Nonsense-mediated mRNA decay (NMD) represents a phylogenetically widely conserved splicing- and translation-dependent mechanism that eliminates transcripts with premature translation stop codons and suppresses the accumulation of C-terminally truncated peptides. Elimination of frameshifted transcripts that result from faulty splicing may be an important function of NMD. To test this hypothesis directly, this study used the IVS1 + 5 G>A thalassemia mutation of the human β-globin gene as a model system. We generated β-globin gene constructs with this mutation and an iron-responsive element in the 5′ untranslated region, which allowed specific experimental activation and inactivation of translation and, hence, NMD of this transcript. Premessenger RNAs with IVS1 + 5 G>A were spliced at normal sites and cryptic sites, enabling a direct comparison of the effect of NMD on the accumulation of normal and frameshifted messenger RNAs. In transfected HeLa cells, the predominant frameshifted transcript was degraded under conditions of active NMD, whereas accumulation to high levels occurred under conditions of specifically disabled NMD, thereby indicating an important physiologic function of NMD in the control of the splicing process. An unexpected finding was that accumulation of a second aberrant transcript remained unaffected by NMD. The IVS1 + 5 G>A mutation thus revealed the presence of an unknown cis-acting determinant that influences the NMD sensitivity of a putative NMD substrate. It can therefore serve as a useful tool for defining the mechanisms that permit specific transcripts to circumvent the NMD pathway.
Introduction
Genetic disorders are commonly caused by nonsense or frameshift mutations that introduce premature translation stop codons (PTCs).1 Synthesis of large amounts of C-terminally truncated polypeptides encoded by such transcripts is avoided by a splicing- and translation-dependent mechanism termed nonsense-mediated decay (NMD).2-6 The medical importance of NMD is well documented in β-thalassemia. If PTCs are located at positions that activate NMD, the common recessive mode of inheritance with asymptomatic heterozygous carriers results. In contrast, PTCs at positions that do not activate NMD yield a stable messenger RNA (mRNA) that is available to direct the synthesis of truncated polypeptides. Such aberrant translation products can act in a dominant-negative fashion, resulting in a symptomatic form of β-thalassemia in heterozygotes and a dominant mode of inheritance.7 8
Although NMD can be envisioned to have beneficial effects in many medically important genes, it seems likely that the wide phylogenetic conservation from yeast to humans indicates an important role for NMD in the control of basic molecular mechanisms. One such basic function might be represented by the elimination of mRNAs that result from errors in the normal splicing process.1,2 9 Such errors are thought to occur at a low rate per gene, but in cells with active gene expression, this may amount to a considerable number of mRNA molecules that direct the synthesis of false polypeptides. Because of the low levels of faulty splice products that result from the normal gene-expression pathway, this hypothesis is difficult to address directly. We therefore established an experimental system that allows systematic analysis of the effect of NMD on aberrantly spliced transcripts with frameshifted open reading frames (ORFs).
Our experimental model is based on the IVS1 + 5 G>A mutation of the human β-globin gene, which is a rare cause of β-positive thalassemia and which was first observed in a patient with homozygous β-thalassemia intermedia.10 As a result of this mutation, about 20% of the mutant pre-mRNAs are processed at the normal splice site, whereas the remaining 80% are spliced at cryptic sites.11 Constructs based on this mutation should therefore express high levels of incorrectly spliced mRNAs with PTCs in phase-shifted ORFs. An important component of the experimental system is based on the translation dependence of NMD. Translation of these mRNAs can be regulated specifically in an iron-dependent manner by insertion of an iron-responsive element (IRE) into the 5′ untranslated region (5′-UTR)12 that allows monitoring of mRNA expression under conditions of enabled or disabled NMD without major general perturbations of cellular metabolism and translation.
Material and methods
Constructs
The human β-globin gene construct with the IRE in the 5′-UTR (wild type [WT]) was described previously.12Construct IVS1 + 5 was generated by means of a G>A site–directed mutagenesis of the WT construct at this position.13 In construct Δ−16 IVS1 + 5, the cryptic splice site 16 nucleotides (nts) 5′ of the intron 1 splice donor was inactivated by a GT>CC mutation at this position, which does not affect the ORF.
In constructs −38 ΔIVS1 and −16 ΔIVS1, the first intron of the WT construct was deleted and the coding regions of the 2 natural exons 1 and 2 were joined at the cryptic splice sites 38 nts or 16 nts 5′ of the canonical site. These deletions were generated by first removing anNcoI/AccI fragment encompassing the entire coding region of exon 1, intron 1, and exon 2 up to the AccI restriction site 212 nts 5′ of the intron 2 splice donor site. This DNA segment was replaced by reverse transcriptase–polymerase chain reaction (RT-PCR) products that were amplified from the mRNA of cells transfected with construct IVS1 + 5. For −38 ΔIVS1 and −16 ΔIVS1, the respective RT-PCR fragments were gel purified in 8% polyacrylamide, digested with NcoI and AccI, and inserted. Construct NS39 ΔIVS1 was generated similarly, except that the exon 1–exon 2 RT-PCR fragment was inserted in the respectiveNcoI and AccI restriction sites of construct PTC 39.12
The series of constructs with inactivating mutations of possible AUG translation-reinitiation sites at positions 66 nts, 74 nts, 99 nts, and 129 nts 3′ of the exon 1–exon 2 junction (ΔAUG 1-4, ΔAUG 1, ΔAUG 2, ΔAUG 3, and ΔAUG 4) were generated by T>A or A>C site–directed mutagenesis of the parent −38 ΔIVS1 construct. In constructs ΔAUG 1-4, ΔAUG 1, ΔAUG 3, and ΔAUG 4, the 2 PTCs further downstream in one of the alternative ORFs 90 nts and 174 nts 3′ of the exon 1–exon 2 junction were also functionally inactivated by site-directed mutagenesis (TGA>AGA).
The WT-ΔIVS1-chloramphenicol acetate transferase (CAT) plasmid that was cotransfected to control for transfection efficiency contained a human β-globin–Escherichia coli CAT hybrid gene in a pCI-neo vector (Promega, Madison, WI). In this construct, the 660-base-pair ORF of the CAT gene without the translation-initiation and termination codons was inserted into the EcoRI site in exon 3 of the WT-ΔIVS1 construct that was identical to NS39 ΔIVS1 except that it did not contain the NS39 mutation. All modifications were confirmed by DNA sequencing.
Cell culture and transfections
HeLa cells were grown in Dulbecco modified Eagle medium under standard conditions and transiently transfected by calcium phosphate precipitation. Thirty μg test constructs was used for ribonuclease protection assays (RPAs). For Northern blot analyses, 25 μg test plasmids was cotransfected with 25 μg WT-ΔIVS1-CAT, which served as a control for transfection efficiency. The cells were washed after 16 hours. Specific regulation of translation of the IRE-containing mRNAs was achieved by supplementing the culture medium with 100 μM heme arginate 8 hours after washing or by iron depletion with 100 μM desferrioxamine added to the culture medium 4 hours after washing. Cells were harvested 24 hours after washing.
RNA analysis
For the RPAs, total cellular RNA was extracted with Trizol reagent (Gibco-BRL, Gaithersburg, MD). Then, 3-6 μg RNA was analyzed by using an excess of a 589-nt complementary RNA β-globin probe spanning exon 1 including the IRE and exon 2 up to the BamHI site. A specific probe was generated for the Δ−16 IVS1 + 5 mutant. Hybridization was carried out at 60°C overnight, ribonuclease treatment was done for 30 minutes at 30°C, and the protected fragments were analyzed on a 8% denaturing polyacrylamide gel. Northern blot analysis was done with 3 to 4 μg total cytoplasmic RNA as described previously.12 Autoradiographic signals were quantified by imaging in a molecular imager (GS-250; Bio-Rad, Hercules, CA).
RT-PCR analysis
Reverse transcription was done with 1 μg mRNA, an oligo-d(T) primer, and 200 U RT. PCR amplification was carried out by using the following human β-globin–specific primers: sense, 5′-TTT TCT CGA GAC ACC ATG GTG CAC CTG ACT CCT G-3′; and antisense, 5′-CTT AGG GTT GCC CAT AAC AG-3′.
Immunoblotting
Immunoblotting and immunostaining were done with an N-terminal β-globin–specific antibody.12
Results
Identification of possible NMD substrates
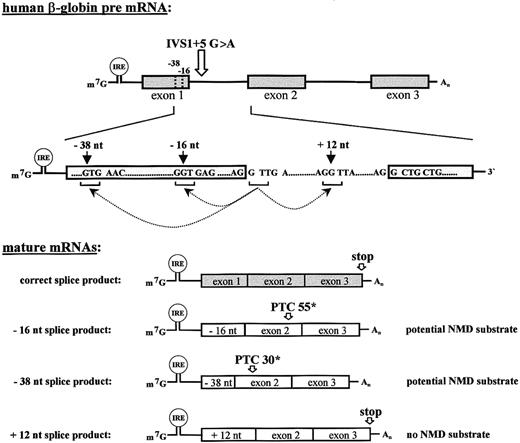
The IVS1 + 5 G>A mutation of the human β-globin gene is a rare cause of β+ thalassemia. This mutation decreases the consensus value of the splice donor site14,15 and has been shown to reduce, though not to abolish completely, the efficiency of proper splice-site selection. In affected homozygous patients, considerable amounts of β-globin and hemoglobin A are synthesized.10 This corresponds to results of RNA analyses in transfected HeLa cells, which showed substantial processing of the mutant pre-mRNAs at the normal splice site. However, most of the pre-mRNA was spliced at the cryptic sites −38, −16, and +12 (Figure1).11
Aberrant splicing of the IVS1 + 5 G>A mutated human β-globin results in 2 potential NMD substrates.
Structure of human β-globin pre-mRNAs and mature mRNAs resulting from splicing at the normal splice site and at 3 cryptic splice sites at positions −38 nts, −16 nts, and +12 nts (arrows) relative to the normal intron 1 splice donor. The ORFs of the −16 and −38 transcripts are phase-shifted at the respective intron 1 splice sites, and translation terminates at PTC 55* and at PTC 30* in exon 2, respectively. In contrast, the +12 transcript is extended by 12 nts and translation terminates at the normal stop codon. The 5′UTR of the mRNAs contains an IRE that confers a specific regulation of translation under conditions of iron depletion and repletion.
Aberrant splicing of the IVS1 + 5 G>A mutated human β-globin results in 2 potential NMD substrates.
Structure of human β-globin pre-mRNAs and mature mRNAs resulting from splicing at the normal splice site and at 3 cryptic splice sites at positions −38 nts, −16 nts, and +12 nts (arrows) relative to the normal intron 1 splice donor. The ORFs of the −16 and −38 transcripts are phase-shifted at the respective intron 1 splice sites, and translation terminates at PTC 55* and at PTC 30* in exon 2, respectively. In contrast, the +12 transcript is extended by 12 nts and translation terminates at the normal stop codon. The 5′UTR of the mRNAs contains an IRE that confers a specific regulation of translation under conditions of iron depletion and repletion.
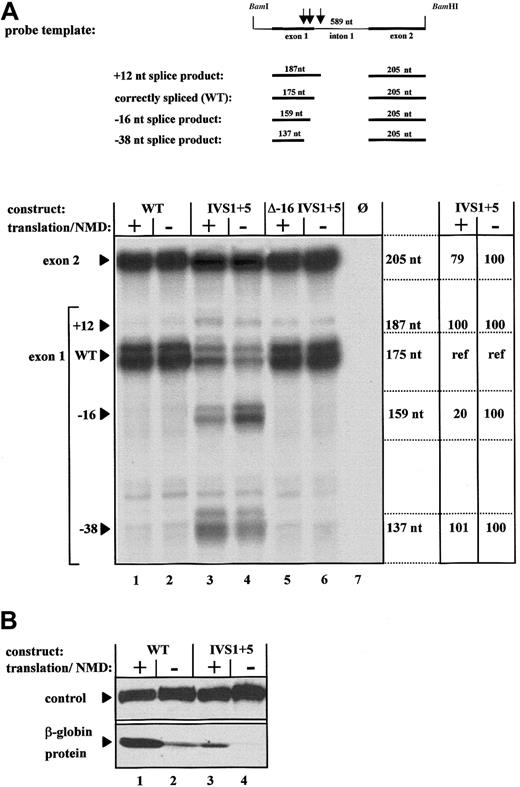
RPA of the mRNA expressed from the WT construct generated 2 major protected fragments of 175 nts and 205 nts, which represented the normal exon 1 and exon 2, respectively. A third 187-nt fragment, which was generated in trace amounts, resulted from splicing at a cryptic site 12 nts 3′ of the correct intron 1 splice donor site (Figure2A, lanes 1 and 2). RPA of the mRNA expressed from the IVS1 + 5 construct showed expression of 2 additional mRNA species represented by exon 1–specific protected fragments of 159 nts and 137 nts (Figure 2A, lanes 3 and 4). The 175-nt RPA fragment represented the only signal specific for the normally spliced mRNA. The 205-nt fragment was protected by both the normal and the abnormally spliced transcripts, whereas the 159-nt fragment and the 137-nt fragment were specific for the −16 and the −38 transcripts, respectively. Therefore, the 175-nt fragment was an ideal internal control because it reflected the normal mRNA not affected by NMD but was derived from the same pre-mRNA as the abnormally spliced transcripts that are possible NMD substrates.
Expression analysis of the aberrantly spliced human β-globin transcripts under conditions of enabled and disabled NMD.
(A) RPAs using total RNA of cells transfected with either the normal human β-globin gene (WT, lanes 1 and 2), a gene with the IVS1 + 5 G>A mutation (IVS1 + 5, lanes 3 and 4), or a gene with the IVS1 + 5 G>A mutation and inactivation of the cryptic splice site at position −16 (Δ −16 IVS1 + 5, lanes 5 and 6). NMD was specifically enabled by iron treatment of transfected cells (lanes 1, 3, and 5) or specifically disabled by iron depletion (lanes 2, 4, and 6). Lane 7 represents the analysis of RNA from untransfected HeLa cells. The protected fragments of 205 nts, 187 nts, 175 nts, 159 nts, and 137 nts result from splicing at the normal splice site or the 3 cryptic splice sites, respectively. The 175-nt exon 1 fragment is specific for the normally spliced transcript, is unaffected by activation or inactivation of NMD, and serves as an internal reference for quantification of the aberrant transcripts (lanes 3 and 4) specifically represented by the 187-nt, 159-nt, and 137-nt fragments, respectively. The 205-nt exon 2 fragment reflects the cumulative expression of all 4 mRNA species derived from the parent pre-mRNA. Expression levels of the different species of mature transcripts of the IVS1 + 5 pre-mRNA are indicated in percentages relative to the internal reference (ref). (B) Immunoblot analysis of cells transfected with constructs WT and IVS1 + 5 under conditions of enabled and disabled translation.
Expression analysis of the aberrantly spliced human β-globin transcripts under conditions of enabled and disabled NMD.
(A) RPAs using total RNA of cells transfected with either the normal human β-globin gene (WT, lanes 1 and 2), a gene with the IVS1 + 5 G>A mutation (IVS1 + 5, lanes 3 and 4), or a gene with the IVS1 + 5 G>A mutation and inactivation of the cryptic splice site at position −16 (Δ −16 IVS1 + 5, lanes 5 and 6). NMD was specifically enabled by iron treatment of transfected cells (lanes 1, 3, and 5) or specifically disabled by iron depletion (lanes 2, 4, and 6). Lane 7 represents the analysis of RNA from untransfected HeLa cells. The protected fragments of 205 nts, 187 nts, 175 nts, 159 nts, and 137 nts result from splicing at the normal splice site or the 3 cryptic splice sites, respectively. The 175-nt exon 1 fragment is specific for the normally spliced transcript, is unaffected by activation or inactivation of NMD, and serves as an internal reference for quantification of the aberrant transcripts (lanes 3 and 4) specifically represented by the 187-nt, 159-nt, and 137-nt fragments, respectively. The 205-nt exon 2 fragment reflects the cumulative expression of all 4 mRNA species derived from the parent pre-mRNA. Expression levels of the different species of mature transcripts of the IVS1 + 5 pre-mRNA are indicated in percentages relative to the internal reference (ref). (B) Immunoblot analysis of cells transfected with constructs WT and IVS1 + 5 under conditions of enabled and disabled translation.
RT-PCR analysis of the mature mRNAs generated by processing of the mutated pre-mRNA revealed 4 different transcripts (data not shown). Sequencing of these 4 RT-PCR products confirmed that the 2 smaller mRNAs were spliced at positions 16 nts (−16-nt splice product) and 38 nts (−38-nt splice product) 5′ of the normal site, respectively. The other 2 products corresponded to the normal transcript and the +12 transcript, respectively. There were no RT-PCR products corresponding to the fainter bands visible in Figure 2, which therefore probably represented unspecific RPA bands, although they were absent from RPAs of nontransfected cells (Figure 2, lane 7).
The −16 and the −38 splice products were frameshifted and expected to terminate translation at PTCs at positions 55 (PTC 55*) and 30 (PTC 30*) relative to the translation-initiation codon (Figure 1). A single pre-mRNA was therefore spliced to generate both normal mRNA and PTC-mutated mRNAs representing possible NMD substrates. Insertion of an IRE into the 5′-UTR of the β-globin gene with the IVS1 + 5 G>A mutation allowed us to regulate translation specifically and to compare directly the influence of NMD requiring cytoplasmic translation on expression of these different transcripts (Figure 1).
Our analysis of protein expression found that iron depletion resulted in a significant decrease in translation of WT mRNA (Figure 2B, lanes 1 and 2), thereby confirming the specific iron-dependent regulation of translation and the validity of the experimental system. The correctly spliced mRNA expressed by the WT and IVS1 + 5 constructs was translated in iron-replete cells, and the amount of protein detected correlated with the level of correctly spliced (WT) mRNA expression (Figure 2A and 2B, lanes 1 and 3). In contrast, the C-terminally truncated peptides encoded by the cryptically spliced transcripts were not detected by the β-globin–specific antibody, possibly because of a different fold of these peptides or altered protein stability.
NMD reduces accumulation of the major aberrant splice product
The influence of NMD on the aberrantly spliced products was internally controlled by comparison of the respective expression levels relative to the protected 175-nt fragment specific for the correctly spliced product and not affected by NMD. Under conditions of enabled translation and active NMD, the correctly spliced mRNA represented the major transcript expressed from construct IVS1 + 5 (Figure 2A, lane 3). In contrast, under conditions of disabled translation and inactive NMD, the −16-nt splice product predominated and accounted for about 80% of the transcripts (Figure 2A, lane 4). Thus, NMD resulted in an at least 5-fold decrease of the major aberrant and frameshifted transcript. Notably, inhibition of translation specifically up-regulated expression of the −16 PTC-containing transcript but not of the normal mRNAs resulting from processing of the same pre-mRNA. This result strongly suggests that the −16 transcript is down-regulated by NMD rather than other translation-dependent, but NMD-independent, mechanisms of mRNA turnover.16
Identification of an NMD-resistant, PTC-mutated splice variant
Under conditions of enabled or disabled translation, the ratio between the normal internal-control transcript indicated by the 175-nt fragment and the minor incorrectly spliced −38-nt product (137 nts) remained unchanged (Figure 2A, lanes 3 and 4). In view of current hypotheses about the mechanism of NMD,1-3,9,17,18 this finding is surprising, because the termination codon at position PTC 30* (Figure 1) is located more than 55 nts 5′ of the final exon-exon junction and is thus expected to trigger NMD.12 19
It is known that splicing recruits several RNA binding proteins with a likely role in NMD to a region of about 20 nts 5′ of a splice junction.20 We therefore tested the hypothesis that a possible interference between the strong −16 cryptic splice site and the weak −38 cryptic splice site abolishes the NMD competence of the −38 transcript. The cryptic −16 splice site in construct IVS1 + 5 was therefore inactivated by GT>CC mutagenesis. Surprisingly, the pre-mRNA encoded by this construct was processed in a similar fashion as the WT construct, with only traces of splicing at the −16 and the −38 sites (Figure 2A, lanes 5 and 6). This result suggests that the sequence at the −16 site is necessary for recruitment of splicing activity to this region.
The NMD competence of the −38 transcript was analyzed specifically in a set of constructs with deletions of intron 1 (Figure3A), which encode mRNAs with either the normal exon 1–exon 2 junction and a nonsense mutation at codon 39 (NS39 ΔIVS1) as a positive control, the −38 (−38 ΔIVS1), or the −16 exon 1–exon 2 junction (−16 ΔIVS1). Expression analysis of these constructs allowed analysis of NMD competence without splicing at the respective exon 1–exon 2 junctions. These constructs encoded mRNAs that protected the expected fragments in an RPA (data not shown). Specific analysis of NMD competence revealed the expected reduction in cytoplasmic mRNA accumulation under conditions of enabled translation for the NS39 ΔIVS1 and the −16 ΔIVS1 constructs (Figure 3B, lanes 1 and 2 and lanes 3 and 4). In contrast, expression of the −38 ΔIVS1 construct was not affected by activation or inactivation of translation and showed an expression pattern similar to that of the WT construct (Figure 3B, lanes 5 and 6 and lanes 7 and 8), thereby confirming the results obtained with IVS1 + 5 (Figure 2).
Expression analysis of human β-globin constructs with deletions of intron 1.
(A) Schematic representation of the constructs used for transfection. Construct NS39 ΔIVS1 contains a nonsense mutation at position 39 (arrow). In constructs −16 ΔIVS1 and −38 ΔIVS1, intron 1 and 16 nts or 38 nts of the exon 1 sequence are deleted, resulting in a transcript with termination codons at positions 55 (PTC 55*) and 30 (PTC 30*), respectively. (B) Northern blot analysis with a β-globin–specific exon 3 probe (top panel) of total cytoplasmic RNA extracted from cells transfected with constructs NS39 ΔIVS1 (lanes 1 and 2), −16 ΔIVS1 (lanes 3 and 4), −38 ΔIVS1 (lanes 5 and 6), and the normal β-globin construct (WT, lanes 7 and 8) under conditions of enabled and disabled translation. The WT-ΔIVS1-CAT plasmid was cotransfected to control for transfection efficiency. The ratio of WT-RNA expression under conditions of active and inactive translation (lanes 7 and 8) was used to control for an unspecific, translation-dependent slight increase in mRNA expression. Values are the mean results from 3 independent experiments after normalization for transfection efficiency and after considering the unspecific (∼ 20%) reduction in RNA expression under conditions of translation inhibition. The immunoblot (bottom panel) shows the specific suppression of translation under conditions of iron depletion.
Expression analysis of human β-globin constructs with deletions of intron 1.
(A) Schematic representation of the constructs used for transfection. Construct NS39 ΔIVS1 contains a nonsense mutation at position 39 (arrow). In constructs −16 ΔIVS1 and −38 ΔIVS1, intron 1 and 16 nts or 38 nts of the exon 1 sequence are deleted, resulting in a transcript with termination codons at positions 55 (PTC 55*) and 30 (PTC 30*), respectively. (B) Northern blot analysis with a β-globin–specific exon 3 probe (top panel) of total cytoplasmic RNA extracted from cells transfected with constructs NS39 ΔIVS1 (lanes 1 and 2), −16 ΔIVS1 (lanes 3 and 4), −38 ΔIVS1 (lanes 5 and 6), and the normal β-globin construct (WT, lanes 7 and 8) under conditions of enabled and disabled translation. The WT-ΔIVS1-CAT plasmid was cotransfected to control for transfection efficiency. The ratio of WT-RNA expression under conditions of active and inactive translation (lanes 7 and 8) was used to control for an unspecific, translation-dependent slight increase in mRNA expression. Values are the mean results from 3 independent experiments after normalization for transfection efficiency and after considering the unspecific (∼ 20%) reduction in RNA expression under conditions of translation inhibition. The immunoblot (bottom panel) shows the specific suppression of translation under conditions of iron depletion.
The expression levels of the mRNAs encoded by the ΔIVS1 series of constructs with or without truncated ORFs were influenced by the activity of translation in a similar fashion as the mRNAs encoded by the intron 1–containing series of constructs (Figure 2A). This finding suggests that the −38 transcript is insensitive to NMD and that this feature is independent of possible differences in spliceosome recruitment to the −16 or −38 site, thereby indicating that the unexpected mRNA accumulation of the −38 transcript is not influenced by differences in RNA processing. Moreover, our analysis of cytoplasmic mRNA showed comparable levels of expression of the −16-nt and the −38-nt transcripts under conditions of inactivated NMD (Figure 3B, lanes 4 and 6), a finding suggesting that the observed NMD insensitivity of the −38-nt transcript was unlikely to have been caused by differences in nucleocytoplasmic transport.
Translational read-through or reinitiation of translation may be alternative explanations for the lack of NMD sensitivity of the −38 transcript. However, translational read-through appears unlikely because the −38 transcript contains multiple PTCs at NMD-competent positions.12 Although NMD requires only a low level of premature termination of translation,21 reinitiation at downstream AUG codons was previously found to stabilize PTC-mutated transcripts.22,23 The ORF of the −38 transcript terminates at codon 30 relative to the translation-initiation codon (Figure 1). The normal β-globin ORF contains an AUG at a position 34 nts 3′ of the PTC in the −38 transcript, and this might be used for reinitiation. This putative transcript would be expected to terminate at the physiologic termination codon. In addition, there are 3 other AUG sites—26 nts, 59 nts, and 89 nts 3′ of the −38 PTC in one of the alternative ORFs—that are expected to terminate at an NMD-incompetent position in the 3′ region of exon 2.12
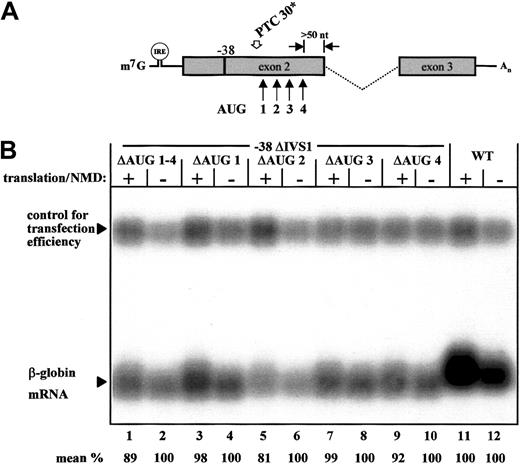
We tested directly the hypothesis that NMD resistance of the −38 transcript is caused by reinitiation of translation at downstream AUG codons by using mutagenesis of the respective sites individually (Figure 4; ΔAUG 1, ΔAUG 2, ΔAUG 3, and ΔAUG 4) or together (Figure 4; ΔAUG 1-4). This approach specifically assessed the effect of downstream AUG codons on the observed NMD resistance of the −38 transcript. Northern blot analysis found that neither a combined inactivation of these AUG sites (Figure4B, lanes 1 and 2) nor any of the single AUG inactivations restored NMD competence (Figure 4B, lanes 3-10). We therefore conclude that the AUG sites 3′ in the PTC in the −38 transcript do not inhibit NMD and that translational reinitiation at downstream AUG codons does not confer resistance to NMD of this transcript.
Expression analysis of −38 ΔIVS1 constructs with inactivation of downstream AUG codons.
(A) The 4 downstream AUG codons that may be used as translation-reinitiation sites in the −38 transcript are indicated by arrows. (B) Northern blot analysis with a β-globin–specific exon 3 probe of the normal construct (WT) and the −38 ΔIVS1 constructs that lack either all 4 downstream AUG codons (ΔAUG 1-4, lanes 1 and 2) or single AUG codons (ΔAUG 1, lanes 3 and 4; ΔAUG 2, lanes 5 and 6; ΔAUG 3, lanes 7 and 8; and ΔAUG 4, lanes 9 and 10). Translation and NMD were regulated specifically by iron depletion and repletion. The WT-ΔIVS1-CAT plasmid was cotransfected to control for transfection efficiency. The ratio of WT-RNA expression under conditions of active and inactive translation (lanes 11 and 12) was used to control for an unspecific translation dependent slight increase in mRNA expression. Values are the mean results from 3 independent experiments after normalization for transfection efficiency and after considering the unspecific (∼ 20%) reduction in RNA expression under conditions of translation inhibition.
Expression analysis of −38 ΔIVS1 constructs with inactivation of downstream AUG codons.
(A) The 4 downstream AUG codons that may be used as translation-reinitiation sites in the −38 transcript are indicated by arrows. (B) Northern blot analysis with a β-globin–specific exon 3 probe of the normal construct (WT) and the −38 ΔIVS1 constructs that lack either all 4 downstream AUG codons (ΔAUG 1-4, lanes 1 and 2) or single AUG codons (ΔAUG 1, lanes 3 and 4; ΔAUG 2, lanes 5 and 6; ΔAUG 3, lanes 7 and 8; and ΔAUG 4, lanes 9 and 10). Translation and NMD were regulated specifically by iron depletion and repletion. The WT-ΔIVS1-CAT plasmid was cotransfected to control for transfection efficiency. The ratio of WT-RNA expression under conditions of active and inactive translation (lanes 11 and 12) was used to control for an unspecific translation dependent slight increase in mRNA expression. Values are the mean results from 3 independent experiments after normalization for transfection efficiency and after considering the unspecific (∼ 20%) reduction in RNA expression under conditions of translation inhibition.
Discussion
The function of the NMD pathway has been shown to be important in humans,7,8 mice,1,24 andCaenorhabditis elegans.5,25 NMD contributes to the quality control of gene expression by monitoring mRNAs for the presence of PTCs and subsequent degradation of affected mRNAs.2-4,9 In human genetic disorders, PTCs are commonly caused by nonsense and frameshift mutations. PTCs can also be introduced by errors in the normal gene-expression pathway, such as unproductive DNA rearrangements in the immune system,26,27or by aberrant splicing. About 15% of all point mutations causing human genetic diseases result in mRNA splicing errors,28-30 and about 30% of all mutations in the β-globin gene result in errors of mRNA processing.31Mutations both in exons32,33 and in introns34can induce activation of cryptic splice sites, intron retention, and exon skipping, thereby creating possible NMD substrates. Moreover, during normal mRNA synthesis, splicing errors are likely to occur at a low frequency. In this study, we assessed directly the specific capacity of the NMD surveillance mechanism to recognize and eliminate faulty splice products as a physiologic function of NMD in the quality control of RNA processing.
Our data show that when NMD is disabled, expression of a human β-globin gene with the IVS1 + 5 G>A mutation causes the predominant accumulation of a faulty and frameshifted mRNA expected to encode anomalous and C-terminally truncated β-globin chains. The specific effect of NMD in RNA surveillance was illustrated in our model system by the at least 5-fold reduction in accumulation of one of the abnormal mRNAs and a relative increase in normal mRNA. In β-thalassemia, accumulation of mRNAs that encode C-terminally truncated β-globin chains can act in a dominant-negative fashion and cause symptoms in heterozygous carriers.7,8 Elimination of frameshifted mRNAs that result from cryptic splicing events is therefore likely to represent an important function of NMD in thalassemia and other genetic disorders.35
In contrast, the second possible NMD substrate, the −38-nt splice product, escaped RNA surveillance. This was an unexpected finding because previous studies showed that inefficiently and alternatively spliced transcripts are surveyed by NMD.25,36,37Translational read-through is unlikely to stabilize this transcript because of the presence of several downstream PTCs. Moreover, our data indicate that neither abnormalities of splicing nor translation reinitiation at downstream AUG codons is a plausible explanation for the NMD resistance of this transcript (Figures 2-4). Contrary to results predicted by the generally used model,1-3,9,17 18our data showed that aberrantly spliced transcripts are not always targeted by the NMD machinery. This finding suggests the existence of additional, unidentified determinants that function to modulate the NMD sensitivity of these transcripts.
Identification of possible NMD targets that are resistant to NMD has contributed substantially to the mechanistic understanding of NMD. Examples are nonsense mutations in the 3′ terminal exon and the final 50 or so nts of the penultimate exon of the pre-mRNA that have permitted identification of the splicing dependence of NMD.12,19,38,39 In the example of an NMD-resistant nonsense mutation of nibrin gene mRNA, reinitiation of translation by means of an internal ribosomal entry site23underlined the dependence of NMD on the function of a hypothetical posttermination surveillance complex.3
In this context, identification of the NMD-resistant −38 transcript generated from the same pre-mRNA as the NMD-sensitive −16 transcript is particularly intriguing. NMD-resistant, PTC-mutated mRNAs were previously observed in several genes, including the β-globin, von Willebrand factor, cystic fibrosis transmembrane conductance regulator, and low-density lipoprotein receptor.40-43 Furthermore, NMD resistance is likely to be important for expression of physiologic transcripts. Apolipoprotein (APO) B mRNA represents an interesting example for this class of transcripts in that RNA editing of an upstream stop codon enables expression of the APO-B100 isoform.44 It is possible that some genes containcis-acting sequences that confer resistance to NMD. Such sequences have been identified in yeast,45-47 but are unlikely to explain differences in NMD efficiency of transcripts with different PTC mutations that result from processing of the same pre-mRNA (as reported here). An intriguing possibility for the NMD resistance of the −38 transcript is that the 22 nts included in the NMD-sensitive −16 transcript but missing from the −38 transcript are required to enable the NMD pathway. If this is true, the 22-nt region would be a necessary, but not sufficient, exonic element for occurrence of NMD. Alternatively, the differences in NMD sensitivity of the −38 and the −16 transcripts may be related to their translation in different ORFs. In either case, our model system offers a useful tool for defining the mechanisms that allow specific transcripts to circumvent the NMD pathway.
Supported by the Deutsche Forschungsgemeinschaft and the Fritz Thyssen Stiftung.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andreas E. Kulozik, Department of Pediatric Oncology, Hematology and Immunology, University of Heidelberg and University of Heidelberg–EMBL Molecular Medicine Partnership Unit, Germany; e-mail: andreas_kulozik@med.uni-heidelberg.de; or Matthias W. Hentze, EMBL and University of Heidelberg–EMBL Molecular Medicine Partnership Unit, Im Neuenheime Feld 150, Heidelberg, Germany; e-mail:matthias.hentze@embl-heidelberg.de.