A single nucleotide polymorphism (SNP) at position 196 in the β3 integrin causes a Leu33Pro substitution in the mature protein. Alloimmunization against the β3Leu33 form (human platelet antigen [HPA]-1a, PlA1, Zwa) in patients who are β3Pro33 homozygous (HPA-1b1b, PlA2A2, Zwbb) causes neonatal alloimmune thrombocytopenia, posttransfusion purpura, or refractoriness to platelet transfusion. Studies with recombinant proteins have demonstrated that amino acids 1 to 66 and 288 to 490 of the β3 integrin contribute to HPA-1a epitope formation. In determining the HPA-1a status of more than 6000 donors, we identified a donor with an HPA-1aweak phenotype and an HPA-1a1b genotype. The platelets from this donor had normal levels of surface αIIbβ3 but reacted only weakly with monoclonal and polyclonal anti–HPA-1a by whole blood enzyme-linked immunosorbent assay (ELISA), flow cytometry, and sandwich ELISA. We reasoned that an alteration in the primary nucleotide sequence of the β3Leu33 allele of this donor was disrupting the HPA-1a epitope. In agreement with this hypothesis, sequencing platelet RNA-derived αIIb and β3 cDNA identified a novel G/A SNP at position 376 of the β3 integrin that encodes for an Arg93Gln replacement in the β3Leu33 allele. Coexpression of the β3Leu33Gln93 encoding cDNA in Chinese hamster ovary cells with human αIIb cDNA showed that the surface-expressed αIIbβ3 reacted normally with β3 integrin–specific monoclonal antibodies but only weakly with monoclonal anti-HPA-1a. Our results show that an Arg93Gln mutation in the β3Leu33 encoding allele disrupts the HPA-1a epitope, suggesting that Arg93 contributes to the formation of the HPA-1a B-cell epitope.
Introduction
The β3 integrin subunit (glycoprotein [GP] IIIa, CD41) forms a heterodimeric complex with the αIIb integrin subunit (GPIIb) on the surface of platelets (αIIbβ3, GPIIb/IIIa, CD61/41) and functions as a major fibrinogen receptor. Activation of αIIbβ3 occurs through so-called inside-out signaling that follows the binding of platelet receptors to components of the subendothelial cell matrix (eg, the binding of α2β1 and GPVI to collagen) or soluble ligands (eg, adenosine diphosphate and thrombin). The activated conformation of αIIbβ3 binds fibrinogen, fibronectin, and vitronectin and has a pivotal role in clot formation after blood vessel damage.1 2
The gene encoding β3 integrin has several single nucleotide polymorphisms (SNPs) that result in single amino acid substitutions of immunologic, and possibly functional, consequence.3Platelet alloantigen systems encoded by SNPs in the β3 integrin gene are of clinical relevance. The C196T SNP, encoding for a Leu33Pro substitution, is the most immunogenic human platelet alloantigen (HPA) system.4 In β3Leu33-negative (ie, β3Pro33 homozygous), HLA-DRB3*0101–positive persons, exposure to the β3Leu33 form is highly immunogenic and alloimmunization causes neonatal alloimmune thrombocytopenia, posttransfusion purpura, and platelet refractoriness.3,5-7 Alloimmunization against β3Leu33 occurs in 1 in 365 pregnant women, and β3Leu33-specific maternal alloantibodies (anti–HPA-1a) cause severe thrombocytopenia in 1 in 1100 neonates.8,9 In such cases, the treatment of choice is β3Leu33-negative donor platelets, the provision of which requires the phenotyping of large numbers of donors.5,10We have previously reported on a recombinant human immunoglobulin (Ig)G1 specific for β3Leu33, which can be used for large-scale donor phenotyping.11-13 In the process of phenotyping more than 6000 donors using this assay, we identified one donor with a β3Leu33weak phenotype but a heterozygous genotype. Here we describe the molecular basis of this unique phenotype, suggesting that Arg93 of the β3 integrin contributes to the formation of the HPA-1a B-cell epitope.
Patients, materials, and methods
Donor samples
More than 6000 EDTA-anticoagulated whole blood donor samples were β3Leu33 phenotyped by enzyme-linked immunosorbent assay (ELISA) with our recombinant antibody CAMTRAN-007 as described previously.11 A single donor (Donor A) with a β3Leu33weak phenotype and a heterozygous genotype was identified. Genomic DNA samples from healthy apheresis donors were from the National Blood Service donor DNA repository. Informed consent was obtained for all samples.
Antibodies
Mouse monoclonal antibodies (mAbs) specific for platelet integrins and glycoproteins were obtained as follows. Anti-αIIbβ3 integrin clones RFGP56 and NIBSC-85/661 have been reported in detail elsewhere14,15; anti-β3 integrin clone Y2/51 from DAKO (Cambridge, United Kingdom); anti-β3 integrin mAb P37 was a kind gift from Dr J. Gonzalez-Rodriguez (Instituto de Quimica Fisica, Madrid, Spain). Recombinant human IgG1 anti-β3Leu33 clone CAMTRAN-00711,13 and mouse mAb clone 9E10 specific for the c-myc tag were provided by The International Blood Group Reference Laboratory (Bristol, United Kingdom). Recombinant human IgG1 anti-β3Leu33 clones 19-7 and 23-15 were a kind gift from Dr Louis Thiobault (Héma-Québec, Canada).16 Human polyclonal sera were from the National Blood Service serum archives and were obtained from patients previously referred for investigation of neonatal alloimmune thrombocytopenia.
β3Leu33 typing
Whole blood phenotyping using the recombinant human IgG1 anti-β3Leu33 CAMTRAN-007 was performed as described previously.11 Results were interpreted as β3Leu33 negative if the optical density (O.D.) was less than 0.2 and as positive if the O.D. was more than 1.2. Any O.D. between these values was considered indeterminate, and repeat testing was performed. Polymerase chain reaction with sequence-specific primers (PCR–SSP) was performed according to the method of Cavanagh et al.17
Monoclonal antibody immobilization of platelet antigens
The binding of human polyclonal anti-β3Leu33 and anti-β3Pro33 was studied using monoclonal antibody immobilization of platelet antigens (MAIPA) with platelets from healthy donors and Donor A.18 19 MAIPA was performed using platelet-rich plasma obtained from citrate-anticoagulated donor blood samples and the mAb NIBSC-85/661 to specifically capture αIIbβ3 from lysed platelets. Bound human IgG was revealed with an alkaline-phosphatase–labeled goat anti–human IgG (Jackson Immunoresearch, West Grove, PA) using Sigma-104 phosphatase substrate. O.D. was read on an ELISA plate reader (Tecan Spectra) at 405 nm. Sera from nontransfused group AB male blood donors were used as negative controls.
Platelet immunofluorescence test
Binding of antibodies to platelets was detected using the platelet immunofluorescence test.20 Stained platelets (10 000) were analyzed on a Coulter XL running System II software (Beckman-Coulter, High Wycombe, United Kingdom). Binding of human and murine antibodies was detected using fluorescein isothiocyanate (FITC)–labeled rabbit anti–human IgG (DAKO) and rabbit anti–mouse IgG (DAKO), respectively. Whole blood HPA-1a phenotyping was performed with FITC-labeled CAMTRAN-007 as described previously.13
cDNA amplification and sequencing
Total platelet RNA was prepared from 109 platelets using 1 mL RNA STAT-60 following the manufacturer's protocol (AMS Biotechnology, Witney, United Kingdom). Isolated RNA was resuspended in 100 μL diethyl-pyrocarbonate (DEPC)–treated water and used as a template for cDNA synthesis, as follows. Random hexamers (3 μg) and 20 μL platelet RNA were incubated at 70°C for 10 minutes and then immediately transferred to ice. Forty units SuperRT reverse transcriptase, 80 U RNAsin, 1 mM each dNTP, and DEPC-treated water to give a total volume of 50 μL were added, and the mixture was incubated at 42°C for 40 minutes. Resultant cDNA was used as a template for PCR amplification of both αIIb and β3 integrins. Amplification reactions were performed in a total volume of 50 μL containing 200 μM each dNTP, 1.5 mM MgCl2, 15 pmol each primer, 5 U Taq polymerase, and 5 μL cDNA. The mixture was incubated at 95°C for 5 minutes, and then 30 cycles consisting of 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute were performed. Four and 5 overlapping fragments spanning the complete open-reading frames (ORFs) of αIIb and β3 integrins were amplified, respectively.
Amplified DNA was purified from agarose gels using the QIAquick gel extraction kit (Qiagen, Crawley, United Kingdom) and was directly sequenced using the Thermosequenase dye terminator cycle sequencing kit (Amersham Life Science, Cleveland, OH). Sequences obtained were compared to published αIIb and β3 integrin sequences (accession numbers J02764 and M20311, respectively).21 22 The β3 integrin PCR product obtained with primers 5′-GGCGGACGAGATGCGAGC-3′ and 5′-GCATCTCGGTTCCGTGACAC-3′ containing the SNPs C196T and G376A was cloned into the TA vector according to the manufacturer's protocol (Invitrogen BV, Groningen, The Netherlands). Recombinant clones were sequenced to confirm the presence of the observed SNPs.
Construction of the mutant β3 integrin cDNA
For generation of the pcDNA 3.1(−)Zeo β3Leu33Gln93 construct, a 500-bp XbaI/KpnI wild-type (WT; Leu33Arg93) fragment was replaced with the fragment encoding Gln93. The β3Pro33Arg93 construct was generated by site-directed mutagenesis of the β3Leu33Arg93 construct using the Altered Sites in vitro mutagenesis kit and the mismatched primer 5′-TGGTGCTCTGATGAAGCTTTGCCTCCGGGCTCA-3′ according to the manufacturer's instructions (Promega, Southampton, United Kingdom). The above primer also introduces a silent mutation encoding aHindIII restriction site (underlined) that allows the rapid identification of recombinant mutant clones. The full-length β3 integrin cDNA thus obtained (Pro33Arg93) was excised from the pAlter phagemid and cloned into the pBJ1 mammalian cell expression vector. All constructs were verified by nucleotide sequencing before transfection.
Transfection and selection of stable cell clones
Plasmids for transfection were mixed with 40 μg LipofectAMINE (Life Technologies, Merelbeke, Belgium) in a final volume of 200 μL Iscoves modified Dulbecco medium (IMDM). The mixture was added to either nontransfected Chinese hamster ovary (CHO) cells or cells that had been pretransfected with human αIIb integrin cDNA and grown to 60% confluence in 100-mm tissue culture plates. Twenty-four hours after transfection, fetal calf serum was added to the culture medium; 48 hours after transfection, the medium was replaced with selective medium (IMDM containing 10% fetal calf serum and 0.8 mg/mL zeocin [Invitrogen]). Positive transfectants were analyzed with the anti-β3 integrin mAb P37 for cell surface expression of the recombinant human β3 integrin, associated with either the endogenous hamster αv or with human αIIb integrins. Stable transfectants were subcloned by limiting dilution and controlled for cell surface expression of human β3 integrin.
RT-PCR and cDNA sequencing of Chinese hamster ovary transfectants
Total RNA was isolated from 5 × 106 transfected cells according to the method of Chomczynski and Sacchi.23First-strand cDNA synthesis from 2 μg total RNA was directed with oligo(dT) primer using an RNA-PCR kit (Perkin Elmer). The coding sequence, corresponding to the mutated β3 integrin region, was amplified using β3-specific primers, and products were analyzed by agarose gel electrophoresis and directly sequenced using thefmol DNA sequencing kit (Promega).
Western blot analysis
Platelets or cultured CHO cells were washed and lysed in 300 μL lysis buffer (150 mM NaCl, 20 mM Tris, pH 8, 1 mM CaCl2, 1 mM MgCl2, 1% Triton X-100, 10 μg/mL leupeptin, 10 μg/mL pepstatin A, 50 μM AEBSF). Lysates were cleared by centrifugation at 12 000 rpm for 10 minutes at 4°C, and the protein concentration was determined using the BCA protein assay (Pierce, Rockford, IL). Fifty micrograms total cell lysate was then resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and was transferred onto a nitrocellulose membrane. The membrane was blocked for 1 hour in blocking buffer (TBS containing 0.1% [vol/vol] Tween and 5% [wt/vol] nonfat dry milk) and was incubated overnight with primary antibody diluted in blocking buffer. After several washes, the membrane was incubated for 1 hour with horseradish peroxidase–conjugated sheep anti-mouse IgG diluted in blocking buffer (Amersham Pharmacia Biotech, Roosendaal, The Netherlands). Membranes were then washed in TBS, and bound antibody was visualized using enhanced chemiluminescence according to the manufacturer's instructions (Pierce).
Immunofluorescence and flow cytometric analysis of CHO transfectants
Flow cytometry was used to detect antibody binding to transfected CHO cells. Briefly, selected transfectants were detached from culture plates with EDTA buffer and were washed twice in incubation buffer (137 mM NaCl, 5 mM KCl, 50 mM HEPES, 1 mg/mL glucose, pH 7.4). Transfected cells (5 × 105) were incubated on ice for 1 hour with directly labeled antibodies. Cells were then washed once, resuspended in incubation buffer, and analyzed on an Epics XL flow cytometer (Beckman-Coulter). Phycoerythrin-labeled anti–human CD61 (PharMingen, San Diego, CA) was used to determine total β3 expression, whereas expression of the HPA-1a epitope was determined by staining with FITC-labeled CAMTRAN-007.
Taqman-based genotyping for the Gln93-encoding allele
Genomic DNA samples were genotyped for the WT Arg93 and novel Gln93-encoding alleles using the primers 5′-TCAAGTCAGTCCCCAGAGGATT-3′ and 5′-AGGTCTCTCCCCGCAAAGAG-3′ with the FAM-labeled WT probe 5′-TCCGGCTCCGGCCAGGTAG-3′ and the VIC-labeled Gln93-specific probe 5′-CTCCGGCTCCAGCCAGGTAGG-3′. The polymorphic nucleotide is highlighted in bold. Amplification reactions were performed with 900 nM each primer and 50 nM each probe at an annealing temperature of 64°C. Allelic discrimination was subsequently determined by a post-PCR plate read using a Perkin Elmer 7700 (Applied Biosystems, Warrington, United Kingdom).
Results
Donor screening
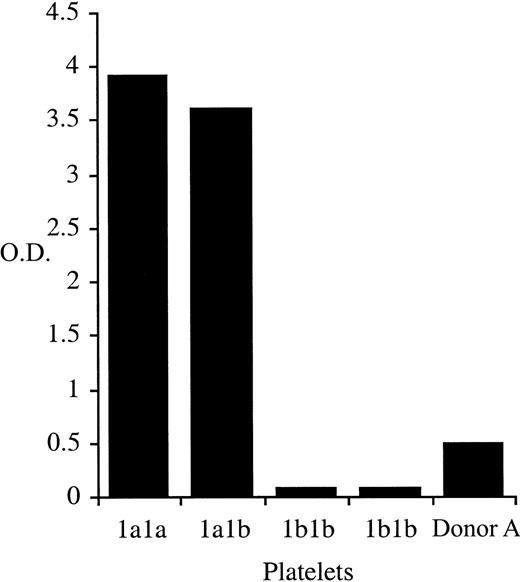
One hundred β3Leu33-negative blood donors and one donor (Donor A) with a repeatedly indeterminate phenotype were identified after the automated phenotyping of 6311 donor samples using whole blood phenotyping ELISA (Figure 1). Of the 100 β3Leu33-negative donors, 54 were anti-cytomegalovirus negative and therefore were eligible for enrollment on the β3Leu33 (HPA-1a)–negative therapeutic platelet panel. Genomic DNA was obtained from these 54 donors and from Donor A. Genotypes of these 55 samples were determined using PCR-SSP; 54 were homozygous for the β3Pro33-encoding allele (data not shown), but Donor A genotyped as β3Leu33Pro33 heterozygous by PCR-SSP (Figure2). This heterozygous genotype was confirmed by Taqman-based genotyping and direct sequencing of β3 integrin cDNA (data not shown).
Whole blood β3Leu33 phenotyping results for donor Donor A.
Whole blood samples were phenotyped for the presence of β3Leu33 as described. Samples obtained from Donor A repeatedly gave an HPA-1aweak phenotype. Control samples of 2 β3Pro33 homozygous (HPA-1b1b) and one each of β3Leu33Pro33 heterozygous (HPA-1a1b) and β3Leu33 homozygous (HPA-1a1a) were included in each assay.
Whole blood β3Leu33 phenotyping results for donor Donor A.
Whole blood samples were phenotyped for the presence of β3Leu33 as described. Samples obtained from Donor A repeatedly gave an HPA-1aweak phenotype. Control samples of 2 β3Pro33 homozygous (HPA-1b1b) and one each of β3Leu33Pro33 heterozygous (HPA-1a1b) and β3Leu33 homozygous (HPA-1a1a) were included in each assay.
Genotype of Donor A by PCR-SSP.
Genomic DNA samples from Donor A, his mother (Donor C), and his son (Donor B) were HPA-1 genotyped by PCR-SSP. The 3 control samples (HPA-1a1a, 1a1b, and 1b1b) show the expected amplicons of 90 bp for HPA-1 (filled arrows) and control amplicons obtained with primers specific for human growth hormone (429 bp; open arrows). The PCR-SSP genotypes of the test samples are HPA-1a1b, HPA-1a1a, and HPA-1a1b for Donor A, Donor C, and Donor B, respectively.
Genotype of Donor A by PCR-SSP.
Genomic DNA samples from Donor A, his mother (Donor C), and his son (Donor B) were HPA-1 genotyped by PCR-SSP. The 3 control samples (HPA-1a1a, 1a1b, and 1b1b) show the expected amplicons of 90 bp for HPA-1 (filled arrows) and control amplicons obtained with primers specific for human growth hormone (429 bp; open arrows). The PCR-SSP genotypes of the test samples are HPA-1a1b, HPA-1a1a, and HPA-1a1b for Donor A, Donor C, and Donor B, respectively.
Characterization of surface expression of αIIbβ3 and β3Leu33 epitope on Donor A's platelets
The cell surface level of αIIbβ3 was estimated by flow cytometry using saturating concentrations of mAb Y2/51 and a commercial phenotyping kit (ADIAflo; American Diagnostica, Greenwich, CT). Reactivity with mAb Y2/51, which recognizes a linear β3 epitope, was comparable to that obtained with control platelets indicating normal levels of β3 on Donor A's platelets (Table1). The level of αIIbβ3 expression was within the normal range of the ADIAflo phenotyping kit (data not shown).
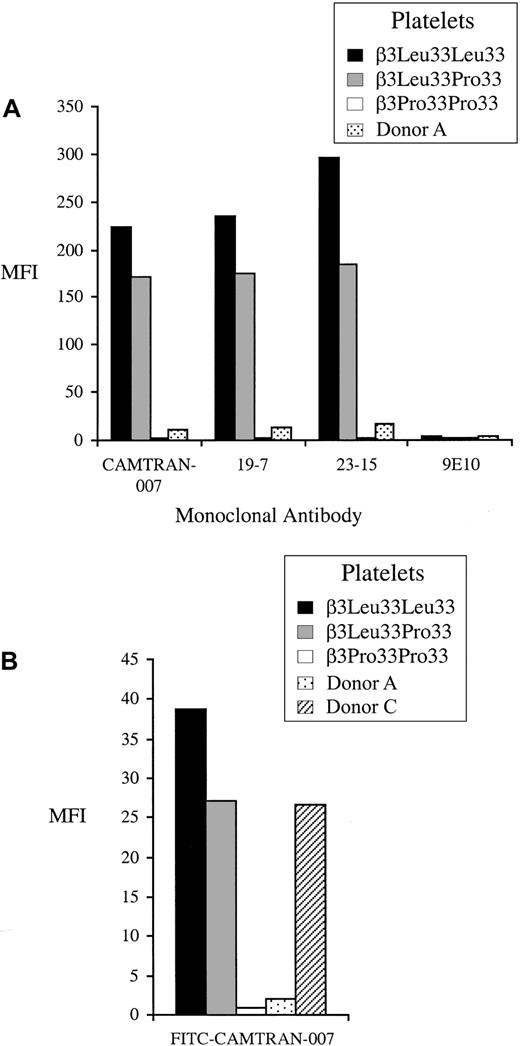
Expression of the β3Leu33 (HPA-1a) epitope on Donor A's platelets was studied in detail by flow cytometry using 3 recombinant human IgG1 β3Leu33 antibodies (CAMTRAN-007, 19-7, and 23-15).13 16Median fluorescence intensity obtained with these mAbs (Figure3A) was significantly reduced in comparison to that observed with platelets from control β3Leu33Pro33 heterozygous donors. A reduced reactivity of Donor A's platelets with FITC-labeled CAMTRAN-007 was also observed in the whole blood, flow cytometry–based phenotyping assay (Figure 3B). The reduced reactivity of Donor A's platelets with anti-β3Leu33 was also observed with polyclonal antisera in MAIPA, suggesting that the epitope recognized by polyclonal and monoclonal anti-β3Leu33 is disrupted (Figure4). Normal reactivity was observed with 2 polyclonal β3Pro33-specific antisera with Donor A's platelets when compared to those obtained with a control heterozygous donor, indicating normal expression of the β3Pro33 epitope on Donor A's platelets.
Reactivity of platelets from Donor A with monoclonal anti-β3Leu33 in flow cytometry.
(A) Washed platelets from Donor A and control donors were stained with 3 human β3Leu33-specific mAbs (CAMTRAN-007, 19-7, and 25-13) in a platelet immunofluorescence test. All 3 mAbs show significantly reduced binding to the platelets from Donor A compared to the heterozygous control. (B) Whole blood β3Leu33 phenotyping was performed using FITC-labeled CAMTRAN-007 as described. Reactivity for Donor A is reduced compared to the β3Leu33 homozygous and heterozygous controls. Reactivity for Donor C is reduced to the level observed with a heterozygote. Median fluorescence intensity (MFI) is presented in each case.
Reactivity of platelets from Donor A with monoclonal anti-β3Leu33 in flow cytometry.
(A) Washed platelets from Donor A and control donors were stained with 3 human β3Leu33-specific mAbs (CAMTRAN-007, 19-7, and 25-13) in a platelet immunofluorescence test. All 3 mAbs show significantly reduced binding to the platelets from Donor A compared to the heterozygous control. (B) Whole blood β3Leu33 phenotyping was performed using FITC-labeled CAMTRAN-007 as described. Reactivity for Donor A is reduced compared to the β3Leu33 homozygous and heterozygous controls. Reactivity for Donor C is reduced to the level observed with a heterozygote. Median fluorescence intensity (MFI) is presented in each case.
β3Leu33 phenotyping by MAIPA with polyclonal antisera.
MAIPA was performed using platelets from Donor A and control donors with 3 β3Leu33-specific antisera (CP, MC, RF) and 2 β3Pro33 antisera (CH, DW). Sera from 2 nontransfused group AB male blood donors (AB1 and AB2) were used as negative controls. Donor A shows normal reactivity with both β3Pro33 antisera but strongly reduced reactivity with all 3 β3Leu33 antisera (HPA-1aweak).
β3Leu33 phenotyping by MAIPA with polyclonal antisera.
MAIPA was performed using platelets from Donor A and control donors with 3 β3Leu33-specific antisera (CP, MC, RF) and 2 β3Pro33 antisera (CH, DW). Sera from 2 nontransfused group AB male blood donors (AB1 and AB2) were used as negative controls. Donor A shows normal reactivity with both β3Pro33 antisera but strongly reduced reactivity with all 3 β3Leu33 antisera (HPA-1aweak).
αIIb and β3 cDNA sequence analysis
Nine cDNA fragments encoding the complete ORFs of αIIb and β3 integrins were amplified from RNA extracted from Donor A's platelets (data not shown). Sequencing of PCR products revealed a single G376A SNP resulting in a β3Arg93Gln substitution, for which Donor A was heterozygous (Figure 5). The presence of the G376A SNP was confirmed by reverse transcription (RT)–PCR using 2 separate platelet RNA preparations (data not shown) and by sequencing the PCR product after cloning it into the TA vector. Moreover, both clones with the 376A nucleotide, encoding Gln93, also encoded Leu at position 33.
A G376A polymorphism was identified in the Leu33 encoding β3 integrin allele from Donor A.
The complete ORFs of both αIIb and β3 integrins were sequenced from cDNA obtained from the platelets of Donor A. Sequencing identified a single, novel polymorphism in the β3 cDNA with adenine or guanine at position 376, for which Donor A is heterozygous. This SNP, indicated by the arrow, results in the replacement of arginine with glutamine at position 93 in the β3Leu33 allele.
A G376A polymorphism was identified in the Leu33 encoding β3 integrin allele from Donor A.
The complete ORFs of both αIIb and β3 integrins were sequenced from cDNA obtained from the platelets of Donor A. Sequencing identified a single, novel polymorphism in the β3 cDNA with adenine or guanine at position 376, for which Donor A is heterozygous. This SNP, indicated by the arrow, results in the replacement of arginine with glutamine at position 93 in the β3Leu33 allele.
Expression of the recombinant mutant β3 integrin subunits in Chinese hamster ovary cells
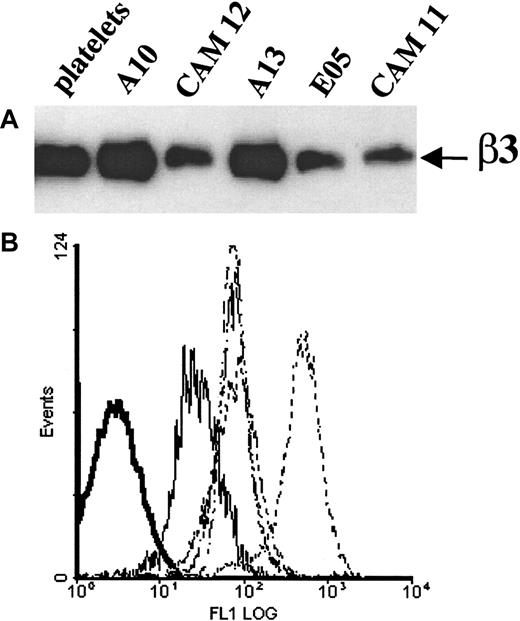
After stable transfection of the mutant β3Leu33Gln93 encoding cDNA into CHO cells, 2 cell lines were produced expressing β3Leu33Gln93 complexed with either hamster αv or human αIIb integrins, Cam11 and Cam12, respectively. The presence of the correct β3 integrin (Arg93 or Gln93) in transfected cell lines was confirmed by RT-PCR and direct sequencing of the amplified cDNA fragment (data not shown). Analysis of the expression of the recombinant β3 integrin subunits by Western blot with mAb P37 showed that the Leu33Gln93-encoding β3 integrin was expressed in both Cam11 and Cam12 clones. In addition, Western blotting showed that β3Leu33Gln93 migrated with an identical electrophoretic mobility to recombinant WT β3Leu33Arg93 and native, platelet-derived β3 integrin (Figure6A). Cell surface expression of the Leu33Gln93 mutant β3 integrin was confirmed in both cell lines by staining with mAb P37 (Figure 6B).
Expression of recombinant β3 integrins in CHO cells.
(A) Western blot analysis of recombinant β3 integrin expression in CHO cells. Cell lysates of transfected CHO cells were prepared, and protein concentration was determined as described in “Materials and methods.” Equal amounts of protein from β3-transfected CHO cells (50 μg) were resolved by 8% SDS-PAGE under nonreducing conditions, transferred onto nitrocellulose, and immunoblotted with a mAb to human β3 (P37). Platelet lysate (5 μg protein) was run in parallel as a positive control. Clone A10, CHO-αIIbβ3Leu33Arg93; clone CAM12, CHO-αIIbβ3Leu33Gln93; clone A13, CHO-αvhamsterβ3Leu33Arg93; clone CAM11, CHO-αvhamsterβ3Leu33Gln93; clone E05, CHO-αvhamsterβ3Pro33Arg93. (B) FACS analysis of CHO cells in suspension after indirect immunofluorescence labeling with the anti-β3 integrin mAb P37. Negative control cells (bold solid line), CAM11 (solid line), CAM12, A13, E05 (dotted lines), A10 (dashed line).
Expression of recombinant β3 integrins in CHO cells.
(A) Western blot analysis of recombinant β3 integrin expression in CHO cells. Cell lysates of transfected CHO cells were prepared, and protein concentration was determined as described in “Materials and methods.” Equal amounts of protein from β3-transfected CHO cells (50 μg) were resolved by 8% SDS-PAGE under nonreducing conditions, transferred onto nitrocellulose, and immunoblotted with a mAb to human β3 (P37). Platelet lysate (5 μg protein) was run in parallel as a positive control. Clone A10, CHO-αIIbβ3Leu33Arg93; clone CAM12, CHO-αIIbβ3Leu33Gln93; clone A13, CHO-αvhamsterβ3Leu33Arg93; clone CAM11, CHO-αvhamsterβ3Leu33Gln93; clone E05, CHO-αvhamsterβ3Pro33Arg93. (B) FACS analysis of CHO cells in suspension after indirect immunofluorescence labeling with the anti-β3 integrin mAb P37. Negative control cells (bold solid line), CAM11 (solid line), CAM12, A13, E05 (dotted lines), A10 (dashed line).
Reactivity of the β3Leu33-specific mAb CAMTRAN-007 with CHO transfectants expressing β3Leu33Arg93 or β3Leu33Gln93
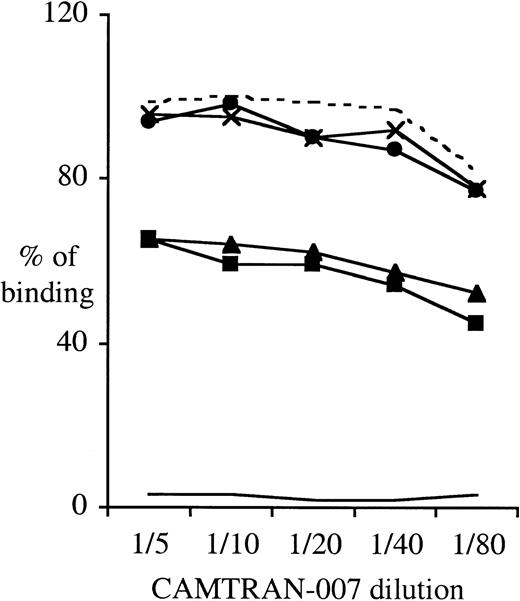
To study the expression of the β3Leu33 (HPA-1a) epitope on the Leu33Gln93-encoding recombinant β3 integrin expressed in CHO cells, we performed flow cytometry using FITC-conjugated CAMTRAN-007. In these studies, the relative binding of CAMTRAN-007 to the β3Leu33Gln93 mutant was reduced to 60% of that observed with the WT (Leu33Arg93) β3 integrin, indicating that the Arg93Gln mutation has a modifying effect on the HPA-1a epitope (Figure7). Interestingly, the reduction in reactivity of CAMTRAN-007 was independent of the association of the β3 integrin with either human αIIb or hamster αv integrins (Figure 7). CAMTRAN-007 did not react with the E05 cell line that expresses β3Pro33Arg93 (HPA-1b), confirming that the mAb is allospecific (Figures 6B, 7).
The Arg93Gln mutation does impair the β3Leu33-specific mAb CAMTRAN-007 binding to β3 integrins expressed in CHO cells.
Adherent CHO cells were detached with EDTA buffer, washed, and directly labeled with the anti-CD61–PE or CAMTRAN-007–FITC for 30 minutes on ice. Cells were washed and analyzed by flow cytometry. To control for the variations in β3 expression between the different cell clones, CAMTRAN-007 binding was normalized to the total β3 integrin expression determined using a β3-integrin–specific mAb (anti-CD61–PE). This ratio, CAMTRAN-007:CD61, was then expressed as a percentage of the ratio obtained for the CHO cell clone A10 with a CAMTRAN-007 dilution of 1:50 (100%). Data are representative of 4 different experiments. Clones as per Figure 6, plus clone A06, CHO-αvβ3Leu33Arg93. - - - indicates A10; ×, A13; ●, A06;___, E05; ▴, Cam12; and ▪, Cam11.
The Arg93Gln mutation does impair the β3Leu33-specific mAb CAMTRAN-007 binding to β3 integrins expressed in CHO cells.
Adherent CHO cells were detached with EDTA buffer, washed, and directly labeled with the anti-CD61–PE or CAMTRAN-007–FITC for 30 minutes on ice. Cells were washed and analyzed by flow cytometry. To control for the variations in β3 expression between the different cell clones, CAMTRAN-007 binding was normalized to the total β3 integrin expression determined using a β3-integrin–specific mAb (anti-CD61–PE). This ratio, CAMTRAN-007:CD61, was then expressed as a percentage of the ratio obtained for the CHO cell clone A10 with a CAMTRAN-007 dilution of 1:50 (100%). Data are representative of 4 different experiments. Clones as per Figure 6, plus clone A06, CHO-αvβ3Leu33Arg93. - - - indicates A10; ×, A13; ●, A06;___, E05; ▴, Cam12; and ▪, Cam11.
Genomic analysis
A clear differentiation between the WT and mutant alleles was obtained by Taqman-based β3G376A SNP genotyping (data not shown). Typing of 300 genomic DNA samples from random donors did not identify additional examples of the Gln93-encoding allele. However, typing of Donor A's immediate family members showed the presence of the Gln93 allele in his mother (Donor C; Figure 8). The Taqman Gln93-positive genotype of Donor C was confirmed by direct sequencing of genomic β3 integrin DNA (data not shown).
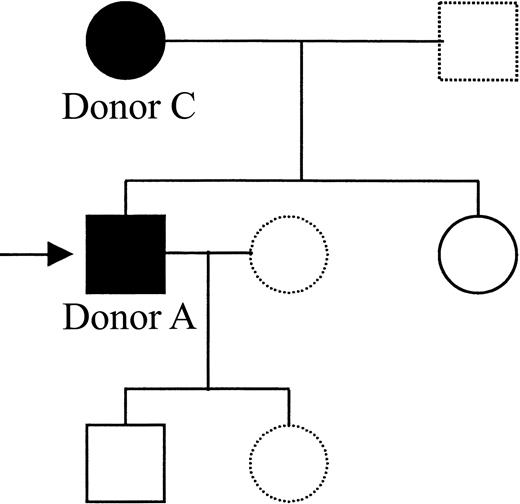
Pedigree of Donor A's family showing members positive for the Gln93-encoding allele.
Genomic DNA samples were obtained from members of Donor A's immediate family and were genotyped for the presence of the Gln93 encoding β3 integrin allele. A complete HPA genotype was also obtained by PCR-SSP (data not shown). The mutant β3Gln93 integrin allele was found in the mother (Donor C) and in the proband. Black symbols represent β3Gln93-positive members, open symbols represent β3Gln93-negative members, and the arrow indicates the proband (Donor A). Samples were not available from members represented by broken symbols.
Pedigree of Donor A's family showing members positive for the Gln93-encoding allele.
Genomic DNA samples were obtained from members of Donor A's immediate family and were genotyped for the presence of the Gln93 encoding β3 integrin allele. A complete HPA genotype was also obtained by PCR-SSP (data not shown). The mutant β3Gln93 integrin allele was found in the mother (Donor C) and in the proband. Black symbols represent β3Gln93-positive members, open symbols represent β3Gln93-negative members, and the arrow indicates the proband (Donor A). Samples were not available from members represented by broken symbols.
Discussion
The β3 integrin is associated with αIIb integrin in a noncovalent and cation-dependent manner on the platelet surface, where it binds fibrinogen, fibronectin, and vitronectin and mediates the aggregation of platelets and subsequent thrombus formation. The genes encoding αIIb and β3 integrins are polymorphic, and 8 SNPs are at the basis of HPA alloantigens.3,6 The bi-allelic HPA-1 system, which is based on a Leu33Pro polymorphism in β3 integrin, is clinically the most significant HPA system.9
The exact molecular nature of the HPA-1 epitope has been studied in some detail. Site-directed mutagenesis studies have confirmed that amino acid 33 of the β3 integrin is essential for the formation of the HPA-1 epitopes.24 Additional studies using recombinant β3 integrin fragments suggest that the HPA-1 epitope is expressed within the N-terminal 66 amino acids.25,26 However, the reactivity of HPA-1a antisera with recombinant fragments was variable and only involved investigations with a small number of samples. More recently, the human HPA-1a epitope has been introduced into mouse β3 integrin by substituting human amino acids into the mouse sequence. The reactivity of anti-β3Leu33 with 42 amino acid recombinant fragments (residues 9 to 50) demonstrated that amino acids 30, 32, and 39, in addition to 33, are critical for allo-antibody binding.27
Further studies have investigated the role of disulfide bonds and noncontiguous sequences in the formation of the HPA-1a epitope. Alanine replacement experiments with β3 integrin, designed to investigate the role of the various disulfide bonds in HPA-1a epitope formation, suggested 2 types (type 1 and type 2) of anti–HPA-1a that were split by their difference in reactivity with the Cys435Ala β3 isoform.28 It is assumed that Cys435 forms a disulfide bond with Cys5 linking the presumed cloverleaf-like, HPA-1a epitope, to the Cys-rich β3 core.29 Both type 1 and type 2 anti–HPA-1a required an intact, conformationally native αIIbβ3 because the replacement of Cys for Ala at N-terminal positions 5, 23, 26, and 38 abrogated reactivity.28 Inhibition of anti–HPA-1a binding by the mouse mAb LK-4 also demonstrates a split in allo-antibody reactivity.30,31 However, the 2-epitope model, proposed on the basis of these experiments, remains in dispute because most antisera were from patients with posttransfusion purpura that are known to contain αIIbβ3 autoantibodies in addition to the HPA-1 alloantibodies.32 There is ample evidence that HPA-1a antibodies do not bind β3 integrin–derived oligopeptides spanning the Leu33Pro33 polymorphism,27,33 findings that are in agreement with those obtained with chimeric β3 molecules.34 Studies with the latter suggested that sequences flanking the Cys435 position that encompassed amino acids 288-490 were important in epitope formation and that these sequences were brought into proximity with the Cys26-Cys38 loop by long-range disulfide bonds, such as the Cys5-Cys435 bond.34
Here we report on a unique donor with a normal level of platelet β3 integrin, as indicated by the reactivity of the mAb Y2/51 (Table 1), but with a severely reduced reactivity with monoclonal and polyclonal anti-β3Leu33 (anti–HPA-1a; Figures 1, 3, 4). The 3 β3Leu33-specific monoclonals show more than 80% reduction in binding to Donor A platelets compared with the heterozygous control (Figure3A), a level of reduction also seen with polyclonal anti-β3Leu33 in the MAIPA assay (Figure 4). A previously reported similar discrepancy between HPA-1 phenotype and genotype was attributed to the donor identified as a carrier of Glanzmann thrombasthenia.35However, several lines of evidence suggest this was not the case with Donor A. First, he does not carry a silent β3Leu33 allele, as judged from the β3Leu33 mRNA level and from sequencing data (Figure 5). Second, the platelet membrane β3 integrin copy number was normal (Table 1). A significant reduction in mAb Y2/51 reactivity would have been observed in the case of a silent β3 allele (eg, in carriers of type 1 Glanzmann thrombasthenia). Finally, sequencing β3 and αIIb integrin cDNA identified a novel G/A SNP at position 376 of the β3 cDNA that encodes for an Arg93Gln substitution linked with the β3Leu33 allele (Figure 5). Expression of the mutant β3Leu33Gln93 integrin cDNA in CHO cells with hamster αv and human αIIb integrins confirmed that the Arg93Gln mutation was responsible for the reduced reactivity with anti-β3Leu33. Reactivity with β3 integrin–specific mAbs demonstrated that the Leu33Gln93-encoded β3 integrin was expressed at the cell surface and was of the correct size (Figure 6A-B). However, CAMTRAN-007 (anti-β3Leu33) showed a 40% reduction in binding to β3Leu33Gln93 relative to β3Leu33Arg93 (Figure 7). That a greater reduction of anti-β3Leu33 binding was seen with the platelets of Donor A and his mother compared with the CHO transfectants was attributed to the homozygous nature of the transfectants in contrast to the heterozygous platelets, to differences in glycosylation, or both.
Arg93 of the β3 integrin is outside the first 66 amino acids and amino acids 288-490 that have previously been shown to be involved in the formation of the HPA-1a epitope.25,26 34 However, replacement of Arg93 with Gln disrupts the binding of anti-β3Leu33 and thus identifies a region of the β3 integrin not previously thought to be involved in HPA-1a epitope formation. That a residue 60 amino acids from the allelic residue has such a dramatic effect on the HPA-1a B-cell epitope is surprising, and an alternative explanation could be a major structural change in β3Leu33Gln93. However, several observations argue against this. First, Donor A's platelets show normal reactivity with β3-specific mAbs. Second, his platelets show normal reactivity with polyclonal anti–HPA-3a in MAIPA, confirming both that the allo-epitope defined by an Ile843Ser substitution in αIIb and that the epitope recognized by the capture monoclonal are intact (data not shown). Third, the observation that all 3 human mAbs derived from HPA-1a allo-immunized patients were minimally reactive with Donor A's platelets strongly suggested that residue 93 provides a critical contact residue for anti-β3Leu33 binding (Figure 3B). It is interesting that 2 of the 3 human monoclonal anti-HPA-1a (19-7 and 23-15) react with the N-terminal 66-amino acid fragment of the β3 integrin but that CAMTRAN-007 does not (N.A.W., unpublished observations, August 2001). We interpret this as evidence that the former 2 are possibly representative of type 1 HPA-1a antibodies and that the latter are representative of type 2 antibodies. That all 3 human monoclonal β3Leu33 antibodies did not bind Donor A's platelets (Figure 3A) suggests that his platelets would not define this split in antibody types.
Finally, family studies indicated cosegregation of the Leu33 and Gln93 codons (Figure 8). Genotyping of family members showed that the Leu33Gln93 β3 integrin allele was inherited from the mother. The mother (Donor C) had a β3Leu33 homozygous PCR-SSP genotype (Figure2). When tested for reactivity with CAMTRAN-007 in whole blood, however, platelet immunofluorescence gave a signal similar to that of the HPA-1a1b heterozygous control (Figure 3B). We were unable to identify any other related or unrelated persons with a β3Gln93 allele after testing 300 DNA samples from random blood donors (data not shown). Taken together, these data suggest that we have identified a private allele unique to this family.
In conclusion, we have identified a rare but informative SNP in the β3 integrin that encodes a glutamine at position 93 instead of the normal arginine. The presence of Gln93, with Leu33, is coupled with a strong reduction in the binding of monoclonal and polyclonal β3Leu33 (HPA-1a) allo-antibodies. Amino acid 93 had not previously been thought to be involved in the formation of the HPA-1 epitope, but our findings indicate that the conformation of the Leu33Pro33-containing loop (residues 26 to 38) or that of the Cys-rich core is conformationally changed by this mutation. If the latter is the correct explanation, it would support the hypothesis that the HPA-1a epitope is discontinuous and that residues from several loops (including a loop containing residue 93) are involved in allo-antibody binding. Continuation of high throughput HPA-1a phenotyping might identify additional persons with unique β3Leu33 alleles, allowing a further unraveling of the molecular structure of the HPA-1a B-cell epitope.
We thank the staffs of the National Blood Service, Oxford and Cambridge Centres, for collecting and testing samples. We also thank Dr A. H. Goodall, University of Leicester, and Prof A. E. G. Kr. von dem Borne, Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, for their kind gifts of monoclonal antibodies.
N.A.W. is supported by a research grant from DiaMed AG, Switzerland. E.S.R. and N.H.C.B. are supported by grants from CRP-Santé, Luxembourg.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Willem H. Ouwehand, Dept of Haematology, Division of Transfusion Medicine, University of Cambridge, Cambridge CB2 2PT United Kingdom; e-mail: who1000@cam.ac.uk.