Clonally expressed T-cell receptor αβ heterodimers are able to bind many different major histocompatibility complex/peptide combinations. This promiscuity is thought to be required for adequate surveillance against microbial and malignancy-associated antigens. After transplantation, T cells may react with nonself structures, contributing to graft-versus-host disease, in the case of hematopoietic stem cell transplantation, or graft failure, when the host immune system is preserved. We describe 2 distinct HLA A*0201–restricted, cytotoxic CD8 T-cell responses to the prevalent chronic pathogen, herpes simplex virus type 2, that cross-react with cells bearing specific alleles of the common HLA B44 family. Transfection of human or primate renal epithelial cells with HLA class I complementary DNA confirmed these results. Given the prevalence of this viral infection and the HLA alleles involved, it is possible that this cross-reactivity may be involved in clinically significant events.
Introduction
Viral infections are associated with graft-versus-host disease (GVHD) and graft failure in human transplant recipients.1 The cellular infiltrate in skin and gut GVHD is CD8 T-cell predominant,2,3 and CD8 T cells can be overrepresented in rejecting solid organs as well.4 The use of acyclovir after bone marrow transplantation, at doses expected to be far more active against herpes simplex virus (HSV) than cytomegalovirus, has been clinically associated with decreased GVHD and increased survival.5 Treatment with the antiherpesviral compound valacyclovir lowered the incidence of graft rejection in kidney transplant recipients.6 Because immunosuppressive therapy, which may worsen viral infection, may be administered in response to GVHD or rejection, it is difficult to establish a causal relationship between infection, allogeneic responses, and GVHD or graft rejection.
HSV type 2 (HSV-2) infection occurs in 22% of adults in the Untied States. HSV frequently reactivates in immunosuppressed transplantation patients. Despite suppressive or episodic therapy with antivirals, reactivation, with peripheral lytic replication, still occurs in many patients, either as a consequence of antiviral drug resistance or after the cessation of routine antiviral prophylaxis.7 HSV provokes a significant CD8 and CD4 response, despite multiple virally encoded immune evasion genes. The precursor frequency HSV-specific CD8 cytotoxic T lymphocytes (CTLs) among CD8 peripheral blood mononuclear cells (PBMCs) is on the order of 1 × 103 by limiting dilution assays8 and higher by tetramer assays (D.M.K. et al, unpublished data, June 2001). We report 2 examples of HLA A2–restricted CD8 CTL cross-reactivity between HSV-2 and prevalent HLA class I molecules in the HLA B44/45 family, and we outline clinical situations in which these molecular interactions may be medically significant.
Study design
Patients and specimens
Patients gave informed consent in accordance with the Helsinki protocol. Protocols were approved by the University of Washington Institutional Review Board. HSV serostatus was tested by type-specific serology.9 Biopsies of recurrent, HSV-2 culture–positive10 lesions were performed as described.11 PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation and cryopreserved.
Cell lines and viruses
Derivation of CD8+ CTL clone 1874.1991.22, specific for HSV-2 protein VP13/14 amino acids 551-559, and clone 5101.1999.23, specific for VP13/14 amino acids 289-298, from recurrent genital HSV-2 skin lesions has been described.12 PBMC-derived clones were made after 2 cycles of peptide restimulation.12CD8-bearing cells were positively selected (Miltenyi, Auburn, CA) and cloned at 1 cell/well.13 Clones with HSV-specific lytic activity were expanded12 by using anti-CD3 monoclonal antibody. Cos-7, an African green monkey renal epithelial cell,14 293, a human renal epithelial cell,11,15 and Epstein-Barr virus (EBV)–transformed lymphocyte cell lines (EBV-LCLs) were cultured as described.16 EBV-LCLs of known HLA type obtained from G. T. Nepom (Virginia Mason Research Center, Seattle, WA) or from J. Hansen (Fred Hutchinson Cancer Research Center, Seattle, WA) included PITOUT, HERLUF, ARENT,17 and IHW09385. C1R18 and C1R cell lines transfected with HLA B*4402, B*4403, B*4405, or B*4407 were kindly provided by Dr A. W. Purcell (Melbourne, Australia). Transfectants were maintained in RPMI 1640, 10% fetal calf serum, 2 mM L-glutamine, 1% pen-strep media containing 500 μg/mL G418 (Sigma, St Louis, MO) for B*4403 or 500 μg/mL hygromycin (Sigma) for the others. HSV-1 strain E11519 and HSV-2 strain 33320 were raised and titered in Vero cells.10
Lymphocyte functional assays
Four-hour 51Cr release cytotoxicity assays with EBV-LCL targets were done as described16 with viral infections at a multiplicity of infection of 10 for 18 hours. Screens of candidate clones were singlicate; confirmatory and alloreactivity assays were triplicate at effector-to-target ratios of 20:1. To assess T-cell activation by cytokine release, Cos-7 or 293 cells were seeded at 9000/well in 96-well flat-bottom plates and transfected in duplicate the next day with 50 ng/well HLA class I heavy chain complementary DNA (cDNA) and Fugene 6 (Boerhinger Mannheim-Roche, Indianapolis, IN) per the manufacturer. Cell-surface expression of human HLA B44 or B45 class I was checked by flow cytometry with fluorescein isothiocyanate-labeled monoclonal antibody B12 as described12 and was positive on 25% to 35% of cells at 48 hours (not shown). Two days after transfection, 5 × 104 to 1 × 105 cloned T cells were added in 150 μL fetal calf serum–based medium. After an additional 24 hours, supernatants were collected and assayed in duplicate for interferon γ (IFN-γ) by enzyme-linked immunosorbent assay as described.12 HLA cDNA for transfection, cloned into pcDNA3.0 (−) or pcDNA3.1 (+) (Invitrogen, Carlsbad, CA), was from low endotoxin kits (Qiagen, Valencia, CA). HLA B*4501 cDNA was cloned as described12; B*4402 and B*4403 were gifts from Dr S. R. Riddell (Fred Hutchinson Cancer Research Center, Seattle, WA). Flow cytometry to characterize lymphocyte clones was performed by using antibodies from Becton Dickinson (San Jose, CA) as described.11
HLA typing
Patients were initially typed serologically at the Puget Sound Blood Center. For definition of HLA B44 alleles, direct sequencing was performed as described.21
Results and discussion
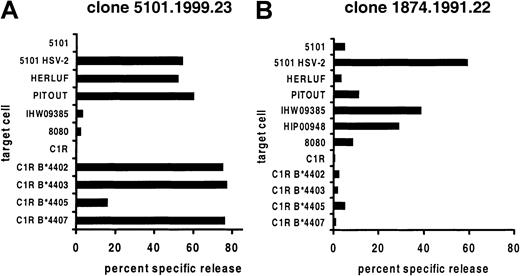
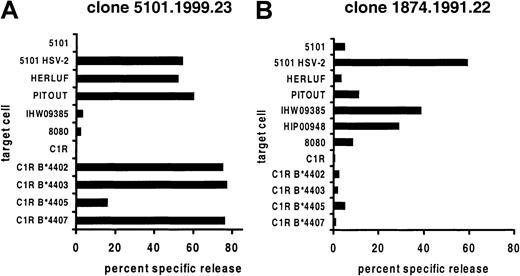
CD8+ CTL clones 1874.1991.22 and 5101.1999.23 were cultured from biopsies of HSV-2 culture–positive genital lesions of patients 1874 and 5101 (Table 1), without secondary in vitro stimulation with antigen.12 The clones were CD3+ and T-cell receptor αβ (TCR-αβ)+ by flow cytometry, each lysed autologous HSV-2 infected cells, and were proven to be HLA A*0201–restricted by release of IFN-γ after coculture with Cos-7 cells cotransfected with A*0201 cDNA and infected with HSV-2.12 For each clone, lysis of certain HLA class I–mismatched, uninfected EBV-LCLs was observed during initial workup (not shown). Cytolytic activity was tested against an extended panel (Table 1) of allogeneic target cells (Figure 1). The observed patterns of reactivity were consistent with recognition of a different, specific set of B44 alleles by each clone.
Cross-reactivity between A*0201+viral peptide and allogeneic cells.
VP13/14-specific CD8 CTL clones derived from genital HSV-2 lesions were used as effector cells against EBV-LCL targets. Target cells from patient 5101 only were tested after infection with HSV-2. Results are representative of at least 2 experiments for each effector and target cell combination.
Cross-reactivity between A*0201+viral peptide and allogeneic cells.
VP13/14-specific CD8 CTL clones derived from genital HSV-2 lesions were used as effector cells against EBV-LCL targets. Target cells from patient 5101 only were tested after infection with HSV-2. Results are representative of at least 2 experiments for each effector and target cell combination.
Clone 5101.1999.23 is specific for HSV-2 VP13/14 amino acids 289-298 (FLVDAIVRVA) in the context of HLA A*0201.12 This clone also lysed uninfected wild-type EBV-LCLs expressing HLA B*4402, B*4403, or B*4407 but not B*4404 or B*4405 (Figure 1). These results were confirmed with a panel of B44 transfectants of the class I–negative cell line C1R. Clone 1874.1999.22 is specific for amino acids 551-559 (GLADTVVAC) of the same HSV-2 protein and is again restricted by A*0201.12 It displayed a different pattern of alloreactivity, lysing only B*4404-expressing cells. A C1R transfectant was not available, but lysis of 2 separate B*4404 cell lines was noted.
To begin to examine alloreactivity among HSV-2 VP13/14-specific CD8 CTLs in the population, we generated 2 CD8 CTL clones from the PBMCs of another HSV-2–infected, A*0201-bearing patient (9383, Table 1) by using peptide 289-298. Clones were screened for lysis of A*0201-bearing, HSV-2 infected cells. Each HSV-2–reactive clone recognized VP13/14 289-298 in the context of A*0201 (Table2). Clone 33 also recognized EBV-LCL bearing the B*4402 allele. Clones 31 and 32 did not lyse wild-type LCLs expressing known B44 alleles, indicating that the response to VP13/14 289-298 for patient 9383 is heterogeneous and contains both cross-reactive and non–cross-reactive clonotypes. However, clone 31 lysed a cell line bearing the related B*4501 allele. Unfortunately, these clones could not be recovered from cryopreservation, so reactivity with defined C1R transfectants or extended panels of wild-type EBV-LCLs could not be performed. It is also possible, therefore, that T-cell clones 9383.33 and 9383.31 recognize other class I or class II determinants expressed by the target cells listed in Table 2.
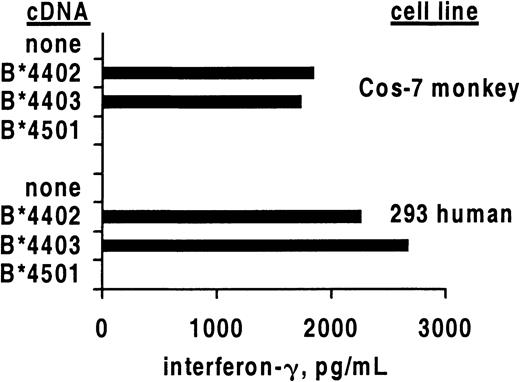
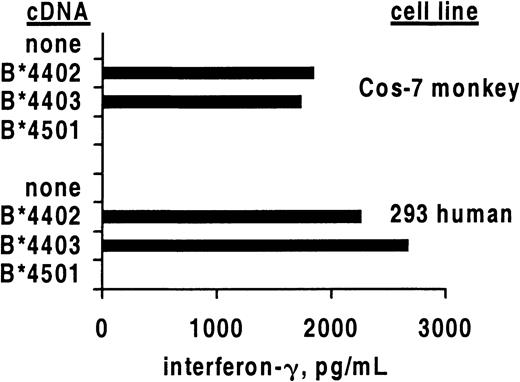
Allo–cross-reactive T cells may recognize 2 different HLA molecules, each combined with a unique peptide, or alternatively may recognize allogeneic HLA in a peptide-independent manner.22 23 To study whether the alloreactivity of HSV-2–specific CTLs required the recognition of a species-specific peptide, we compared the recognition of human and monkey cells transfected with HLA class I heavy chain cDNA (Figure 2). After translation, the foreign polypeptide assembles with endogenous β2-microglobulin and peptides and is transported to the cell surface. Specific secretion of IFN-γ was noted after contact of clone 5101.1999.23 with either human or monkey renal epithelial cells bearing B*4402 or B*4403 but not negative control B*4501. These results confirm the cytolysis data (Figure 1) for this T-cell clone. If allorecognition of B*4402 and B*4403 by 5101.1999.23 is peptide dependent, an identical or structurally similar peptide is expressed in cells of renal origin from 2 different primate species.
Cross-reactive CD8 T-cell clones secrete IFN-γ in response to allogeneic HLA class I.
Mean secretion of IFN-γ by CD8 clone 5101.1999.23 in response to stimulation by the indicated primate renal epithelial cell lines transiently transfected with the indicated HLA class I heavy chain cDNA molecules. Results are representative of 3 experiments.
Cross-reactive CD8 T-cell clones secrete IFN-γ in response to allogeneic HLA class I.
Mean secretion of IFN-γ by CD8 clone 5101.1999.23 in response to stimulation by the indicated primate renal epithelial cell lines transiently transfected with the indicated HLA class I heavy chain cDNA molecules. Results are representative of 3 experiments.
These data suggest that several unique HLA A*0201–restricted, VP13/14-specific CD8 CTL clonotypes have the ability to recognize allogeneic human cells. In each case, the allogeneic target structures probably contain a member of the HLA B44 family. HLA B44 alleles, although distinct, have related predicted amino acid sequences and peptide-binding motifs.24 Although TCR can recognize HLA molecules in the absence of bound peptides,25well-described examples of murine cross-reactive T cells document the importance of both nonself major histocompatibility complex (MHC) and peptide.26 At this time, the molecular identity of the peptide or peptides bound to HLA B44 or 45 and recognized by HSV-2–reactive CD8 are not known. The peptide(s) is likely to have a broad tissue distribution, as both B cells and renal epithelium–derived cells are reactive, and to be conserved between African green monkeys and humans.
T-cell cross-reactivity between self-MHC/viral and allogeneic determinants is well known. Examples of alloreactivity for HSV-specific CD8 and CD4-specific human T-cell clones have been described27,28 but have not defined to high resolution the molecular nature of the viral or HLA target structures. Comparison of the estimated sizes of the TCR-αβ T-cell repertoire (approximately 108) and the set of possible peptides of the size recognized by T-cells (approximately 1013) has lead to the hypothesis that T-cell cross-reactivity to many different MHC/peptide combinations is ubiquitous.29,30 A thoroughly analyzed example of virus/allo–cross-reactivity in the CD8 CTL response to EBV26 has revealed that the EBV infection status of HLA B8–bearing persons has a substantial influence on the precursor frequency of alloreactive, B*4402-specific CD8 CTLs. T-cell clones cross-reactive with B8 and a defined EBV peptide, and also B*4402 and an as-yet undefined “self” peptide, occur with high prevalence and magnitude. As predicted by self-tolerance, B8/B*4402 heterozygous persons do not develop these T-cell clonotypes after EBV infection.26
Several predictions based on our findings could be tested in further investigations. PBMCs from A*0201/B44 persons infected with HSV-2 ought not to give rise to B44-specific alloreactive T cells after in vitro restimulation with HSV-2 peptide. We would also predict that allogeneic stimulation of PBMCs from A*0201-bearing donors by B44-bearing cells would result in CTLs with activity against A*0201-infected cells sensitized by HSV-2 infection or HSV-2 peptides. Positive results from such a reciprocal experiment have been reported in defined EBV systems.31 Our laboratory plans to address these predictions in future experiments.
Our observations may have clinical consequences in 2 distinct clinical scenarios. In the first scenario, hematopoietic stem cells and/or naive or T cells are transplanted into a HLA A*0201– and HLA B*44–bearing recipient. If the recipient has HSV-2 infection, potentially cross-reactive T cells will be stimulated that may react against recipient structures and cause GVHD. A variation would occur if memory CD8 CTLs from a HLA A*0201–bearing, HSV-2 immune donor were transferred along with the stem cells; these CTLs could be restimulated by HSV-2 antigen in the context of A*0201 or B44-“housekeeping” peptide structures, or both, and attack parenchymal tissues such as gut or skin. The second scenario might occur after transplantation of a B44-bearing organ into a HLA A*0201+, HSV-2–infected recipient. In response to changing levels of immune suppression, antiviral therapy, and viral antigen load, preexisting A*0201-restricted memory/effector CD8 CTLs originally primed by HSV-2 infection would recognize B44 structures on the transplanted organ. The A*0201/B44 recipients of transplanted B44 organs might be protected from graft rejection mediated by cross-reactive T cells, as potentially B44-reactive T cells in the recipient would be reduced by negative selection during development. Future clinical studies may be able to address these predictions and disease models.
We thank Sigrid N. Reymond for technical assistance; Eric Mickelson, Dr Jei Pi, Dr John Hansen, Dr Anthony Purcell, and Dr Gerald Nepom for HLA-typed cell lines; Dr Karen Nelson, Dr Anna Wald, and Dr Lawrence Corey for helpful discussions; Dr Mark Gavin for technical advice; and our patients and the clinicians at the Virology Research Clinic, Seattle, WA, for their participation and assistance in obtaining research specimens.
Supported by grants AI30713 and AI50132 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David M. Koelle, Harborview Medical Center Box 359690, 325 Ninth Ave, Seattle, WA 98104; e-mail:viralimm@u.washington.edu.