The mammalian β-globin locus is a multigenic, developmentally regulated, tissue-specific locus from which gene expression is regulated by a distal regulatory region, the locus control region (LCR). The functional mechanism by which the β-globin LCR stimulates transcription of the linked β-like globin genes remains unknown. The LCR is composed of a series of 5 DNaseI hypersensitive sites (5′HSs) that form in the nucleus of erythroid precursors. These HSs are conserved among mammals, bind transcription factors that also bind to other parts of the locus, and compose the functional components of the LCR. To test the hypothesis that individual HSs have unique properties, homologous recombination was used to construct 5 lines of mice with individual deletions of each of the 5′HSs of the endogenous murine β-globin LCR. Here it is reported that deletion of 5′HS1 reduces expression of the linked genes by up to 24%, while deletion of 5′HS4 leads to reductions of up to 27%. These deletions do not perturb the normal stage-specific expression of genes from this multigenic locus. In conjunction with previous studies of deletions of the other HSs and studies of deletion of the entire LCR, it is concluded that (1) none of the 5′HSs is essential for nearly normal expression; (2) none of the HSs is required for proper developmental expression; and (3) the HSs do not appear to synergize either structurally or functionally, but rather form independently and appear to contribute additively to the overall expression from the locus.
Introduction
The multigene β-globin locus has been intensively studied as a model for the tissue-specific and developmental regulation of gene expression.1 Transcription of the locus is restricted to erythroid cells, and each gene is expressed during specific developmental stages. Analysis of naturally occurring deletions in the human locus revealed that regions far upstream of the β-like globin genes are important for regulation of the locus.2-4 These regions encompass major DNase I hypersensitive sites (5′HS1 to 5 in human, 5′HS1 to 6 in mouse) located upstream of the embryonic globin genes.5-9 The smallest and most well studied of these deletions, the Hispanic thalassemia deletion,4 removes 5′HS2 to 5 as well as 27 kilobases (kb) upstream of this region and leads to an alteration of the generalized DNase I sensitivity of the locus, alterations in the timing of replication and origin used, and the transcriptional silencing of the locus.10
Linkage of large restriction fragments containing these 5′HSs to human β-globin genes leads to high-level expression in transgenic mice (reviewed in Grosveld et al11 and Townes and Behringer12) and contrasts with earlier studies in which isolated globin transgenes were expressed at low levels or not at all.13 This region, the locus control region (LCR), is defined by its ability to mediate high-level, erythroid-specific expression of linked transgenes and to significantly reduce the influence of integration site on the expression of linked genes. Extensive analysis has attempted to determine which sequences within the LCR are necessary for activity and how these sequences may interact. The DNA sequences associated with each HS contain a several hundred base pair (bp) core region that is highly conserved in evolution, suggesting that these HS cores may be important for LCR function (reviewed in Hardison et al14).
Individual HSs and combinations of HSs have been assayed for their ability to direct high-level expression of linked genes in erythroid cells in transient and stable expression assays in tissue culture and in transgenic mice.15-17 Most of this work has used sequences from the human β-globin LCR, which is highly homologous to the murine LCR. Study of the human locus in a transgenic mouse has the advantage of analyzing human sequences, but the disadvantage of analyzing the function of the human locus at ectopic sites that are not the normal genomic location of the locus, and the human locus is therefore potentially sensitive to position effects. In addition, the interaction of the milieu of murine regulatory factors and the human locus may not be fully analogous to the interaction between the human regulatory factors and the human locus. To circumvent these complications, and to preserve the spatial relationships between the genes and HSs, we have analyzed the endogenous mouse β-globin locus by generation of specific LCR mutations using homologous recombination (HR) in embryonic stem (ES) cells followed by the generation of mutant mice. Previously, we and collaborators have reported on the deletions of 5′HS2, 3, and 5/6.9,18 19 Deletion of 5′HS1 or 4 from the endogenous locus has not been reported previously.
Published studies of human 5′HS1 and 4 using several assay systems have been performed. Unlike 5′HS2, neither 5′HS1 nor 5′HS4 has enhancer activity in transient transfection assays.15 The 5′HSs from the human LCR have been studied in transgenic mice in a variety of contexts, including linkage of individual 5′HSs to globin transgenes.17 When compared with 5′HS1, 5′HS4 led to an increase of expression and had a predominant effect on the β gene. In addition to these gain-of-function assays, single 5′HSs have been deleted from large transgenes that contain the remainder of the LCR and all the gene-coding segments in their normal juxtaposition. The transcriptional phenotype observed varies among studies,20-24 possibly because deletions of any 5′HSs increase susceptibility to position effects.20 Presumably, these variable results are due to a combination of these position effects and the different deletions generated.
Here we report the generation and analysis of mice with deletions of 5′HS1 and 4 from the endogenous murine LCR. This completes the set of deletions of the individual 5′HSs of the murine β-globin LCR. The results from this study, as well as the results from our former individual 5′HS deletions, support a model in which none of the HSs is developmentally stage specific and the HSs contribute in a simple additive manner to the overall activity of the LCR.
Materials and methods
Production and screening of mutant mice
Targeted deletion of 5′HS1 and 4 were done as described by means of a loxP flanked PGK-neo cassette and AK-7 ES cells.9,18,19 These deletions remove nucleotides −4675 to −6999, and −19 849 to −22 561, relative to the Ey cap site, respectively. Correct structure of the targeted alleles was confirmed by Southern blotting by means of a BglII/SpeI digest for Δ1 and Δ1neo mice, and a SphI digest for Δ4 and Δ4neo mice. Mice carrying targeted deletions were generated by injection of the targeted ES cells into the blastocysts of C57BL/6 mice by means of standard techniques. After germline transmission of the ES cell genome, selectable markers were excised in vivo as described.9 Homozygous mutants were bred to C57BL/6 mice to generate mice heterozygous for the HbbS and HbbD haplotypes.
Reverse transcriptase–polymerase chain reaction assays
RNA isolation, reverse transcriptase–polymerase chain reaction (RT-PCR), and quantitation was done as described previously.9,18 19 Analysis was done with yolk sac from embryonic day 10.5 postconception (dpc), fetal liver from 15.5 dpc, and adult blood. The data points are derived from at least 2 independent reverse transcription–reactions, and messenger RNA (mRNA) was obtained from at least 4 mutant animals.
Chromatographic quantitation of the globin chains
High-pressure liquid chromatography (HPLC) analysis was performed on adult peripheral blood from heterozygotic mice as previously described by means of a 3-step acetonitrile gradient ranging from 35% to 55% after treatment of samples with cystamine (Sigma, St Louis, MO).25
Results
Production of mice with deletions of 5′HS1 or 5′HS4
To investigate the contribution of 5′HS1 and 5′HS4 to the activity of the endogenous murine β-globin LCR, we have produced mice with individual deletions of each of these sites. The 2.3-kb 5′HS1 deletion was designed to remove the core binding sites, the region to which the HS maps, and all regions of homology with human 5′HS1. A similar approach was taken for the 2.7-kb deletion of 5′HS4, except that 3 small blocks of homology (of 7, 11, and 7 bp) upstream of the HS core remain after deletion of the HS. These small regions of homology have not been shown to bind any known transactivators (see “Discussion”). The overall strategy was the same as that employed for prior deletions of 5′HS2, 3, and 5/6.9,18,19Previously, we determined that the presence of a selectable marker within the LCR significantly affects expression of the linked globin genes.18,19 Therefore, we used a selectable marker (PGK-neo) flanked by loxP sites, the recognition sequence for the Cre recombinase. We have previously demonstrated the utility of using site-specific recombinases to excise selectable markers after HR events (reviewed in Fiering et al27). After targeted deletion of the HSs in ES cells, mice were generated (Δ1neo and Δ4neo), and selectable markers were excised in vivo by means of the Cre recombinase, as previously described, to produce the Δ1 and Δ4 lines of mice. Figure 1 diagrams this sequence of events, demonstrates how the locus was modified, and provides examples of the Southern blots used to recognize the appropriate genotypes.
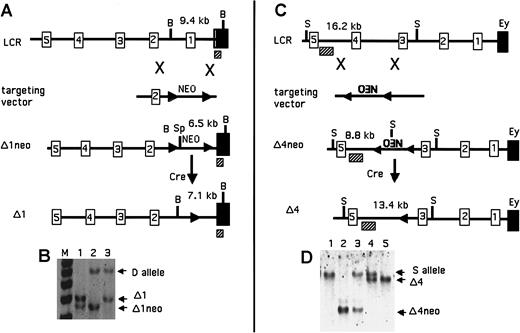
Map of the deletions of 5′HS1 and 5′HS4.
(A) Modifications of 5′HS1 as labeled. The numbered boxes are 5′HSs; the solid boxes are Ey; the hatched boxes are the probe used for Southern analysis. B denotes BglII sites, and Sp denotes SpeI. Numbers above the map are the expected restriction fragment sizes. (B) Southern blot demonstrating targeted deletions of 5′HS1. Genotypes of the mice are as follows: lane 1, Δ1neo/Δ1; lane 2, Δ1neo/D; and lane 3, Δ1/D. (C) Modifications of 5′HS4 as labeled. Elements are as in panel A. S denotes SphI sites. (D) Southern blot demonstrating targeted deletions of 5′HS4. Genotypes are as follows: lane 1, S/S; lane 2, Δ4neo/Δ4neo; lane 3, Δ4neo/S; lane 4, Δ4/S; and lane 5, Δ4/Δ4.
Map of the deletions of 5′HS1 and 5′HS4.
(A) Modifications of 5′HS1 as labeled. The numbered boxes are 5′HSs; the solid boxes are Ey; the hatched boxes are the probe used for Southern analysis. B denotes BglII sites, and Sp denotes SpeI. Numbers above the map are the expected restriction fragment sizes. (B) Southern blot demonstrating targeted deletions of 5′HS1. Genotypes of the mice are as follows: lane 1, Δ1neo/Δ1; lane 2, Δ1neo/D; and lane 3, Δ1/D. (C) Modifications of 5′HS4 as labeled. Elements are as in panel A. S denotes SphI sites. (D) Southern blot demonstrating targeted deletions of 5′HS4. Genotypes are as follows: lane 1, S/S; lane 2, Δ4neo/Δ4neo; lane 3, Δ4neo/S; lane 4, Δ4/S; and lane 5, Δ4/Δ4.
Deletion of 5′HS1 and 5′HS4 affects the level but not the pattern of expression
To determine whether the deletion of either HS has an effect on the level, timing, or tissue specificity of β-globin gene expression, mice and embryos were analyzed by internally controlled quantitative RT-PCR assays that exploit restriction fragment length polymorphisms between the 2 major β-globin alleles in mice, HbbS and HbbD. Primer pairs specific for the Ey, the βh1, and the adult β-globin genes (β-major and β-minor) coamplify mRNA from HbbS and HbbD isotypes equally, and cleavage at polymorphic restriction enzyme sites allows quantitative determination of expression from one allele compared with the other. Targeted mutations were made on an HbbD allele in ES cells, and the resultant mutant mice were bred to mice carrying a wild-type HbbS allele, allowing comparison of expression from the mutant HbbD allele to that of the internal control HbbS allele in these heterozygous animals. For developmental analysis, the 3 sets of primers were used to examine RNA from yolk sac from dpc 10.5 embryos, liver from dpc 15.5 fetuses, and peripheral blood from adults.
Analysis of expression in heterozygotes carrying the Δ1neo mutation is summarized in Table1. A moderate decrease was observed at all developmental stages. Analysis of mice lacking 5′HS1 and without the selectable marker (Δ1) is shown in Figure2 and is quantitated in Table 1. Expression of all genes at all developmental stages is reduced slightly, and no abnormal developmental expression is detected. As seen previously, the presence of the PGK-neo gene exacerbates the reduction in expression caused by the deletion itself.
Expression of β-like globin genes in Δ1/wild-type heterozygous mice.
The HbbD allele (D) carries the targeted mutation, while the HbbS allele (S) is wild type. RNA was analyzed from dpc 10.5 yolk sac, dpc 15.5 fetal liver, and adult peripheral blood by means of RT-PCR assays for Ey, βh1, and adult primers (top, middle, and bottom panels). WT indicates wild-type D/S animals; Δ1 indicates mutant Δ1/S heterozygous mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
Expression of β-like globin genes in Δ1/wild-type heterozygous mice.
The HbbD allele (D) carries the targeted mutation, while the HbbS allele (S) is wild type. RNA was analyzed from dpc 10.5 yolk sac, dpc 15.5 fetal liver, and adult peripheral blood by means of RT-PCR assays for Ey, βh1, and adult primers (top, middle, and bottom panels). WT indicates wild-type D/S animals; Δ1 indicates mutant Δ1/S heterozygous mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
The presence of selectable markers in the LCR has been shown to decrease transcription of linked globin genes in both the α and the β loci18,19,26 28; however, the mechanism by which this occurs remains unknown. Therefore, it was of interest to determine whether the interference was dependent on the orientation of the selectable marker within the locus. To address this question, mice with the identical deletion as the Δ1neo mice but with the PGK-neo gene in the opposite transcriptional orientation were generated and analyzed as above (Δ1neo−). Table 1 demonstrates that regardless of orientation, the presence of the marker has a substantial suppressive effect on expression of all genes at all stages.
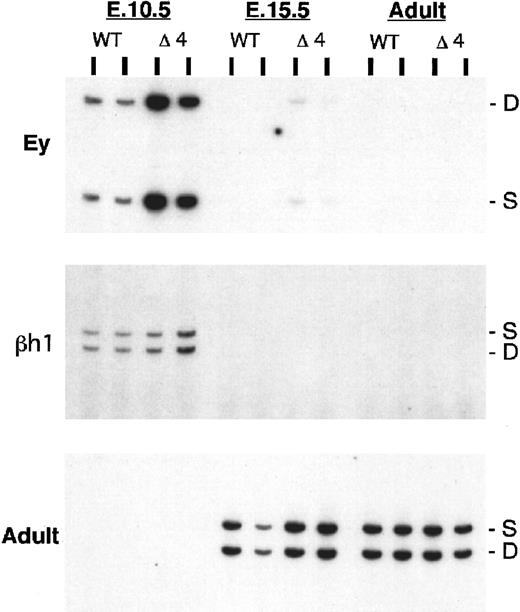
After 5′HS4 was deleted from the endogenous locus, expression of mRNA in the Δ4neo and Δ4 mice was assayed as described above for the Δ1 mice. Analysis of expression in the mice with a replacement of 5′HS4 by the PGK-neo marker (Δ4neo) shows a moderate to severe reduction in expression of all genes at all developmental stages (Table1). In contrast, upon removal of the selectable marker, the transcription is decreased predominantly in the adult globin genes (Figure 3; Table 1). While the embryonic genes are not significantly affected, a moderate decrease in the expression of β-major and β-minor is observed. The developmental timing of expression of all genes remains unchanged.
Expression of β-like globin genes in Δ4/wild-type heterozygous mice.
The HbbD allele (D) carries the targeted mutation, while the HbbS allele (S) is wild type. RNA was analyzed from dpc 10.5 yolk sac, dpc 15.5 fetal liver, and adult peripheral blood by means of RT-PCR assays for Ey, βh1, and adult primers (top, middle, and bottom panels). WT indicates wild-type D/S animals; Δ4 indicates mutant Δ4/S heterozygous mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
Expression of β-like globin genes in Δ4/wild-type heterozygous mice.
The HbbD allele (D) carries the targeted mutation, while the HbbS allele (S) is wild type. RNA was analyzed from dpc 10.5 yolk sac, dpc 15.5 fetal liver, and adult peripheral blood by means of RT-PCR assays for Ey, βh1, and adult primers (top, middle, and bottom panels). WT indicates wild-type D/S animals; Δ4 indicates mutant Δ4/S heterozygous mice. S and D mark the RT-PCR products from the S and D alleles, respectively.
Deletion of 5′HS1 and 5′HS4 have a greater effect on β-minor than on β-major
Previously, HPLC analysis demonstrated that changes in β-globin protein paralleled changes in mRNA in mice lacking 5′HS2, 3, and 5/6, and that this assay could be used to accurately quantitate the levels of β-major and β-minor.25 Levels of β-major and β-minor in the peripheral blood of heterozygous Δ1 and Δ4 mice were determined and are shown in Table2. The Δ1 mice had 88% of the normal level of total β (as compared with expression from the single isotype), similar to our RT-PCR results. Analysis of the β-major and β-minor proteins individually revealed that while β-major was mildly reduced to 90% of normal, β-minor was more severely reduced to 78% of normal. The protein levels of total β in the Δ4 mice was 86% of normal, with β-major reduced to 90% of normal, and β-minor more severely reduced to 70% of normal. Finally, for both the deletions, presence of the selectable marker further reduced the ratio of β-minor to β-major (data not shown).
Discussion
Influence of the selectable marker
Previously, we demonstrated that replacement of LCR HSs with a PGK-neo gene significantly reduces expression from the locus. When deletion of an HS reduces expression, as is seen with deletion of 5′HS1, 2, 3, or 4 (see below), the presence of a selectable marker exacerbates this reduction, potentially owing to promoter interference.18,19 Regardless of mechanism, this effect is independent of the orientation of the marker. Mice with 5′HS1 replaced with the marker in both orientations have a similar transcriptional phenotype. In contrast to deletion of 5′HS1, 2, 3, or 4, deletion of 5′HS5/6 did not significantly reduce expression with or without the marker.9 The lack of effect of the marker at 5′HS5/6 could be due to its location upstream of the sites shown to affect transcription directly, to the fact that deletion of 5′HS5/6 does not suppress expression, or to the fact that this is the one deletion that used the hygromycin gene as a selectable marker.
Effect of 5′HS deletion after removal of the selectable marker
After removal of the selectable marker, we observed that deletion of individual 5′HS1, 2, 3, or 4 measurably reduced expression of one or more genes. None of the deletions perturbed the developmental timing of expression or mediated more than a 40% reduction in expression. Finally, all of these deletions preferentially reduced the most distal adult gene, β-minor, more than the proximal adult gene, β-major.
While the deletion of 5′HS4 deletes the core binding sites, 3 small evolutionarily conserved regions of homology (7, 11, and 7 bp) were not removed. A search for conserved consensus binding sites for factors known to bind HSs or activate globin gene transcription revealed a single YY1 site. In theory, these remaining regions could lead to the formation of a 5′HS and complement the function of the 5′HS4 deletion. However, as demonstrated previously, analysis of the chromatin structure of the 5′HS4 deletion reveals that 5′HS1, 2, 3, and 5/6 form normally, but no HS forms at or immediately surrounding the site of the 5′HS4 deletion.29
Previously, it was suggested that individual 5′HSs had unique effects limited to a specific stage of development or specific target gene.17 While both the deletion of 5′HS4 presented here and prior transgenic studies demonstrate that 5′HS4 predominantly affects adult gene expression, this correlation in transgenic and targeted deletion results does not hold true for the other LCR deletions. For example, while transgenic studies suggested that 5′HS3 preferentially affects embryonic/fetal gene expression, this was not borne out in our targeted deletion from the endogenous locus.19 This discrepancy is likely to be due to differences between assaying expression from the human locus as a randomly integrated transgene in mice and assaying expression from the endogenous mouse locus (see below).
The finding that deletion of a 5′HS has a greater affect on transcription of the adult globin genes is not unique to 5′HS4; both the 5′HS2 and the 5′HS3 deletions mediated greater reduction of the adult genes than of the embryonic genes.18,19 Thus, the adult genes at the endogenous locus are more dependent on the LCR for their activity. This conclusion is supported by our analysis of mice with a deletion of the entire LCR in which the greatest residual expression is in the embryonic βh1 gene.30,31 While these findings could be explained by stage-specific differences in the milieu of trans-acting factors or differences in promoter sequences, it is also simply explained by tracking or linking models of LCR-mediated gene activation (reviewed in Bulger and Groudine32). In these models, the LCR is responsible for generating a wave of complexes or epigenetic modification that spreads from the LCR through the locus. Mutations within the LCR may decrease this activity, preferentially affecting the more distal genes. This mechanistic possibility is consistent with the finding that the distal adult gene, β-minor, is always reduced more by LCR mutations than the proximal adult gene, β-major.
How do these results compare with previous analysis of 5′HS1 and 5′HS4 function?
The LCR has been extensively studied in transgenic mice containing portions of the human β-globin locus. Since transgenes are by definition studied in ectopic genomic locations, a relevant question is whether a particular cis element in a transgene stimulates transcription, suppresses position effects, or does both. In practice, distinguishing these possibilities is not simple, since transcription levels are affected by position effects. Large transgenes with the entire human β-globin locus demonstrate a greater than 2-fold range of expression per copy33 34; thus, the ability to recognize a modest change in expression caused by a change in the construct depends on having sufficient numbers of transgenic lines to allow the level of expression to be compared in a statistically significant manner. Since this requirement is rarely met, transgenic systems are most useful for analyzing how regulatory elements influence position effects or developmental specificity of expression.
Studies in the endogenous locus, such as ours, cannot address the influence of the HSs on position effects, but since there is no background of position effects to complicate analysis of expression levels, such studies can more sensitively assay the influence of an HS deletion on expression levels. Examination of the single 5′HS deletions reveals decreases in adult gene expression of 0% to 40%, free of position effects. The results of Milot et al20 are consistent with our results since they demonstrate that large transgenes containing the locus can function normally in some genomic locations with a single 5′HS deleted. We show that one of those locations is the endogenous location.
In contrast to Milot et al,20 additional studies using large transgenes with individual HS deletions demonstrated that deletion of 5′HS4 can severely reduce expression levels and affect formation of other 5′HSs.21,35 These conclusions differ from the transcriptional analysis presented here and from our prior studies of the mouse locus showing that deletion of one 5′HS does not prevent formation of the remaining LCR 5′HSs.29These differences may be due to inherent differences in comparing deletions in a human transgene with different sized deletions in the endogenous mouse locus. Alternatively, it can be argued that, as concluded by Milot et al,20 deletion of 5′HS4 simply increases susceptibility to position effects.
Previously, a patient with a small deletion that removed 5′HS1 was described.36 The patient was a compound heterozygote with a thalassemic allele in trans to the 5′HS1 deletion. In contrast to other patients with the same thalassemic allele, no transcriptional phenotype could be ascribed to the deletion in this patient. Owing to difficulties in quantitation in this situation, the mild decrease in adult expression we observe in the adult mouse would probably have been missed. Thus, results obtained in the analysis of 5′HS1 deletions in a human and in mice are in agreement: the endogenous locus, whether mouse or human, is unimpaired or slightly impaired by deletion of 5′HS1.
Mechanistic implication
What do these studies tell us about the mechanism by which the LCR influences expression of the linked genes? In addition to demonstrating that no 5′HS has a unique activity and that each 5′HS contributes to LCR activity, our results support a model in which each 5′HS contributes in an additive manner to LCR activity, at least with regard to adult gene expression. While previous studies using transgenes suggested that 5′HSs synergize or interact in a way whereby individual HSs are essential for LCR activity,21,22,24 we demonstrate that in the endogenous murine locus, each site acts independently and none is critical. The sum of reduction in gene expression of the adult genes in peripheral blood from animals carrying targeted deletions of individual 5′HSs is 114% (Δ1, reduced 22%; Δ2, reduced 41%; Δ3, reduced 29%; Δ4, reduced 19%; Δ5/6, reduced 3%), similar to the reduction seen when the entire LCR is deleted30 31and accounting for the full expression activity of the locus. While this is also true of the adult genes at dpc 15.5, as noted before, the embryonic genes are less affected by single site deletions in the LCR. The additive nature of the 5′HSs on adult gene transcription and the observation that deletion of each 5′HS does not disturb structural formation of the other 5′HSs fit with a model in which each 5′HS independently contributes to LCR function.
We initiated these studies with the hypothesis that individual HSs had specific unique activities. We have disproved this hypothesis and instead have shown that all the HSs seem to act in a similar additive fashion to influence expression. The mechanism of this influence remains to be demonstrated.
The authors thank Agnes Telling, Kirsten Robinson, and Michelle Mehaffey for technical assistance; and the Dartmouth Transgenic Mouse Facility and the Fred Hutchinson Cancer Research Center Image Analysis Laboratory and Core Biotech Facility.
Supported by National Institutes of Health grants DK54071 (S.N.F.), DK44746 (M.G.), and P30 HD28834 through the University of Washington Child Health Research Center (M.A.B.); by a Burroughs-Wellcome Career Development Award (S.N.F.); and an American Society of Hematology Scholar Award (S.N.F.). M.A.B. is a Howard Hughes Medical Institute Physician Postdoctoral Fellow and a J. S. McDonnell Foundation Scholar.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mark Groudine, 1100 Fairview Ave N, Seattle WA, 98109; e-mail: markg@fhcrc.org.