The recurrent translocation t(5;11)(q35;p15.5) associated with a 5q deletion, del(5q), has been reported in childhood acute myeloid leukemia (AML). We report the cloning of the translocation breakpoints in de novo childhood AML harboring a cryptic t(5;11)(q35;p15.5). Fluorescence in situ hybridization (FISH) analysis demonstrated that the nucleoporin gene (NUP98) at 11p15.5 was disrupted by this translocation. By using 3′–rapid amplification of complementary DNA ends (3′-RACE) polymerase chain reaction, we identified a chimeric messenger RNA that results in the in-frame fusion of NUP98to a novel gene, NSD1. The NSD1 gene has 2596 amino acid residues and a 85% homology to the murine Nsd1with the domain structure being conserved. The NSD1 gene was localized to 5q35 by FISH and is widely expressed. The reciprocal transcript, NSD1-NUP98, was also detected by reverse transcriptase–polymerase chain reaction. This is the first report in which the novel gene NSD1 has been implicated in human malignancy.
Introduction
Recurring chromosome translocations are common in a wide spectrum of hematologic malignancies.1-3 We have reported the identification of a recurrent cryptic translocation t(5;11)(q35;p15.5) associated with a deletion of the long arm of chromosome 5, del(5q), in de novo childhood acute myeloid leukemia (AML).4 Using fluorescence in situ hybridization (FISH), we localized the breakpoint on 11p15.5 between Harvey ras-1 related gene complex (HRC) and radixin pseudogene(RDPX1). The nucleoporin 98 gene (NUP98) lies inside this region, and its rearrangement has been documented in both de novo and therapy-related myelodysplastic syndrome, AML, and acute lymphoblastic leukemia in children and adults.5-13 We speculated that NUP98 was a good candidate gene for the involvement in the t(5;11) AML.
We now confirm that the chromosome 11 breakpoint gene isNUP98, and we report the cloning of its novel fusion partner, nuclear receptor-binding Su(var), Enhancer of zeste [E(z)], and Trithorax (Trx) (SET) domain protein 1 (NSD1), as the chromosome 5 breakpoint gene in the recurrent t(5;11)(q35;p15.5) in childhood AML.
Study design
Donor samples
We analyzed samples from 2 patients with childhood AML-M2 with t(5;11)(q35;p15.5) and a del(5q). The clinical and cytogenetic data of these 2 cases have been previously reported as patients nos. 1 and 3.4 Detailed molecular analysis was performed on the leukemic sample of patient no. 3 because RNA was available only from this patient.
FISH analysis
For FISH studies PAC 1173K1 (containing exons 10-20 of theNUP98 gene), plasmids p9R1 (containing exons 10-12 of theNUP98 gene),14 and p6G2 (containing exons 13-14 of the NUP98 gene)14 from 11p15.5 were used. A 5p probe, cos113-1,15 was used to identify both chromosomes 5. A 1.2-kilobase (kb) complementary DNA (cDNA) clone of the human NSD1 (IMAGE clone 2621298) was also used as a probe for FISH studies. FISH analysis was performed on bone marrow metaphases as previously described.4
Nucleic acid isolation
Total RNA was extracted from cryopreserved leukemic cell suspensions using the Totally RNA kit (Ambion, Austin, TX). Plasmid DNA was extracted using Qiagen reagents and protocols (Qiagen miniprep kit, Qiagen, Crawley, United Kingdom).
3′-RACE
The 3′-RACE (3′–rapid amplification of cDNA ends) was performed using the SMART-RACE cDNA amplification kit and protocol (Clontech, Palo Alto, CA). Briefly, first-strand cDNA was reverse transcribed from 1 μg total RNA using Superscript II and the 3′-RACE cDNA synthesis primer (3′-CDS) from the kit. An aliquot of the first-strand cDNA was then amplified using a NUP98 gene–specific forward primer (NUP98-1, Table 1) and a universal primer mix (SMART-RACE kit). Polymerase chain reaction (PCR) conditions were as described by the manufacturer. A nested PCR reaction using the nested universal primer (SMART-RACE kit) as the reverse primer and NUP98-2 (internal to NUP98-1, Table 1) as the forward primer was performed according to the manufacturer's instructions. Second-round PCR products were electrophoresed, purified, and subcloned. Colonies with recombinant plasmids containing the PCR products were screened by hybridization using standard protocols.16 An oligonucleotide probe of 116 base pairs (bp) (NUP-ex12) generated using primers NUP98-3 and NUP98-4 was used for screening (Table 1). All positive plasmid clones were selected for sequencing.
Reverse transcriptase–PCR
Reverse transcriptase (RT)-PCR was performed using the RT one-step RT-PCR kit (ABgene, Surrey, United Kingdom) with sense NUP98-5 and antisense NSD1-1 primers (Table 1). The PCR thermal cycling protocol was performed according to the manufacturer's instructions with an annealing temperature of 58°C. Similarly, RT-PCR was performed using sense NSD1-2 and antisense NUP98-6 primers and the same PCR conditions as above (Table 1). The RT-PCR products of both reactions were subcloned and sequenced.
Sequence analysis
Plasmid clones were sequenced using the Autoread Cy-5 sequencing kit and ALF express automated sequencer (Amersham-Pharmacia Biotech, Amersham, United Kingdom). Sequence analysis was performed using the Wisconsin Package Version 10.1 (Genetics Computer Group, Madison, WI).
Northern blot analysis
A cDNA clone (Image clone 2621298) of the human NSD1gene was used to probe human multiple tissue Northern blots according to the manufacturer's instructions (Clontech).
Results and discussion
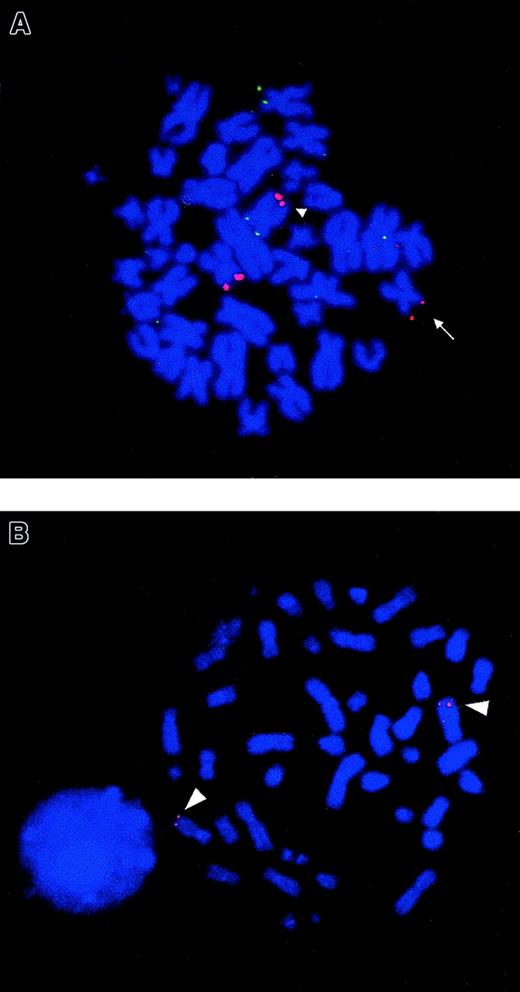
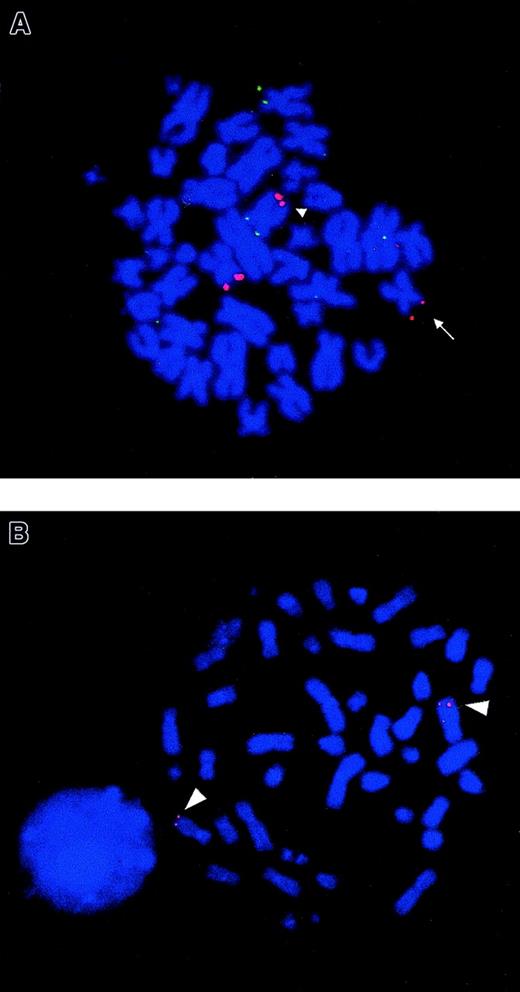
In the current study, we identified NUP98 as the chromosome 11 breakpoint gene by FISH using a PAC clone 1173K1 in 2 t(5;11)(q35;p15.5) AML cases (patients nos. 1 and 3) (Figure1A). Further, FISH analyses using plasmids specific for exons of NUP98 (p6G2 and p9R1) confirmed that the breakpoint was within NUP98, in the intron between exons 12 and 13 of this gene in patients nos. 1 and 3. Previous studies have shown the principal transcript in otherNUP98 translocations fuses the 5′ end of the NUP98gene in-frame to the 3′ end of the partner gene.7-13The 3′-RACE was performed on RNA isolated from patient no. 3 to identify the fusion partner of NUP98.
FISH analysis of NUP98 rearrangement and chromosomal mapping of humanNSD1 gene.
(A) FISH using PAC 1173K1 (containing NUP98 gene) shows that this probe hybridizes to the normal chromosome 11, der(11) (arrow) and the der(5) (arrowhead), seen as red fluorescent signals in bone marrow metaphase from patient no. 3. The probe showing green fluorescent signals is the control 5p probe (cos113-1) used to identify both chromosomes 5. The same FISH result was also seen in metaphases from patient no. 1. (B) FISH using a cDNA clone of human NSD1 (Image clone 2621298) as a probe on normal peripheral blood metaphase shows that this novel gene localizes to 5q35 (red fluorescent signals) (arrowhead).
FISH analysis of NUP98 rearrangement and chromosomal mapping of humanNSD1 gene.
(A) FISH using PAC 1173K1 (containing NUP98 gene) shows that this probe hybridizes to the normal chromosome 11, der(11) (arrow) and the der(5) (arrowhead), seen as red fluorescent signals in bone marrow metaphase from patient no. 3. The probe showing green fluorescent signals is the control 5p probe (cos113-1) used to identify both chromosomes 5. The same FISH result was also seen in metaphases from patient no. 1. (B) FISH using a cDNA clone of human NSD1 (Image clone 2621298) as a probe on normal peripheral blood metaphase shows that this novel gene localizes to 5q35 (red fluorescent signals) (arrowhead).
RACE-PCR products were purified and subcloned into plasmid vectors. A total of 88 recombinant clones were screened by hybridization and 45 clones hybridized to an internal NUP98 oligonucleotide, NUP-ex12. Sequence analysis revealed that 13 of the 3′-RACE clones diverged from the germline NUP98 sequence at nucleotide 1552 (GenBank accession number U41815). A BLAST search showed that the sequence immediately 3′ of this divergence was a novel human gene with 85% homology to the gene encoding murineNsd1 (GenBank accession number NM_008739).
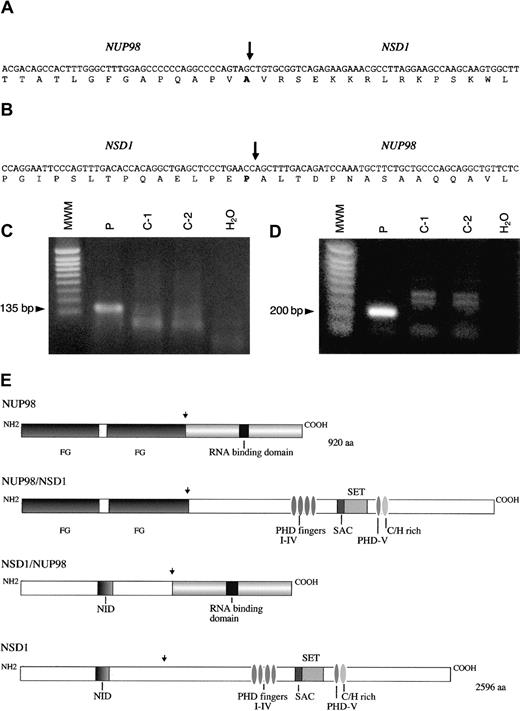
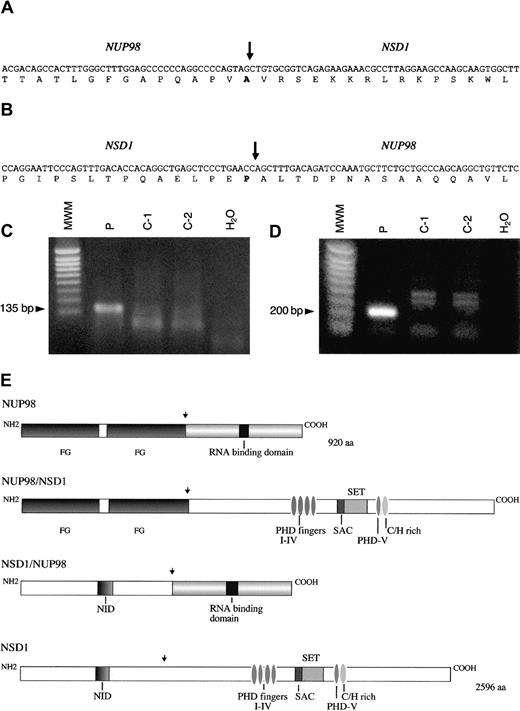
The full coding sequence of this novel human gene was determined by the sequencing of cDNA and genomic clones. The sequence had an open reading frame of 7788 nucleotides and a predicted protein of 2596 amino acids (GenBank accession number AF322907). The coding sequence of the novel gene was compared with the coding sequence of murineNsd1,17 and 85% homology was found at the amino acid level. Therefore, this indicated that the gene was the human homolog of the mouse Nsd1 gene, and it was designatedNSD1. Sequence analysis of 3′-RACE products showed thatNUP98 and NSD1 messenger RNA (mRNA) were fused in-frame joining nucleotide 1552 of NUP98 to the nucleotide 3504 of NSD1 (Figure 2A). The domain structure of the human NSD1 gene product was the same as that of murine Nsd1 protein,17 with conserved SET domain, SET domain–associated cysteine-rich domain (SAC domain), and 5 PHD fingers.
NUP98-NSD1 and NSD1-NUP98 fusion cDNA.
Nucleotide and amino acid sequences around the junction of (A)NUP98-NSD1 fusion transcript and (B) NSD1-NUP98fusion transcript. The NUP98 and NSD1 fusion points are shown by a vertical arrow. (C) RT-PCR assay forNUP98-NSD1 fusion mRNA. Primers NUP98-5 and NSD1-1 were used to amplify the 135 bp fusion mRNA. (D) RT-PCR assay forNSD1-NUP98 fusion mRNA. Primers NSD1-2 and NUP98-6 were used to amplify the 200 bp fusion mRNA. No specific products of the expected size are seen in the normal controls (C1-C2) and negative controls (H2O) in panels C and D. MWM indicates 1 kb ladder molecular weight marker. (E) Schematic representations of the wild-type NUP98, NSD1, and fusion NUP98-NSD1, NSD1-NUP98 proteins. Functional domains indicated for NUP98 are FG repeat regions at the N-terminus and RNA binding domain at the C-terminus. Domains for NSD1 are N-terminus, nuclear receptor interaction domain (NID), PHDI-IV fingers, SET and SAC domains, PHDV finger, and cysteine-histidine (C/H)-rich motif, and C-terminus. Functional domains of NSD1 are similar to the murine Nsd1 as designated in reference 17. NUP98 and NSD1 fusion points are shown by a vertical arrow.
NUP98-NSD1 and NSD1-NUP98 fusion cDNA.
Nucleotide and amino acid sequences around the junction of (A)NUP98-NSD1 fusion transcript and (B) NSD1-NUP98fusion transcript. The NUP98 and NSD1 fusion points are shown by a vertical arrow. (C) RT-PCR assay forNUP98-NSD1 fusion mRNA. Primers NUP98-5 and NSD1-1 were used to amplify the 135 bp fusion mRNA. (D) RT-PCR assay forNSD1-NUP98 fusion mRNA. Primers NSD1-2 and NUP98-6 were used to amplify the 200 bp fusion mRNA. No specific products of the expected size are seen in the normal controls (C1-C2) and negative controls (H2O) in panels C and D. MWM indicates 1 kb ladder molecular weight marker. (E) Schematic representations of the wild-type NUP98, NSD1, and fusion NUP98-NSD1, NSD1-NUP98 proteins. Functional domains indicated for NUP98 are FG repeat regions at the N-terminus and RNA binding domain at the C-terminus. Domains for NSD1 are N-terminus, nuclear receptor interaction domain (NID), PHDI-IV fingers, SET and SAC domains, PHDV finger, and cysteine-histidine (C/H)-rich motif, and C-terminus. Functional domains of NSD1 are similar to the murine Nsd1 as designated in reference 17. NUP98 and NSD1 fusion points are shown by a vertical arrow.
RT-PCR on material from patient no. 3 was performed using primers flanking the NUP98-NSD1 junction and gave a product of expected size (135 bp) (Figure 2C). Sequence analysis of RT-PCR products confirmed that NUP98 and NSD1 mRNA were fused in-frame at the same NUP98 exon as reported in the literature.5-8,10,11 13 A reciprocal fusion mRNA transcript NSD1-NUP98 was also detected by RT-PCR analysis with an expected product size of 200 bp (Figure 2D). Sequence analysis showed that the mRNA was fused in-frame joining nucleotides 3503 ofNSD1 to nucleotide 1553 of NUP98 (Figure 2B). Expression of both NUP98-NSD1 and NSD1-NUP98transcripts suggests that both may have a biological role. The expression pattern of human NSD1 in normal human tissues was analyzed using the novel cDNA clone as a probe and showed that this gene is widely expressed in hematologic and other tissues, and there are 2 transcripts of approximately 10.5 and 12 kb, respectively. FISH analysis using the novel cDNA clone revealed that the humanNSD1 was localized to 5q35 as expected (Figure 1B).
The NUP98 gene encodes a 98-kd component of the nuclear pore complex and localizes to the nucleoplasmic side of the nuclear pore complex.18 It is thought to function as a docking protein through the N-terminal domain of the protein, which contains the conserved multiple phenylalanine-glycine (FG) repeats.18 The FG repeats of NUP98 have been shown to be retained in the various fusion transcripts.5-13 The FG repeats were also retained in the NUP98-NSD1 fusion transcript in the patient reported here.
The fusion partner of NUP98 in the t(5;11) AML isNSD1, which is reported here for the first time and is the human homolog of murine Nsd1. Murine Nsd1 contains 2 distinct nuclear receptor interaction domains and also contains distinct activation and repression domains, and it has been suggested that it may define a novel class of bifunctional transcriptional intermediary factors.17 By comparison to the murine Nsd1 protein, the human NSD1 protein has the domain structure conserved and these domains are found in proteins involved in the epigenetic control of transcription; it therefore has been suggested that Nsd1 is involved in some aspects of transcriptional control at the chromatin level.17 It is thought that the PHD domain is a protein-protein interaction domain and that SET/SAC domains at least in one protein serve as a histone methyltransferase.19,20 The schematic representations of NUP98-NSD1 and NSD1-NUP98 fusion proteins are shown in Figure 2E.
In summary, we have cloned the recurrent t(5;11) chromosomal translocation, which results in the previously undescribed in-frame fusion between NUP98 and a novel gene NSD1 in de novo childhood AML. Future studies based on the structure-function relationship of the NSD1 gene and functional analysis of the 2 predicted chimeric proteins will provide an insight into the mechanism of leukemogenesis.
The authors thank Robin Roberts-Gant for assistance with the figure preparations.
Supported by the Leukaemia Research Fund, United Kingdom (R.J.J., C.F., A.J.S., F.W., J.B., J.S.W.), and the Medical Research Council (K.C, L.K).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Rina J. Jaju, LRF Molecular Haematology Unit, Nuffield Dept of Clinical Laboratory Sciences, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DU, United Kingdom; e-mail:rina.jaju@ndcls.ox.ac.uk.