Abstract
von Willebrand factor (vWF) is synthesized in megakaryocytes and endothelial cells as a very large multimer, but circulates in plasma as a group of multimers ranging from 500 to 10 000 kd. An important mechanism for depolymerization of the large multimers is the limited proteolysis by a vWF-cleaving protease present in plasma. The absence or inactivation of the vWF-cleaving protease results in the accumulation of large multimers, which may cause thrombotic thrombocytopenic purpura. The vWF-cleaving protease was first described as a Ca++-dependent proteinase with an apparent molecular weight of approximately 300 kd. Thus far, however, it has not been isolated and characterized. In this study, the purification of human vWF-cleaving protease from a commercial preparation of factor VIII/vWF concentrate by means of several column chromatographic steps, including 2 steps of heparin-Sepharose column, is reported. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis of the anion exchange and gel filtration column fractions showed that the vWF-cleaving protease activity corresponded to a protein band of 150 kd. After reduction, it migrated with an apparent weight of 190 kd. The amino terminal sequence of the 150-kd band was AAGGIL(H)LE(L)L(D)AXG(P)X(V)XQ (single-letter amino acid codes), with the tentative residues shown in parentheses. A search of the human genome sequence identified the vWF-cleaving protease as a new member of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin type I motif) family of metalloproteinase. An active site sequence of HEIGHSFGLEHE (single-letter amino acid codes) was located at 150 residues from the N terminus of the protein.
Introduction
von Willebrand factor (vWF) plays an essential role in platelet adhesion to damaged blood vessels by forming a bridge between platelet surface glycoproteins and damaged subendothelium. vWF also plays an important role in hemostasis by binding and stabilizing coagulation factor VIII, protecting it from proteolysis.1,2 vWF is composed of subunits of 2050 amino acid residues with the following repeated motifs or domains: D′-D3-A1-A2-A3-D4-B1-B2-B3-C1-C2.3,4 vWF precursor is synthesized as a very large protein in endothelial cells5and megakaryocytes6 and undergoes several posttranslational events; these include removal of a signal peptide and a large propeptide, the formation of intrachain and interchain disulfide bonds, and glycosylation. Dimers are formed in the endoplasmic reticulum through the formation of C-terminal interchain disulfide bonds,7 and these dimers form multimers in the Golgi apparatus by the formation of interdimer disulfide bonds at the N-terminal region of the protein. This structure has been referred to as a head-to-head and tail-to-tail mode.8
The mature vWF is synthesized and stored in endothelial cells9,10 and then slowly released into the circulating blood. Cultured human umbilical vein endothelial cells release only the fully polymerized form of vWF to the condition medium.11Also, normal platelets contain exclusively the intact form of vWF in their granules.12 In plasma, however, vWF exists as a mixture of disulfide-bonded multimers, with sizes ranging from dimers (500 kd) to highly polymerized forms as large as 10 000 kd.1,2 A size distribution of multimers is important for normal hemostasis in that the larger multimers have a higher thrombotic activity than smaller polymers.13 An excess of the very large vWF multimers in circulation, however, can result in platelet clumping, thrombosis, and thrombocytopenia.14 The large polymerized forms of vWF undergo limited proteolytic processing after their release into circulating blood. Accordingly, samples of plasma-derived vWF give bands of 189, 140, and 170 kd after reduction, in addition to the intact 225-kd band on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The 140- and 170-kd bands result from the cleavage of a specific Y(842)–M(843) (single-letter amino acid codes) peptide bond in the A2 domain.15 The cleavage site that generates the 189-kd band has not been identified. Thus, partially cleaved vWF multimers are composed of a variety of these fragments and intact chains, which are linked by many disulfide bonds.15-17
Furlan et al18 and Tsai31 reported the partial purification of the vWF-cleaving protease from human serum in 1996. The apparent molecular weight of the enzyme was approximately 300 kd as estimated by gel filtration. Also, the enzyme required divalent cations for catalytic activity. The enzyme was found to specifically cleave the Y(842)–M(843) bond to produce the 140- and 170-kd bands, which were the same as those present in plasma-derived vWF. EDTA and ethyleneglycotetraacetic acid inhibited the protease activity, while serine protease or thioprotease inhibitors had no effect on the enzyme. This suggested that the protease was a member of the metalloproteinase family. In vitro, the protease cleaved vWF under nonphysiological conditions, such as low ionic strength in the presence of 1 to 1.5 M urea or guanidine. When plasma was placed under shear stress, the very large multimers of vWF were converted to smaller multimers, and the levels of the 170- and 140-kd fragments were increased.19 Therefore, the protease appeared to cleave vWF when it was partially denatured under shear stress.
Recently, a number of patients have been reported who have a complete absence of the vWF-cleaving protease activity, and this correlated with thrombotic thrombocytopenic purpura. Some patients developed antibodies of the immunoglobulin (Ig)–G subclass to the protease, while others carry familial deficiencies of the protease.20-23 These results showed that the vWF-cleaving protease plays an essential role in generating the normal size distribution of vWF multimers in plasma. Despite the importance of this protease, it has not been identified and characterized. In this study, the vWF-cleaving protease has been purified to apparent homogeneity from a commercial factor VIII/vWF concentrate and its N-terminal sequence determined. Computer searches of the human genome sequence identified the protease as a new member of the metalloproteinase family.
Materials and methods
Materials
A commercial preparation of factor VIII/vWF concentrate was generously provided in 1981 by Dr Henry Kingdon of Hyland Therapeutics, now a Division of Baxter International (Deerfield, IL), and has been kept frozen at −20°C. Diisopropyl fluorophosphate (DFP) and iodoacetamide (IAA) were purchased from Sigma (St Louis, MO). Superose-6, Superdex-200, butyl-Sepharose 4 Fast Flow, Q Sepharose, and Sephacryl-500 were obtained from Pharmacia (Piscataway, NJ). Heparin-Sepharose was prepared as described earlier.24 vWF was prepared as described by Shapiro et al25; ethanol was added dropwise to 100 mL factor VIII/vWF concentrate (final 3%) at from 0°C to 2°C under constant stirring. The precipitate obtained was dissolved in 100 mL 50 mM Tris-HCl, pH 7.35, and dialyzed against the same buffer. The sample was then applied to a Q Sepharose column (1.6 × 13 cm), and the proteins were eluted by a 300-mL linear salt gradient (0 M and 0.5 M NaCl). The fractions of the second protein peak contained vWF. Fragment III (frag III), the N-terminal fragment of vWF from S(1) to E(1365) (single-letter amino acid codes), was prepared from vWF as described by Girma et al8 and further purified by gel filtration on Superdex-200 column.
Assay of vWF-cleaving protease activity
Frag III (0.2 to 0.4 μg) was incubated for 20 minutes at room temperature with test samples (2 to 10 μL) in 50 μL of 50 mM Tris-HCl (pH 8.0)/8 mM BaCl2/0.02% Tween-20. The samples were transferred to microdialysis chambers (Hampton Research, Laguna Hills, CA) and dialyzed overnight at 37°C against 5 mM Tris-HCl (pH 8.0)/1 M urea. The samples were then run on 6% or 7.5% SDS-PAGE gel26 after reduction, and proteins were blotted onto Immobilon-P transfer membrane (Millipore, Bedford, MA) according to Towbin et al.27 Rabbit anti-vWF antibodies (rabbit × human factor VIII–related antigen) (Accurate Chemical and Scientific, Westbury, NY) were used as the first antibodies. Anti–rabbit IgG alkaline phosphatase conjugate (Sigma) or affinity-purified goat anti–rabbit IgG peroxidase conjugate (Tago, Burlingame, CA) was employed as the second antibody. Antigen bands were visualized by means of 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (BCIP/NBT) phosphatase substrate (Kirkegaard and Perry Labs, Gaithersburg, MD) or the enhanced chemiluminescent detection reagent (Amersham/Pharmacia Biotech, Buckinghamshire, England).
Purification of vWF-cleaving protease from commercial factor VIII/vWF concentrates
The frozen commercial factor VIII/vWF concentrate (380 mL) was thawed overnight at 4°C. CaCl2 (final, 20 mM) and thrombin (60 μg) were added, and incubated overnight at 4°C. Fibrin clots were removed by centrifugation at 16 000g for 15 minutes. Ammonium sulfate was added to the supernatant (total protein, 354 mg) to 33% saturation and stirred for 20 minutes. The precipitate obtained by centrifugation was suspended in approximately 50 mL of 50 mM Tris-HCl (pH 8.5), treated with DFP (final, 0.1 mM) and IAA (final, 5 mM) for 1 hour, and then dialyzed overnight against 2 L of 50 mM Tris-HCl (pH 7.5)/0.6 M ammonium sulfate. Gelatinous precipitates that appeared during dialysis were removed, and the sample was applied to a butyl-Sepharose column (1.2 × 6 cm) at a flow rate of 6 mL/10 min at ambient temperature. After the column was washed with the same buffer, adsorbed proteins were eluted with 50 mM Tris-HCl (pH 7.5). The eluted fractions were pooled, and ammonium sulfate was added to 40% saturation. The precipitates were collected by centrifugation and dissolved in approximately 5 mL of 50 mM Tris-HCl (pH 8.5). The sample (160 mg) was treated for 1 hour with DFP (final, 1 mM) and IAA (5 mM) and applied to a Sephacryl-500 column (2.5 × 60 cm) at a flow rate of 5 mL/12 min at 4°C. The proteins were eluted with 50 mM Tris-HCl (pH 8.0)/0.2 M NaCl and appeared in a single peak. The major portion of the peak was pooled and dialyzed overnight against 1 L of 50 mM MES-NaOH (pH 6.6)/0.1 M NaCl with one buffer change. The sample (114 mg) was then applied to a heparin-Sepharose column (1.4 × 9 cm) at a flow rate of 0.8 mL/min, and nonadsorbed fractions were pooled and concentrated to approximately 40 mL, followed by dialysis against 2 L of 50 mM MES-NaOH (pH 6.6). The sample (13 mg) was then applied to a second heparin-Sepharose column (1.4 × 3 cm). After the column was washed successively with 50 mM MES-NaOH (pH 6.6) and 50 mM MES-NaOH (pH 6.6)/25 mM NaCl, adsorbed proteins were eluted by a 50-mL linear NaCl gradient (25 mM and 225 mM NaCl). Fractions with protease activity were pooled and dialyzed against 1 L of 50 mM Tris-HCl (pH 8.0)/40 mM NaCl with one buffer change. The sample (3.6 mg) was then applied at a flow rate of 0.5 mL/min to a Waters diethylaminoethanol (DEAE) column (1.0 × 6 cm) attached to a Waters Protein Purification System (Milford, MA). After washing with 50 mM Tris-HCl (pH 8.0)/40 mM NaCl, proteins were eluted by a 40-mL salt gradient (40 mM and 640 mM NaCl) in the buffer. A portion of the active fraction from the DEAE column was applied to Superose-6 column (0.9 × 30 cm) and run with 50 mM Bis-Tris (pH 7.2)/0.15 M NaCl at a flow rate of 0.5 mL/min. Fractions of 0.5 mL were collected.
Sequence and other analysis
After SDS-PAGE, the protein bands were transferred to Immobilon-P membrane, which was stained briefly with Coomassie Blue G. After destaining, the 150-kd band was excised and subjected to sequence analysis. Automated Edman degradation was carried out in an Applied Biosystems 477A Protein Sequencer connected to 120 A Applied Biosystems Analyzer (Foster City, CA). SDS-PAGE was performed on 6% Laemmli gels, which were then stained with Blue stain reagent (Pierce, Rockford, IL) or silver stain reagent (Bio-Rad, Hercules, CA) for proteins and periodic acid–Schiff reagent28 for glycoprotein. Protein concentrations were estimated by absorbance assuming = 10.0.
Results and discussion
Purification of vWF-cleaving protease
A commercial preparation of factor VIII/vWF concentrate was used as the starting material, since it contained a substantial amount of the protease activity with far less protein contamination than plasma. Fibrinogen was initially removed by thrombin addition because it interfered with the chromatographic resolution and was abundant in the starting material. The defibrinated sample was then fractionated by ammonium sulfate precipitation (0% to 33% saturation) followed by chromatography on a hydrophobic interaction column with the adoption of the conditions described by Furlan et al.18 The protease activity was adsorbed and eluted from the column as reported by these authors. Substantial purification was then achieved by successive heparin-Sepharose column chromatography. In the first heparin-Sepharose column, the sample was applied in the presence of 0.1 M NaCl. Under these conditions, the activity did not bind to the column, whereas approximately 90% of contaminant proteins were bound and removed. The nonadsorbed fraction was then applied to a second heparin-Sepharose column in the absence of salt, where it was bound and then eluted by a salt gradient. After the second heparin column, the protease was further purified by DEAE and gel filtration column chromatography.
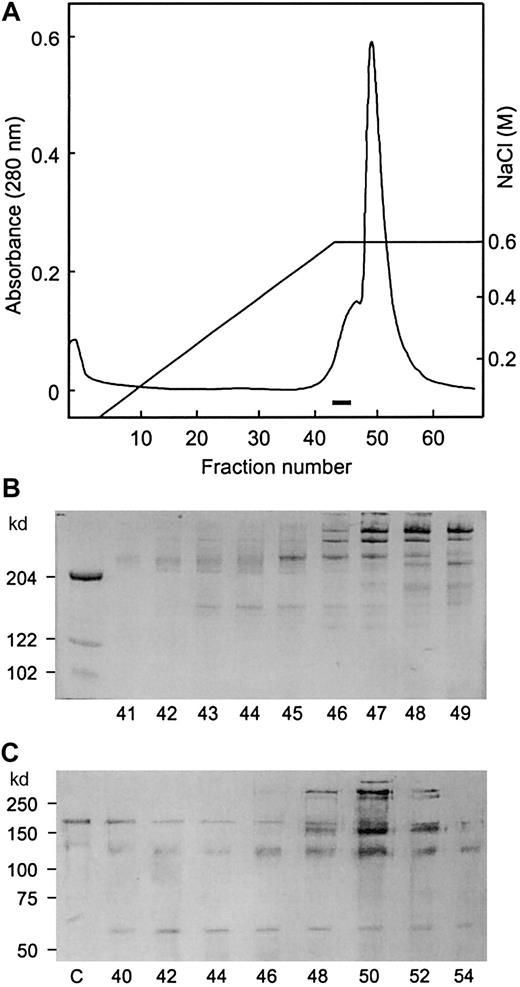
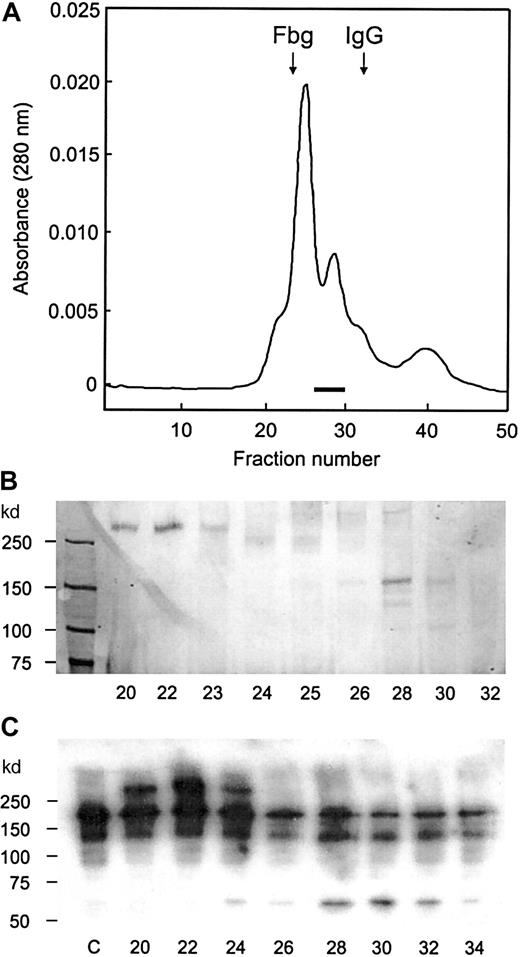
The second heparin column fraction was applied to a DEAE column, and the proteins were eluted in a single peak with an ascending shoulder (Figure 1A). By SDS-PAGE analysis, a discrete 150-kd band appeared to coincide with the ascending shoulder area, trailing into the major protein peak area. A number of large proteins were present in the shoulder area, and several distinct bands with high molecular weights appeared in the major peak. A small protein band was also seen in the major peak area (Figure 1B). The protease activity was assayed by immunoblotting analysis to detect proteolytic fragments of frag III, which extends from S(1) to E(1365) in vWF. The cleavage at Y(842)–M(843) bond generates 2 fragments of 140 kd and 65 kd. The 140-kd fragment, which originates from the N terminus, is identical to the 140-kd fragment produced from plasma vWF. The 65-kd fragment results from the C-terminal portion of frag III. The protease activity was detected in fractions 40 to 54 and was highest in fractions 42 and 46. This activity correlated with the intensity of the 150-kd protein band. With these fractions, the intensity of the 190-kd band (frag III) decreased and that of the 140-kd and 65-kd bands increased when compared with the control. vWF antigen was detected in the major peak, including fractions 48 through 54. In these fractions, vWF was also cleaved by the protease, resulting in the generation of a 170-kd fragment that migrated as a band below intact frag III. Trailing of both the 150-kd protein and the protease activity into the major peak indicated that the 150-kd protease binds to and comigrates with larger proteins. In the following gel filtration column, the larger proteins were completely separated from the 150-kd protease, which eluted in the fractions 26 through 30 (Figure2 A-B). vWF protease activity was detected in fractions 24 through 30, with a higher activity in fractions 28 through 30 (Figure 2C). This was coincident with the intensity of the 150-kd band. Results obtained in the DEAE and gel filtration columns confirmed that the 150-kd protein was the vWF-cleaving protease.
DEAE column chromatography.
The sample (3.6 mg) from the second heparin column was applied to the DEAE column connected to Waters Protein Purification System. (A) Protein elution pattern. The fractions in which the 150-kd band was detected by SDS-PAGE analysis are indicated by a bar. (B) SDS-PAGE analysis. Samples (20 μL) were applied to 6% gel without reduction, and protein bands were stained by Blue stain reagent. Fraction numbers are shown at the bottom. (C) Immunoblotting analysis for the protease activity. After the reactions, samples were run on 7.5% gel with reduction and transferred to membrane. The bands were visualized by alkaline phosphatase substrate. Lane c is a control without the enzyme.
DEAE column chromatography.
The sample (3.6 mg) from the second heparin column was applied to the DEAE column connected to Waters Protein Purification System. (A) Protein elution pattern. The fractions in which the 150-kd band was detected by SDS-PAGE analysis are indicated by a bar. (B) SDS-PAGE analysis. Samples (20 μL) were applied to 6% gel without reduction, and protein bands were stained by Blue stain reagent. Fraction numbers are shown at the bottom. (C) Immunoblotting analysis for the protease activity. After the reactions, samples were run on 7.5% gel with reduction and transferred to membrane. The bands were visualized by alkaline phosphatase substrate. Lane c is a control without the enzyme.
Gel filtration on Superose-6 column.
A portion of DEAE column fraction was applied to the Superose–6 column. (A) Protein elution pattern. The fractions in which the 150-kd band was detected by SDS-PAGE analysis are indicated by the bar, and the elution positions of fibrinogen (330 kd) and IgG (150 kd) are indicated by arrows. (B) SDS-PAGE analysis. The samples (20 μL) were applied to 6% gel without reduction, and protein bands were visualized by silver staining. (C) Immunoblot analysis for the protease activity. After the reaction, samples were run on 6% gel with reduction and transferred to membrane. The bands were detected by the enhanced chemiluminescent reagent. The fraction numbers are shown in the bottom, and lane c is a control without the enzyme.
Gel filtration on Superose-6 column.
A portion of DEAE column fraction was applied to the Superose–6 column. (A) Protein elution pattern. The fractions in which the 150-kd band was detected by SDS-PAGE analysis are indicated by the bar, and the elution positions of fibrinogen (330 kd) and IgG (150 kd) are indicated by arrows. (B) SDS-PAGE analysis. The samples (20 μL) were applied to 6% gel without reduction, and protein bands were visualized by silver staining. (C) Immunoblot analysis for the protease activity. After the reaction, samples were run on 6% gel with reduction and transferred to membrane. The bands were detected by the enhanced chemiluminescent reagent. The fraction numbers are shown in the bottom, and lane c is a control without the enzyme.
One or 2 faint bands below the 150-kd band were also detected in the gel filtration column fractions (Figure 2B). Since these bands were not detected in the DEAE column fractions, it appeared likely they were generated from the 150-kd protein after its separation from the larger proteins. The 150-kd protein was stable, and no accompanying bands were observed in earlier steps, probably because it was protected by the presence of other proteins. A large 280-kd protein, which eluted in the first peak of the gel filtration column, was reactive to anti-vWF antibodies (Figure 2C), indicating that the protease binds to vWF. The binding affinity was not strong, however, since the 2 proteins were readily separated by gel filtration in the presence of 0.15 M NaCl. When an excess amount of vWF was present, the protease activity was completely bound to vWF: the protease coeluted with large proteins from a Sephacryl-500 column used in the early stage of purification.
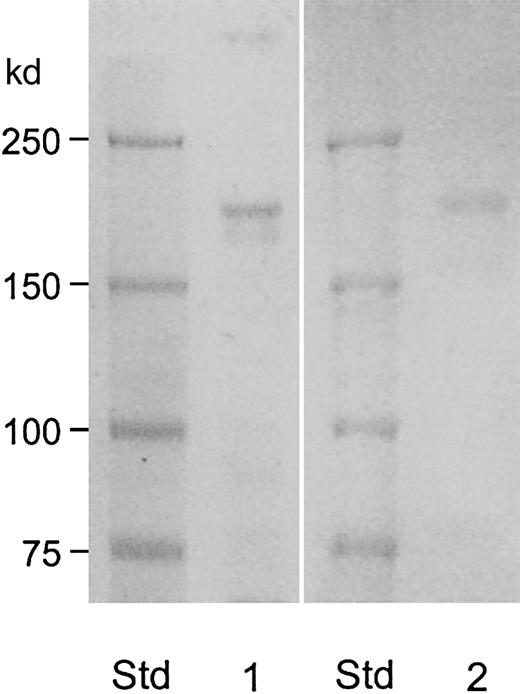
The protease migrated as 150-kd band without reduction, but it migrated more slowly after reduction (Figure 3, lane 1) with molecular weight estimated to be 190 kd. This value was consistent with the elution position from the Superose 6-column: the 150-kd protein eluted between fibrinogen (330 kd) and IgG (150 kd) as shown in Figure 2A. The vWF-cleaving protease was positive by carbohydrate staining (Figure 3, lane 2). These data show that vWF-cleaving protease is a single-chain glycoprotein with an apparent molecular weight of 190 kd.
Molecular weight estimation and detection of carbohydrate moiety of the purified vWF-cleaving protease.
The samples (30 μL) from fraction 28 of the Superose-6 column (described in Figure 2) were applied to 6% gel after reduction. Lane 1 was stained for protein by Coomassie Blue stain reagent and lane 2 was stained for carbohydrate by periodic acid–Schiff reagent.
Molecular weight estimation and detection of carbohydrate moiety of the purified vWF-cleaving protease.
The samples (30 μL) from fraction 28 of the Superose-6 column (described in Figure 2) were applied to 6% gel after reduction. Lane 1 was stained for protein by Coomassie Blue stain reagent and lane 2 was stained for carbohydrate by periodic acid–Schiff reagent.
Furlan et al18 first found vWF-cleaving protease activity in plasma and characterized its enzymatic properties. The enzyme properties of our preparation are consistent with their observations: cation-dependency, fragmentation, and apparent molecular weight (300 kd by gel filtration in an earlier stage of purification). Unique assay (dialysis of vWF/protease against 1 M urea in buffer of low ionic strength) was required to detect the enzyme activity. In addition, we used exactly the same conditions for the butyl-Sepharose column and obtained the same results. Thus, we are confident that the purified protease reported here is the same as the one that Furlan et al18 described.
N-terminal amino acid sequence of vWF-cleaving protease and computer search of the human genome
The 150-kd band from the DEAE column fraction was subjected to sequence analysis, and an N-terminal sequence of AAGGIL(H)LE(L)L(D)AXG(P)X(V)XQ (single-letter amino acid codes) was obtained. Residues in the parentheses were tentatively identified, while X represents uncertain residues. A search of the human genome sequences with this N-terminal amino acid sequence of vWF-cleaving protease indicated that the positively identified residues in the protein were identical to a sequence translated from human chromosome 9q34. Furthermore, the tentatively identified amino acids in positions 7, 10, 12, 16, 18, and 20 were confirmed in the genomic sequence. A probe sequence of 97 bases from chromosome 9q34 encompassing the aligned region was then used to search the human-expressed sequence tags (EST) database. One entry that was identified (GenBank gi 3841324) was 592 bases in length and overlapped with the probe sequence. Translation of the composite genomic and EST sequence showed that it encoded a novel sequence that was homologous to the protease domain of metalloproteinases of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin type I motif) subfamily.29Specifically, it contained the sequence HEIGHSFGLEHD (single-letter amino acid code), 150 residues from the N-terminal AAGG sequence (Figure 4). This matched the consensus extended active site sequence of metalloproteinases in this family.30 This sequence also showed the presence of a precursor or pro piece with a typical furin-processing site.
Composite N-terminal sequence of vWF-cleaving protease.
The sequence is derived from sequential Edman degradation and translation from genomic (Genbank gi 12733516) and EST (3841323) sequences. Residues determined in part by Edman degradation are boxed. Residues 150 to 161, containing the extended active site sequence of metalloproteinases of the ADAMTS subfamily, are double-underlined. Residues −1 to −4, corresponding to a putative furin-processing site at the C terminus of the precursor form, is underlined.
Composite N-terminal sequence of vWF-cleaving protease.
The sequence is derived from sequential Edman degradation and translation from genomic (Genbank gi 12733516) and EST (3841323) sequences. Residues determined in part by Edman degradation are boxed. Residues 150 to 161, containing the extended active site sequence of metalloproteinases of the ADAMTS subfamily, are double-underlined. Residues −1 to −4, corresponding to a putative furin-processing site at the C terminus of the precursor form, is underlined.
The EST sequence was then used as a probe sequence in a subsequent search, which identified an entry in the GenBank database (GenBank gi 12735207) that overlaps with the 3′ end of the EST sequence for 140 nucleotides. This entry (2272 bases) contains an open reading frame derived from chromosome 9 (C9ORF8) predicted by computer analysis of the human genomic sequence in the 9q34 region. Analysis of this predicted open reading frame shows it encodes a disintegrinlike sequence, followed by 3 thrombospondin type I motifs. All overlapping sequences align with the human chromosome 9q34 contig sequence in sequential order in 18 apparent exons. The composite sequence represents an incomplete complementary DNA (cDNA) without the 5′ end coding for the signal peptide and the N-terminal pro region and the 3′ end coding for additional C-terminal thrombospondin motifs. Other inconsistencies, including base variations and reading frame shifts, are also present. These analyses show that the vWF-cleaving protease is a new metalloproteinase that belongs to the ADAMTS subfamily. The gene encoding this enzyme, which has been designatedHGNC, is located in chromosome 9q34. Experiments to clarify the cDNA sequence and identify the 5′ and 3′ regions are in progress. In the accompanying paper (see Gerritsen et al,32 page 1654) the authors also purified vWF-cleaving protease to apparent homogeneity. The amino-terminal sequence was essentially identical to that reported in this paper.
We are grateful to Dr Earl W. Davie for his valuable advice and encouragement throughout this work. We also thank Dr Ling Xie for technical assistance and Jeff E. Harris for assistance in manuscript preparation.
Supported by grant HL16919 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kazuo Fujikawa, Department of Biochemistry, Box 357350, University of Washington, Seattle, WA 98195; e-mail:fujikawa@u.washington.edu.