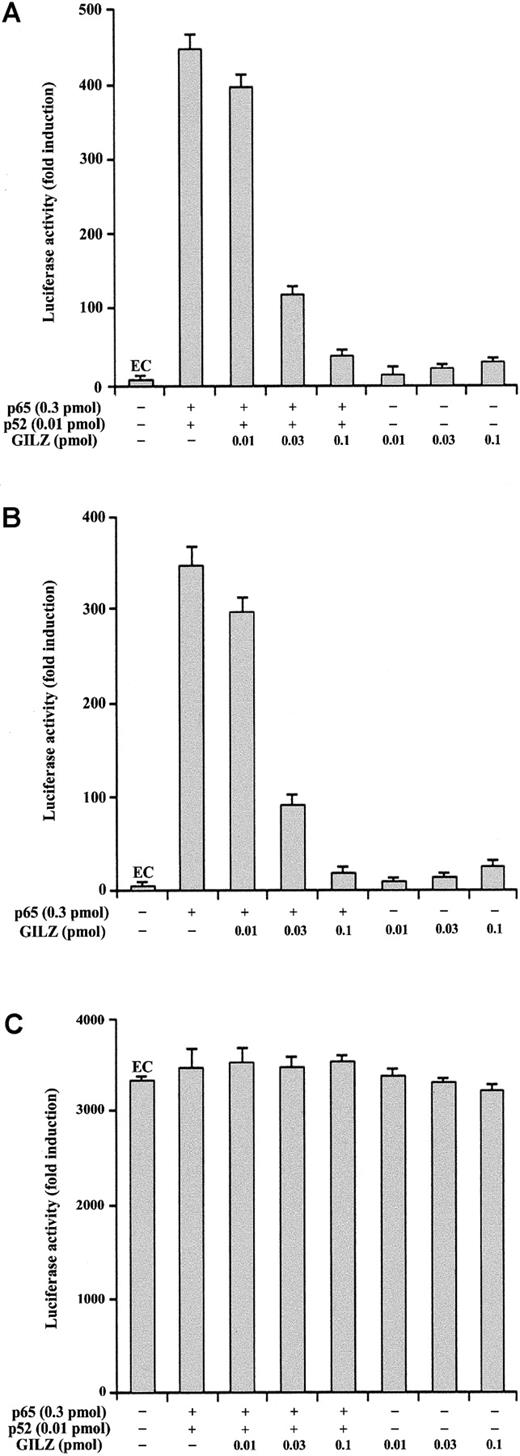
GILZ inhibits NF-κB transcription activity in vivo
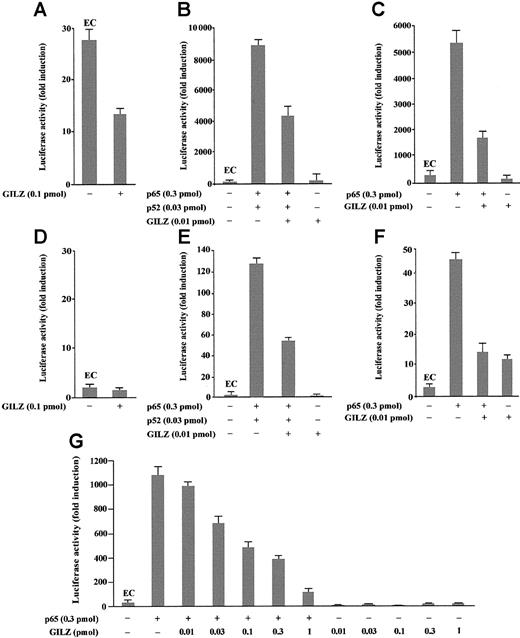
To investigate the role of GILZ in the regulation of NF-κB–driven transcription, we tested the ability of GILZ overexpression to modulate the in vivo transcription using a reporter gene linked to a κB sequence in transient transfection assay. In particular, eukaryotic expression plasmids pMT2T-p65, pMT2T-p52, and pMT2T-GILZ were cotransfected in 293 cells with a target plasmid in which an IgK/HIV-κB (pBIIXLUC) sequence was linked upstream of a minimal c-fos promoter and luciferase coding sequence. Luciferase activity was measured as a function of κB-dependent transcription at 48 hours after transfection as previously described.45 The values, reported in Figure2, are expressed as fold increases of luciferase activity. The value marked EC (endogenous control) represents the value obtained by transfection of the target plasmid along with the control pMT2T empty vector and documents the level of endogenous NF-κB activity in 293 cells (Figure 2A-C,G). In particular, Figure 2A indicates that 293 cells have an intrinsic NF-κB activity and that this transcriptional activity is inhibited by GILZ expression (P < .01). Moreover, Figure 2B indicates that p65 and p52 overexpression greatly enhances luciferase activity in transfected cells and shows that GILZ has the capability to negatively regulate NF-κB–driven transcription (P < .01, column 3 versus column 2) but does not have an intrinsic capability of transactivation when transfected alone (Figure 2B, column 4). This negative effect is dose dependent and augments progressively with GILZ transfectant concentrations ranging from 0.01 pmol to 1 pmol (Figure2G). Furthermore, GILZ has the capability to negatively regulate κB-driven transcription mediated by p65 (Figure 2C).
GILZ inhibits p65/p52-dependent transactivation of a IgG/HIV-κB (pBIIXLUC) site-driven luciferase reporter.
Analysis of transcriptional activity was performed on transiently transfected 293 (A-C, G) or NTera-2 (D-F) cells. The indicated amounts of PMT2T-GILZ (0.1 pmol = 370 ng), PMT2T-p65 (0.3 pmol = 1.4 μg), and PMT2T-p52 (0.01 pmol = 47 ng), alone or in combination, were cotransfected with 15 μg (4 pmol) of the target plasmid pBIIXLUC. The values are expressed as fold increases of luciferase activity. Each transfection was performed in triplicate, and SD bars are shown. EC indicates endogenous control.
GILZ inhibits p65/p52-dependent transactivation of a IgG/HIV-κB (pBIIXLUC) site-driven luciferase reporter.
Analysis of transcriptional activity was performed on transiently transfected 293 (A-C, G) or NTera-2 (D-F) cells. The indicated amounts of PMT2T-GILZ (0.1 pmol = 370 ng), PMT2T-p65 (0.3 pmol = 1.4 μg), and PMT2T-p52 (0.01 pmol = 47 ng), alone or in combination, were cotransfected with 15 μg (4 pmol) of the target plasmid pBIIXLUC. The values are expressed as fold increases of luciferase activity. Each transfection was performed in triplicate, and SD bars are shown. EC indicates endogenous control.
Because undifferentiated NTera-2 cells have no detectable endogenous NF-κB activity, they are ideal for testing functions of exogenously introduced Rel-related or I-κB–related proteins.48 49To investigate whether the inhibitory effect of GILZ on NF-κB transcription activity could be evidenced also in these cells, NTera-2 cells were transiently cotransfected with different plasmid combinations. As expected, low or undetectable luciferase activity was measured in controls or GILZ-transfected NTera-2 cells (Figure 2D). However, measurable luciferase activity was detected in cells transfected with p65 and p52 (Figure 2E) or with p65 alone (Figure 2F) and, similar to the results obtained with 293 cells, GILZ inhibits this transcription activity (P < .01; Figure 2F, column 3 versus column 2). In conclusion, these results indicate that GILZ can inhibit NF-κB activity in vivo and that this effect is independent from other Rel-related or I-κB–related proteins.
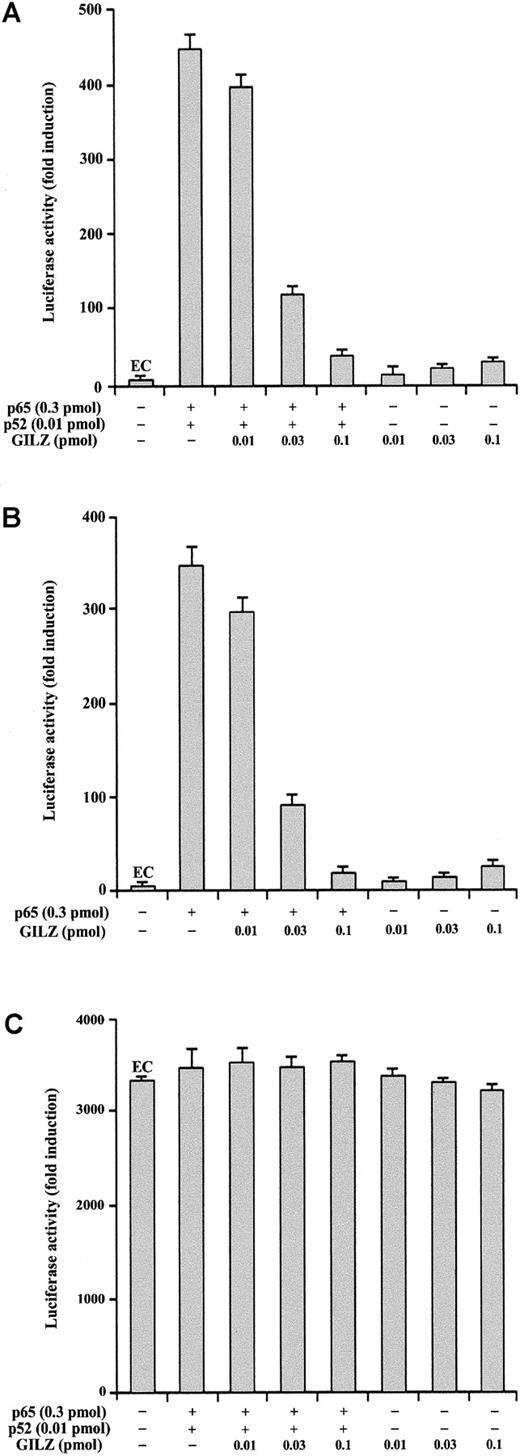
We also performed experiments to analyze whether GILZ could modulate the NF-κB–driven transcriptional activity using a luciferase reporter plasmid under the control of an IL-2 promoter region (pGL3–IL-2). As shown in Figure 3, GILZ inhibits NF-κB–dependent transcription (P < .01; column 4 versus column 2). In particular, Figure 3A indicates that GILZ negatively regulates p65- and p52-driven transcriptional activity but does not have an intrinsic capability of transactivation when transfected alone. This inhibiting effect is dose dependent and augments progressively with GILZ transfectant concentrations ranging from 0.01 to 0.1 pmol (Figure 3A). Furthermore, GILZ has the capability to negatively regulate κB-driven transcription mediated by p65 alone (Figure 3B). No effects were observed using a NF-κB–independent reporter as control (Figure 3C). These results indicate that GILZ is able to counter the NF-κB activity in an IL-2 promoter-driven luciferase activity in vivo.
GILZ inhibits p65/p52-dependent transactivation of a pGL3–IL-2 reporter vector.
Analysis of transcriptional activity was performed on 293 cells transiently transfected with a luciferase reporter gene (pGL3–IL-2) containing an IL-2 promoter region. The indicated amounts of PMT2T-GILZ, PMT2T-p65, and PMT2T-p52, alone or in combination, were cotransfected with 15 μg of the IL-2–driven luciferase plasmid (A-B) or control vector pRc/RSV-luc (C). The values are expressed as fold increases of luciferase activity. Each transfection was performed in triplicate, and SD bars are shown. EC indicates endogenous control.
GILZ inhibits p65/p52-dependent transactivation of a pGL3–IL-2 reporter vector.
Analysis of transcriptional activity was performed on 293 cells transiently transfected with a luciferase reporter gene (pGL3–IL-2) containing an IL-2 promoter region. The indicated amounts of PMT2T-GILZ, PMT2T-p65, and PMT2T-p52, alone or in combination, were cotransfected with 15 μg of the IL-2–driven luciferase plasmid (A-B) or control vector pRc/RSV-luc (C). The values are expressed as fold increases of luciferase activity. Each transfection was performed in triplicate, and SD bars are shown. EC indicates endogenous control.
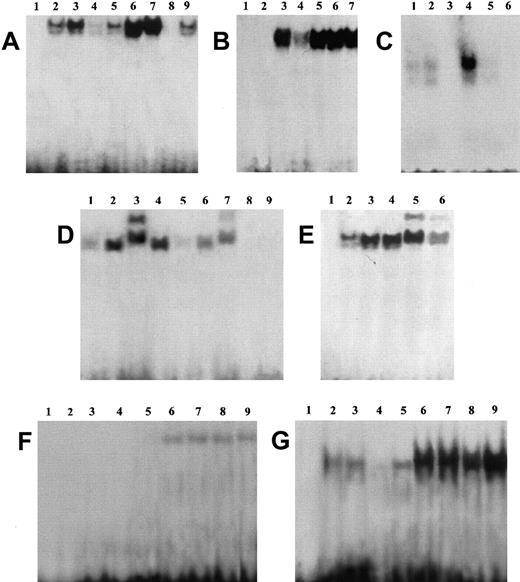
GILZ prevents DNA binding of NF-κB
NF-κB has been shown to be involved in T-cell activation, induction of IL-2 and IL-2R, Fas and FasL gene expression, and apoptosis.19,22,52 53 Because GILZ inhibits IL-2 and IL-2R expression and NF-κB transcriptional activity, we performed experiments to analyze whether GILZ could also interact with the NF-κB–DNA binding.
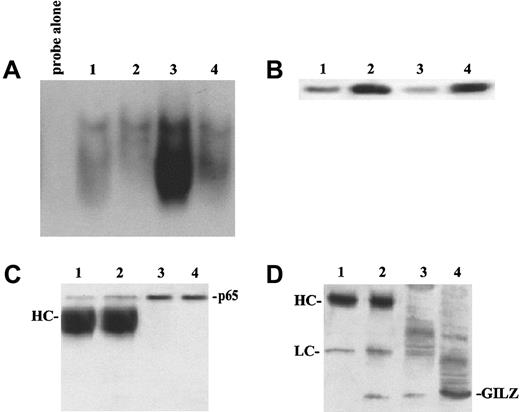
EMSA performed using a specific sequence containing an NF-κB binding site and nuclear extracts prepared from untreated and anti-CD3–treated clones, transfected with GILZ or empty vector, reveals an inhibition of NF-κB–DNA binding activities in clones overexpressing GILZ (Figure4). Empty vector–transfected T-cell clone (PV6) and the parental 3DO cell line display activation of NF-κB upon anti-CD3 stimulation (Figure 4A, lanes 6 and 7 versus lanes 2 and 3). In contrast, anti-CD3–treated clones overexpressing GILZ (ST7 and GIRL19) failed to induce NF-κB activation (Figure 4A, lanes 8 and 9 versus lanes 4 and 5). As control, the presence of a specific competitor (cold NF-κB) prevented complex formation (Figure4C, lane 3 versus lane 4). In addition, GST-GILZ fusion protein was also able to inhibit NF-κB binding when added to nuclear extracts from anti-CD3–activated 3DO cells (Figure 4B, lane 4 versus lane 3). As control, GST-p56lck fusion protein does not interfere with NF-κB binding (Figure 4B, lane 6). However, GST-GILZ alone did not cause any shift effect, indicating that GILZ-mediated inhibition could not be due to competitive binding to the dsDNA NF-κB–specific probe (Figure 4B, lane 2). As further control, no shift inhibition could be detected with GST alone (Figure 4B, lane 7) or the buffer, used for GST-GILZ preparation, alone (Figure 4B, lane 5). We also performed supershift experiments, and the results indicate that pretreatment with antisera specific for p65 (Figure 4D, lane 3) or p52 (Figure 4E, lane 5) supershifted the complex, suggesting that the complex contains heterodimers of p65 and p52. Supershifted binding activity was inhibited in GILZ overexpressing clones (Figure 4D, lane 7 versus lane 3, and Figure 4E, lane 6 versus lane 5). No detectable differences were observed in the pattern of NF-AT and OCT-1 transactivation between empty vector– and GILZ-transfected clones (Figure 4F-G). In conclusion, these results indicate that GILZ inhibits NF-κB–DNA binding activity.
GILZ inhibits NF-κB binding to its DNA motif in an EMSA assay.
(A) EMSA assay performed using nuclear extract from untreated (lanes 2-5) and anti-CD3–treated (2 hours, lanes 6-9) 3DO cells (lanes 3, 7) and empty vector– (PV6, lanes 2, 6) or GILZ-transfected (ST7, lanes 4, 8; GIRL19, lanes 5, 9) clones. Lane 1: probe alone. (B) Nuclear extract from anti-CD3–treated 3DO cells, alone (lane 3) or added with GST-GILZ fusion protein (lane 4), or buffer used for GST-GILZ preparation alone (lane 5), or GST-P56lck fusion protein (lane 6), or GST alone (lane 7) as controls. Lane 1: probe alone. Lane 2: GILZ-GST alone. (C) Nuclear extract from untreated (lanes 1, 2) or anti-CD3–stimulated (lanes 3-5), empty-vector– (PV6, lanes 1, 3, 4), or GILZ-transfected (ST7, lanes 2, 5) cells. Lane 3: control with nuclear extract from anti-CD3–treated empty vector–transfected cells plus competitor cold probe. Lane 6: labeled probe alone. (D) Nuclear extract of untreated (lanes 1, 5) or anti-CD3–treated (lanes 2-4, 6, 7) PV6 cells. Nuclear extract plus anti-p65 antibody (lane 3) or control antibody (lane 4). Nuclear extract plus GILZ-GST fusion protein (lanes 5, 6). Nuclear extract plus anti-p65 and GILZ-GST fusion protein (lane 7). Lane 8: GILZ-GST fusion protein alone. Lane 9: probe alone. (E) Nuclear extract of untreated (lane 2) or anti-CD3–treated PV6 (lanes 3-6) cells. Nuclear extract plus anti-p52 antibody (lanes 5, 6) or control antibody (lane 4). Nuclear extract plus anti-p52 and GILZ-GST fusion protein (lane 6). Lane 1: probe alone. (F,G) EMSA assay performed using as probe NF-AT and OCT-1, respectively. Nuclear extracts from untreated (lanes 2-5) or anti-CD3–treated (lanes 6-9) 3DO,3 7 PV6 (lanes 2, 6), ST7 (lanes 4, 8), and GIRL19 (lanes 5, 9). Lane 1: probe alone.
GILZ inhibits NF-κB binding to its DNA motif in an EMSA assay.
(A) EMSA assay performed using nuclear extract from untreated (lanes 2-5) and anti-CD3–treated (2 hours, lanes 6-9) 3DO cells (lanes 3, 7) and empty vector– (PV6, lanes 2, 6) or GILZ-transfected (ST7, lanes 4, 8; GIRL19, lanes 5, 9) clones. Lane 1: probe alone. (B) Nuclear extract from anti-CD3–treated 3DO cells, alone (lane 3) or added with GST-GILZ fusion protein (lane 4), or buffer used for GST-GILZ preparation alone (lane 5), or GST-P56lck fusion protein (lane 6), or GST alone (lane 7) as controls. Lane 1: probe alone. Lane 2: GILZ-GST alone. (C) Nuclear extract from untreated (lanes 1, 2) or anti-CD3–stimulated (lanes 3-5), empty-vector– (PV6, lanes 1, 3, 4), or GILZ-transfected (ST7, lanes 2, 5) cells. Lane 3: control with nuclear extract from anti-CD3–treated empty vector–transfected cells plus competitor cold probe. Lane 6: labeled probe alone. (D) Nuclear extract of untreated (lanes 1, 5) or anti-CD3–treated (lanes 2-4, 6, 7) PV6 cells. Nuclear extract plus anti-p65 antibody (lane 3) or control antibody (lane 4). Nuclear extract plus GILZ-GST fusion protein (lanes 5, 6). Nuclear extract plus anti-p65 and GILZ-GST fusion protein (lane 7). Lane 8: GILZ-GST fusion protein alone. Lane 9: probe alone. (E) Nuclear extract of untreated (lane 2) or anti-CD3–treated PV6 (lanes 3-6) cells. Nuclear extract plus anti-p52 antibody (lanes 5, 6) or control antibody (lane 4). Nuclear extract plus anti-p52 and GILZ-GST fusion protein (lane 6). Lane 1: probe alone. (F,G) EMSA assay performed using as probe NF-AT and OCT-1, respectively. Nuclear extracts from untreated (lanes 2-5) or anti-CD3–treated (lanes 6-9) 3DO,3 7 PV6 (lanes 2, 6), ST7 (lanes 4, 8), and GIRL19 (lanes 5, 9). Lane 1: probe alone.
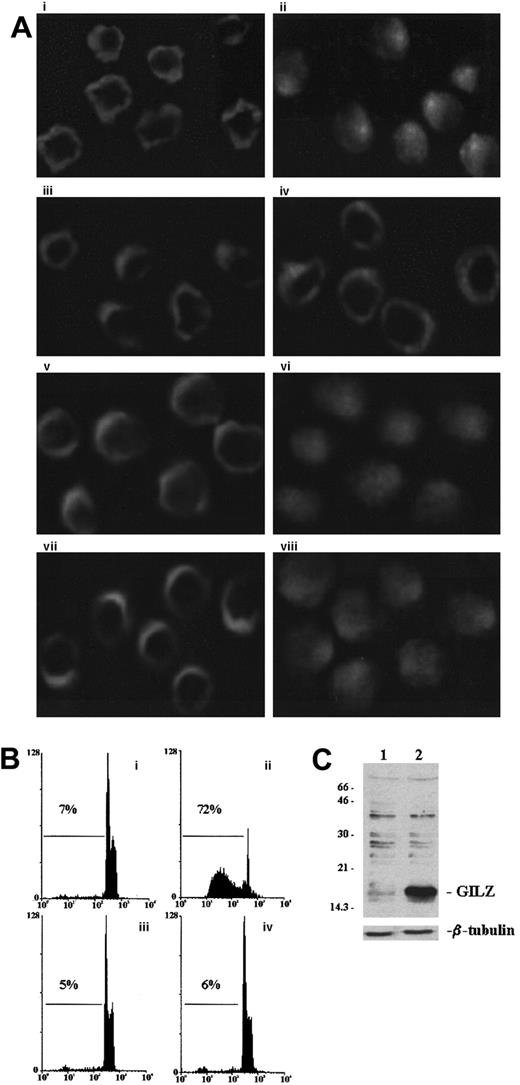
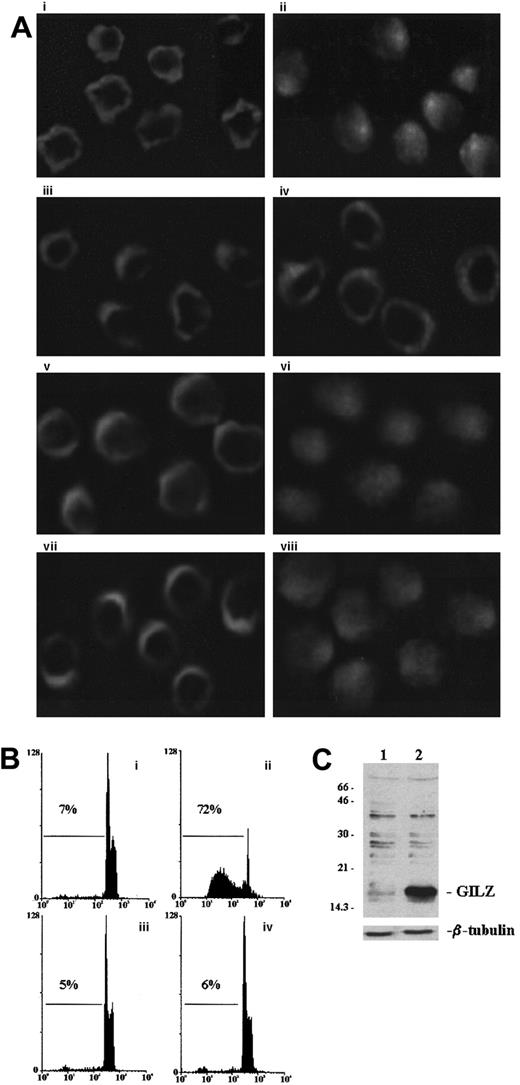
GILZ prevents NF-κB nuclear translocation
We performed experiments to analyze whether GILZ overexpression could modulate the anti-CD3–induced NF-κB nuclear translocation. For that reason empty vector–transfected and GILZ-transfected cells, untreated or treated with anti-CD3 mAb, were stained and processed for immunofluorescence analysis using anti–NF-κB antibodies. Figure5 indicates that anti-CD3 treatment induces NF-κB translocation in empty vector–transfected cells (Figure 5Aii) as compared with untreated control (Figure 5Ai) and that GILZ overexpression inhibits the anti-CD3–induced translocation (Figure 5Aiv versus 5Aiii). In contrast, NF-AT nuclear translocation occurs in both empty vector–transfected (Figure 5Av-Avi) and GILZ-transfected (Figure 5Avii-Aviii) clones. As control, the levels of apoptosis (Figure 5B) and GILZ expression (Figure 5C) were also determined.
GILZ inhibits nuclear traslocation of NF-κB.
(A) Immunofluorescence analysis of empty vector–transfected (PV6; i, ii, v, vi) or GILZ-transfected (GIRL19; iii, iv, vii, viii) clones. Cells untreated (i, iii, v, vii) or treated for 2 hours on plates coated with anti-CD3 (ii, iv, vi, viii) were stained with anti-p65 antibody (i, ii, iii, iv) or anti–NF-AT antibody (v, vi, vii, viii) by paraformaldehyde-saponin procedure. (B) Cytofluorimetric analysis of TCR-induced apoptosis in PV6 (i, untreated; ii, treated on plates coated with anti-CD3) and GIRL19 (iii, untreated; iv, treated with anti-CD3) clones as evaluated at 18 hours by PI assay. (C) Western blot analysis of GILZ protein in PV6 (lane 1) and GIRL19 (lane 2) clones. Original magnification: × 900.
GILZ inhibits nuclear traslocation of NF-κB.
(A) Immunofluorescence analysis of empty vector–transfected (PV6; i, ii, v, vi) or GILZ-transfected (GIRL19; iii, iv, vii, viii) clones. Cells untreated (i, iii, v, vii) or treated for 2 hours on plates coated with anti-CD3 (ii, iv, vi, viii) were stained with anti-p65 antibody (i, ii, iii, iv) or anti–NF-AT antibody (v, vi, vii, viii) by paraformaldehyde-saponin procedure. (B) Cytofluorimetric analysis of TCR-induced apoptosis in PV6 (i, untreated; ii, treated on plates coated with anti-CD3) and GIRL19 (iii, untreated; iv, treated with anti-CD3) clones as evaluated at 18 hours by PI assay. (C) Western blot analysis of GILZ protein in PV6 (lane 1) and GIRL19 (lane 2) clones. Original magnification: × 900.
These results indicate that GILZ inhibits anti-CD3–induced NF-κB nuclear translocation and further confirms previously reported observations42 that induction of GILZ expression contributes to the modulation of TCR-mediated T-cell activation and apoptosis.
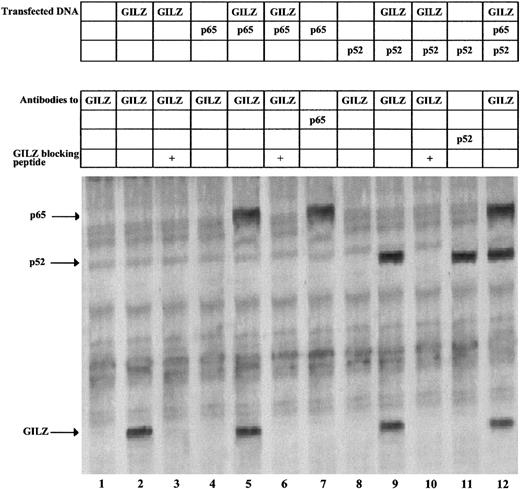
GILZ coimmunoprecipitates with p65 and p52
To investigate the possible in vivo protein-protein interaction between GILZ and NF-κB, we performed coimmunoprecipitation experiments in GILZ and p65/p52 cotransfected cells. For that purpose, NTera-2 cells, which lack the NF-κB complex including I-κB molecules, were cotransfected with expression vectors encoding GILZ, p65, or p52 and labeled with [35S]methionine. As shown in Figure 6, untransfected NTera-2 cells contain no detectable GILZ protein (lane 1), whereas cells transfected with GILZ clearly show the band of GILZ protein (lane 2). As control, the immunoprecipitation is blocked by addition of the fusion protein previously used to generate the GILZ antibody (lane 3). Furthermore, anti-GILZ antibodies coimmunoprecipitate GILZ and p65 (lane 5) or GILZ and p52 (lane 9), and this again is blocked by inclusion of the GILZ peptide (lanes 6 and 10). The identity of the coimmunoprecipitated p65 and p52 in lanes 5 and 9 is confirmed by its comigration with the p65 and p52 proteins immunoprecipitated with anti-p65 and anti-p52 antibodies, respectively (lanes 7 and 11), and by immunoprecipitation from unlabeled extracts followed by Western blotting with p65 and p52 antibodies (data not shown). Finally, immunoprecipitation with anti-GILZ antibodies of extracts from cells cotransfected with GILZ, p65, and p52 results in the detection of GILZ, p65, and p52 (lane 12).
GILZ coimmunoprecipitates with p65 and p52.
NTera-2 cells were transfected with 6 μg of expression plasmids for GILZ, p65, and p52 either alone or in combination. Following transfection, the cells were metabolically labeled and whole-cell extracts used for immunoprecipitations with anti-GILZ, anti-p65, and anti-p52 antibodies, as indicated. Anti-GILZ immunoprecipitations were blocked with 400 ng of the GILZ fusion protein used to generate the antibody. The arrows indicate the immunoprecipitated proteins.
GILZ coimmunoprecipitates with p65 and p52.
NTera-2 cells were transfected with 6 μg of expression plasmids for GILZ, p65, and p52 either alone or in combination. Following transfection, the cells were metabolically labeled and whole-cell extracts used for immunoprecipitations with anti-GILZ, anti-p65, and anti-p52 antibodies, as indicated. Anti-GILZ immunoprecipitations were blocked with 400 ng of the GILZ fusion protein used to generate the antibody. The arrows indicate the immunoprecipitated proteins.
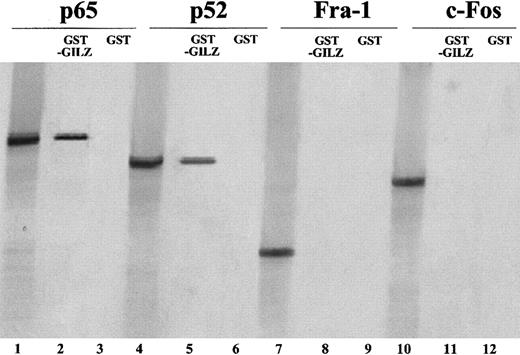
GILZ binds p65 and p52 in vitro
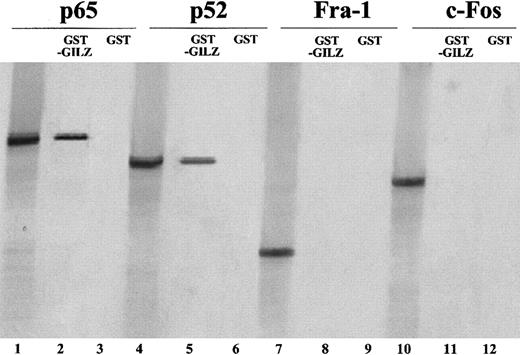
To further investigate if GILZ can physically associate with p65 and p52, partially purified GST-GILZ fusion protein was mixed with radiolabeled p65 or p52 and precipitated with glutathione-coated beads. Figure 7 shows that GST-GILZ fusion protein binds p65 (lane 2) and p52 (lane 5), whereas the GST protein alone (lanes 3 and 6) does not. As control, the in vitro–transcribed p65 (lane 1) and p52 (lane 4) were loaded. Furthermore, to analyze the binding specificity, we also performed similar experiments using other members of the leucine zipper family. In vitro–translated c-fos and Fra-1 proteins were incubated with GST-GILZ. Results indicate that neither Fra-1 (lane 8) nor c-fos (lane 11) proteins bind GILZ. As control, in vitro–translated Fra-1 (lane 7) and c-fos (lane 10) were loaded. These results indicate that there is a direct GILZ–NF-κB interaction.
GILZ physically associate in vitro with p65 and p52.
GST-GILZ fusion protein attached to glutathione Sepharose beads was incubated with 35S-labeled in vitro–transcribed p65, p52, c-fos, and Fra-1. Lanes 1, 4, 7, 10: in vitro–translated proteins (2 μL); lanes 2, 5, 8, and 11: in vitro–translated proteins (5 μL) precipitated by the GST-GILZ fusion protein attached to glutathione beads; lanes 3, 6, 9, 12: in vitro–translated proteins after precipitation with GST protein attached to glutathione beads.
GILZ physically associate in vitro with p65 and p52.
GST-GILZ fusion protein attached to glutathione Sepharose beads was incubated with 35S-labeled in vitro–transcribed p65, p52, c-fos, and Fra-1. Lanes 1, 4, 7, 10: in vitro–translated proteins (2 μL); lanes 2, 5, 8, and 11: in vitro–translated proteins (5 μL) precipitated by the GST-GILZ fusion protein attached to glutathione beads; lanes 3, 6, 9, 12: in vitro–translated proteins after precipitation with GST protein attached to glutathione beads.
GILZ, up-regulated by DEX treatment of thymocytes, coimmunoprecipitates with NF-κB/p65
To address whether GILZ overexpression in transfected clones could mimic some of the biological effects induced by DEX treatment, including NF-κB transactivation, we performed EMSA analysis using nuclei from DEX-treated normal thymocytes. Figure8 shows that an increased GILZ expression, induced by DEX treatment (Figure 8B), correlates with the inhibition of anti-CD3–driven NF-κB activation (Figure 8A). In the attempt to analyze if this effect could be due to a direct GILZ–NF-κB interaction we also performed, in the same experiment, coimmunoprecipitation assay. Figure 8 indicates that anti-p65 antibody immunoprecipitated p65 from untreated and DEX-treated thymocytes (Figure 8C, lanes 1 and 2) and coimmunoprecipitated GILZ from DEX-treated but not from untreated thymocytes (Figure 8D, lane 2 versus lane 1). These data suggest a good correlation between DEX-induced GILZ overexpression, NF-κB inhibition, and GILZ–NF-κB interaction.
Glucocorticoid treatment of thymocytes blocks NF-κB DNA binding, up-regulates GILZ expression, and induces its association with p65.
(A) EMSA analysis of NF-κB binding factors from untreated thymocytes (lane 1), or thymocytes cultured for 4 hours with 100 nM DEX (lane 2), or anti-CD3 mAb 1 μg/mL (lane 3), or the combination of anti-CD3 and DEX (lane 4). (B) Western blot analysis of GILZ. Lane 1: untreated thymocytes; lane 2: thymocytes treated with 100 nM DEX; lane 3: thymocytes treated with 1 μg/mL anti-CD3 mAb; lane 4: thymocytes treated with DEX and anti-CD3 mAb. (C,D) Immunoprecipitation of p65 and coimmunoprecipitation of GILZ. Mouse thymocytes were cultured in the presence or absence of DEX; cellular protein was collected 4 hours after stimulation and immunoprecipitated with anti-p65 antiserum (3 μL). Immunoblotting with anti-p65 antiserum (C) or anti-GILZ (D) was performed as described in the text. Lane 1: immunoprecipitation of untreated thymocytes; lane 2: immunoprecipitation of DEX-treated thymocytes; lane 3: total lysate from untreated thymocytes; lane 4: total lysates from DEX-treated thymocytes.
Glucocorticoid treatment of thymocytes blocks NF-κB DNA binding, up-regulates GILZ expression, and induces its association with p65.
(A) EMSA analysis of NF-κB binding factors from untreated thymocytes (lane 1), or thymocytes cultured for 4 hours with 100 nM DEX (lane 2), or anti-CD3 mAb 1 μg/mL (lane 3), or the combination of anti-CD3 and DEX (lane 4). (B) Western blot analysis of GILZ. Lane 1: untreated thymocytes; lane 2: thymocytes treated with 100 nM DEX; lane 3: thymocytes treated with 1 μg/mL anti-CD3 mAb; lane 4: thymocytes treated with DEX and anti-CD3 mAb. (C,D) Immunoprecipitation of p65 and coimmunoprecipitation of GILZ. Mouse thymocytes were cultured in the presence or absence of DEX; cellular protein was collected 4 hours after stimulation and immunoprecipitated with anti-p65 antiserum (3 μL). Immunoblotting with anti-p65 antiserum (C) or anti-GILZ (D) was performed as described in the text. Lane 1: immunoprecipitation of untreated thymocytes; lane 2: immunoprecipitation of DEX-treated thymocytes; lane 3: total lysate from untreated thymocytes; lane 4: total lysates from DEX-treated thymocytes.
GILZ does not interfere with I-κB
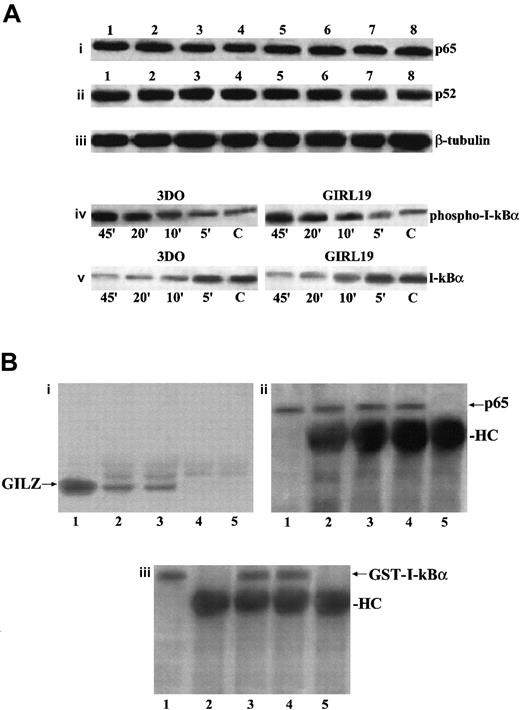
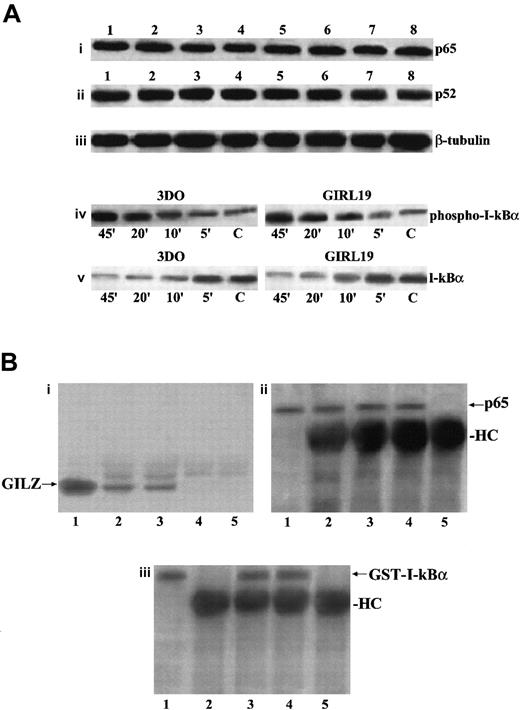
We evaluated if GILZ overexpression could down-regulate p65 and p52 expression or could counter the anti-CD3–induced down-modulation of I-κB expression. Results indicate that there were no differences in p65 or p52 expression between anti-CD3–treated empty vector– and GILZ-transfected clones either at the protein level, as evaluated by Western blotting (Figure9Ai-Aii), or at the transcription level, as evaluated by Northern blotting (data not shown).
GILZ does not affect p65 and P52 expression, I-κBα phosphorylation and degradation, or I-κB/NF-κB binding.
(A) Western blot analysis performed using protein lysates from untreated (lanes 1-4) or anti-CD3–treated (2 hours, lanes 5-8) 3DO cells (lanes 1, 5), empty vector–transfected clone PV6 (lanes 2, 6), GILZ-transfected clones ST7 (lanes 3, 7), and GIRL19 (lanes 4, 8). The nitrocellulose membrane was immunoblotted with anti-p65 antibody (i), or with anti-p52 antibody (ii), or with anti-β–tubuline mAb used as a control (iii). Western blot analysis of protein lysates from 3DO or GIRL19 treated with anti-CD3 mAb for the times indicated (C: untreated cells). The immunoblotting was performed using an mAb that recognizes the phosphorylated form of I-κBα (iv), or with a control antibody anti–I-κBα, phosphorylation-state independent (v). (B) Coimmunoprecipitation of p65/GILZ and p65/GST–I-κBα. In vitro–translated p65 (5 μL) was incubated overnight with GILZ (1 μg) or GILZ and GST–I-κBα (100 μg) and immunoprecipitated with anti-p65 antibody (3 μL). Immunoblotting was performed with antibody anti-GILZ (i, lane 1: nonimmunoprecipitated GILZ control; lane 2: p65 plus GILZ; lane 3: p65 plus GILZ plus GST–I-κBα; lane 4: p65 alone; lane 5: GILZ alone) or antibody anti-p65 (ii, lane 1: nonimmunoprecipitated p65 control; lane 2: p65 plus GILZ; lane 3: p65 plus GILZ plus GST–I-κBα; lane 4: p65 alone; lane 5: GILZ alone) or antibody anti–I-κBα (iii, lane 1: nonimmunoprecipitated GST–I-κBα control; lane 2: p65 plus GILZ; lane 3: p65 plus GILZ plus GST–I-κBα; lane 4: p65 plus GST–I-κBα; lane 5: GILZ plus GST-I-κBα).
GILZ does not affect p65 and P52 expression, I-κBα phosphorylation and degradation, or I-κB/NF-κB binding.
(A) Western blot analysis performed using protein lysates from untreated (lanes 1-4) or anti-CD3–treated (2 hours, lanes 5-8) 3DO cells (lanes 1, 5), empty vector–transfected clone PV6 (lanes 2, 6), GILZ-transfected clones ST7 (lanes 3, 7), and GIRL19 (lanes 4, 8). The nitrocellulose membrane was immunoblotted with anti-p65 antibody (i), or with anti-p52 antibody (ii), or with anti-β–tubuline mAb used as a control (iii). Western blot analysis of protein lysates from 3DO or GIRL19 treated with anti-CD3 mAb for the times indicated (C: untreated cells). The immunoblotting was performed using an mAb that recognizes the phosphorylated form of I-κBα (iv), or with a control antibody anti–I-κBα, phosphorylation-state independent (v). (B) Coimmunoprecipitation of p65/GILZ and p65/GST–I-κBα. In vitro–translated p65 (5 μL) was incubated overnight with GILZ (1 μg) or GILZ and GST–I-κBα (100 μg) and immunoprecipitated with anti-p65 antibody (3 μL). Immunoblotting was performed with antibody anti-GILZ (i, lane 1: nonimmunoprecipitated GILZ control; lane 2: p65 plus GILZ; lane 3: p65 plus GILZ plus GST–I-κBα; lane 4: p65 alone; lane 5: GILZ alone) or antibody anti-p65 (ii, lane 1: nonimmunoprecipitated p65 control; lane 2: p65 plus GILZ; lane 3: p65 plus GILZ plus GST–I-κBα; lane 4: p65 alone; lane 5: GILZ alone) or antibody anti–I-κBα (iii, lane 1: nonimmunoprecipitated GST–I-κBα control; lane 2: p65 plus GILZ; lane 3: p65 plus GILZ plus GST–I-κBα; lane 4: p65 plus GST–I-κBα; lane 5: GILZ plus GST-I-κBα).
Furthermore, because phosphorylation of I-κBα is essential for its degradation and for the release of active NF-κB,38 we stimulated with anti-CD3 mAb, for different times, empty vector– and GILZ–transfected clones and performed Western blotting using an antibody recognizing the phosphorylated I-κBα form and an antibody recognizing both the phosphorylated and nonphopsphorylated forms. Figure 9Aiv-Av shows that phosphorylation and degradation occur either in empty vector– or in GILZ–transfected clones following the same kinetic. We also performed experiments to test whether GILZ and I-κB could compete for binding to p65. Results show that antibody anti-p65 coimmunoprecipitates GILZ, either in the absence or in presence of GST–I-κBα used at a concentration 100 times higher, thus indicating that I-κBα does not prevent the formation of p65/GILZ complex (Figure 9Bi, lanes 2 and 3), although it binds p65 in the presence (Figure 9Biii, lane 3) or in absence (Figure 9Biii, lane 4) of GILZ.
TCR triggering inhibits GILZ expression
We have previously shown that GILZ is normally expressed in freshly isolated thymocytes, spleen, and lymph node cells and that its expression is increased by treatment with DEX.42 Moreover, GILZ overexpression counters the TCR/CD3-activated Fas/FasL up-regulation and AICD,42 suggesting that GILZ could contribute to the DEX-induced inhibition of T-cell activation, Fas/FasL up-regulation, and death.
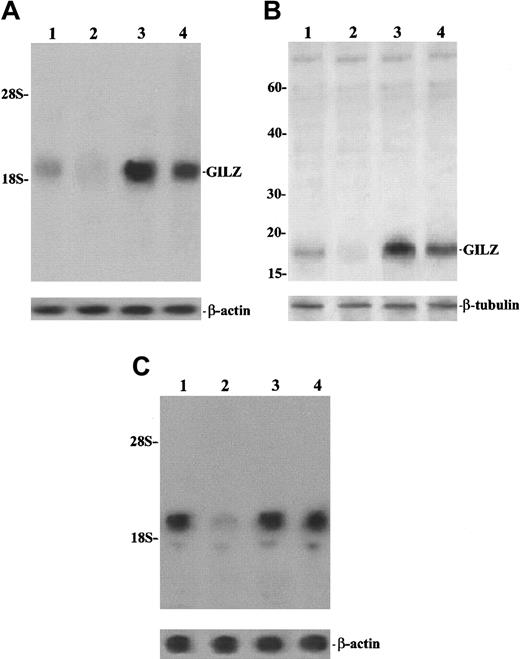
In an attempt to determine the possible physiologic role of GILZ, we performed experiments to analyze the effect of TCR/CD3 triggering on GILZ expression, comparing resting and anti-CD3–treated freshly isolated thymocytes. As shown in Figure10, results obtained with normal thymus cells confirm previous data showing that GILZ is expressed in resting cells and is up-regulated by DEX (Figure 10A and 10B, lane 3). Moreover, the figure also indicates that stimulation of the TCR/CD3 complex, through anti-CD3 mAb, inhibits GILZ expression both at the mRNA and protein level (Figure 10A and 10B, lane 2 versus lane 1) and that this inhibition is countered by DEX treatment (Figure 10A and 10B, lane 4). Moreover, the anti-CD3–induced GILZ inhibition is a consequence of T-cell activation in that it is weakened by cyclosporin A (Figure 10C, lane 4 versus lane 2), a drug that is able to inhibit the TCR-activated transduction pathway.54 These results indicate that GILZ expression inversely correlates with T-cell activation, being higher in resting cells and lower in cells activated by TCR/CD3 triggering.
TCR/CD3 triggering inhibits GILZ expression.
Northern (A,C) and Western (B) blot analysis of GILZ. (A,B) Lane 1: untreated thymocytes; lane 2: thymocytes treated for 6 hours on plates coated with anti-CD3 (1 μg/mL); lane 3: thymocytes treated with 100 nM DEX for 6 hours; lane 4: thymocytes treated for 6 hours with anti-CD3 plus DEX. For the Northern blot analysis, the filter hybridized with GILZ cDNA was exposed for autoradiography for 36 hours. (C) Lane 1: untreated thymocytes; lane 2: thymocytes treated for 6 hours on plates coated with anti-CD3 (1 μg/mL); lane 3: thymocytes treated with cyclosporin A (1 μg/mL) for 6 hours; lane 4: thymocytes treated for 6 hours with anti-CD3 plus cyclosporin A. Filter hybridized with GILZ cDNA was exposed for autoradiography for 3 days.
TCR/CD3 triggering inhibits GILZ expression.
Northern (A,C) and Western (B) blot analysis of GILZ. (A,B) Lane 1: untreated thymocytes; lane 2: thymocytes treated for 6 hours on plates coated with anti-CD3 (1 μg/mL); lane 3: thymocytes treated with 100 nM DEX for 6 hours; lane 4: thymocytes treated for 6 hours with anti-CD3 plus DEX. For the Northern blot analysis, the filter hybridized with GILZ cDNA was exposed for autoradiography for 36 hours. (C) Lane 1: untreated thymocytes; lane 2: thymocytes treated for 6 hours on plates coated with anti-CD3 (1 μg/mL); lane 3: thymocytes treated with cyclosporin A (1 μg/mL) for 6 hours; lane 4: thymocytes treated for 6 hours with anti-CD3 plus cyclosporin A. Filter hybridized with GILZ cDNA was exposed for autoradiography for 3 days.
Discussion
In the present paper we describe a new mechanism by which GCHs inhibit T-cell activation. In particular, we show that the glucocorticoid-induced leucine zipper factor GILZ inhibits NF-κB activity and IL-2/IL-2R expression.
GCHs induce T-cell apoptosis in different experimental systems, including hybridoma T cells, thymocytes, and peripheral lymphocytes.28,29,55 However, a protective effect of GCHs has also been described in T cells, and in particular it has been shown that DEX is able to counter TCR-activated death.33,56,57 This dual effect, apoptosis activation and inhibition, is not surprising because it has been reported that GCHs may activate a caspase cascade, a central mechanism involved in T-cell apoptosis, as well as weaken TCR-induced activation of the NF-κB complex and the consequent Fas/FasL up-regulation and AICD.24,33,53 58
In an attempt to analyze the protective effect of GCHs on TCR-activated apoptosis, we have previously performed experiments aimed at identifying genes whose transcription is induced by DEX.41,44 We showed that overexpression of GILZ, one of the identified DEX-induced genes, inhibits the activation-induced up-regulation of Fas/FasL and consequent apoptosis, suggesting that the GILZ antiapoptotic effect results from the inhibition of T-cell activation.42
In the present study we show that GILZ overexpression is able to inhibit IL-2 production and IL-2R up-regulation induced by T-cell activation. Previous results indicate that GILZ, like DEX, inhibits Fas/FasL expression.42 In this respect, it is interesting that IL-2 is involved in induction of Fas/FasL expression,59 60 suggesting that the GILZ effect on activation-induced Fas/FasL expression and apoptosis may be due, at least in part, to inhibition of IL-2 and IL-2R expression.
It has been previously shown that IL-2/IL-2R and Fas/FasL systems are under the control of a number of TCR-activated transcription factors. In particular, it has been reported that NF-κB activation and nuclear translocation is required for synthesis of a number of molecules involved in T-cell activation and apoptosis, including IL-2/IL-2R and Fas/FasL.19,22,52,53 GCH can interfere with NF-κB either by direct interaction between GR and NF-κB/p65 or by competition for coactivators or by augmentation of I-κB molecules.35-40 These different mechanisms are not always contemporaneously activated following GCH treatment, so that they can act either concomitantly or independently, and inhibition of NF-κB activity has been previously described in the absence of I-κB up-regulation.40
NF-κB has been shown to be involved in apoptosis and can either induce or protect from cell death in different experimental systems.24,61-63 In particular, NF-κB activation promotes Fas/FasL expression and has been described as a proapoptotic mechanism involved in AICD.24,52,53,64 65 Thus, NF-κB contributes not only to T-cell activation but also to AICD.
The experiments described here, to elucidate whether GILZ could be involved in the molecular mechanism of NF-κB regulation, have demonstrated that GILZ inhibits TCR-induced NF-κB activation. In particular, we show that GILZ interferes with p65 and p52 through a protein-to-protein interaction both in vitro and in vivo. As a consequence, in vitro NF-κB–DNA binding and in vivo TCR/CD3-triggered transactivation activity and nuclear translocation are inhibited. GILZ expression does not influence I-κBα phopsphorylation and degradation or I-κB/NF-κB binding. Moreover, coimmunoprecipitation and inhibition of in vivo transactivation activity are also detected in transfected NTera-2 cells, which are defective for the whole NF-κB system, thus suggesting that GILZ-p65/p52 interaction does not require other Rel- or I-κB–related proteins. These results indicate that GILZ, like GCHs, is able to counter T-cell activation and is compatible with the well-known immunosuppressive and anti-inflammatory effect of GCHs and their capability to inhibit T-cell activation and death.31-33,56 66
Finally, we also performed experiments to determine if T-cell activation could inhibit GILZ expression. Results show that T-cell activation does indeed inhibit GILZ expression, indicating that GILZ-mediated modulation of TCR-induced responses is part of a circuit because T-cell activation and GILZ expression are mutually exclusive. However, the in vivo relevance of those observations remains to be proved, and future studies on genetically manipulated mice could better elucidate this aspect. Such studies are currently being pursued in our laboratory.
Supported by Associazione Italiana Ricerca sul Cancro (AIRC), Milan, Italy, and by CNR target project on Biotechnology, Rome, Italy.
E.A. and G.M. contributed equally to this paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Carlo Riccardi, Section of Pharmacology, Dept of Clinical and Experimental Medicine, Via del Giochetto, 06100 Perugia, Italy; e-mail: riccardi@unipg.it.