Abstract
Human herpesvirus 8 (HHV-8; Kaposi sarcoma–associated herpesvirus)–specific cytotoxic T-lymphocyte (CTL) and interferon-γ (IFN-γ) responses to proteins produced during the lytic cycle of HHV-8 replication are mediated by HLA class I–restricted, CD8+ T cells. We have characterized the fine specificity of the CD8+ T-cell response to 25 peptides derived from 5 HHV-8 lytic cycle proteins based on a prediction model for HLA A*0201 binding motifs. One of the 25 HLA A*0201 peptides derived from the glycoprotein B (gB) homolog of Epstein-Barr virus (gB492-500; LMWYELSKI; single-letter amino acid codes) bound to HLA A*0201 and stimulated IFN-γ responses in CD8+ T cells from HHV-8+, HLA A*0201 persons, but not HHV-8–seronegative or non–HLA A*0201 persons. The peptide also induced IFN-γ and CTL reactivity to naturally processed gB protein. The peptide was a major immunogenic epitope of HHV-8 as indicated by induction of IFN-γ responses in peripheral blood mononuclear cells from 5 of 5 HHV-8 seropositive, HLA A*0201 persons when gB492-500 was presented by autologous dendritic cells. T-cell reactivity to gB492-500 was not related to detectable HHV-8 DNA in the blood. These data show that CD8+ T cells recognize an HLA A*0201–restricted epitope for HHV-8 lytic cycle protein gB, particularly when presented by dendritic cells. This epitope may be important in control of HHV-8 infection by CD8+ T cells.
Introduction
Human herpesvirus 8 (HHV-8; also termed Kaposi sarcoma–associated herpesvirus) is a gammaherpesvirus that is considered to be the causative agent of Kaposi sarcoma (KS), body cavity B-cell lymphoma, and multicentric Castleman disease.1 T-cell immunity is thought to play an important role in control of HHV-8 infection and these associated diseases.2-5 In this regard, we2,3 and others4 have demonstrated that major histocompatibility complex (MHC) class I–restricted, CD8+ cytotoxic T lymphocytes (CTLs), and interferon γ (IFN-γ) responses are detectable in peripheral blood mononuclear cells (PBMCs) of HHV-8+ individuals using recombinant vaccinia viruses expressing HHV-8 lytic and latent cycle proteins. These anti–HHV-8 T-cell responses are lower during human immunodeficiency virus type 1 (HIV-1) infection and may be involved in prevention of KS and other HHV-8–associated diseases.
Support for the potential role of CD8+ T-cell immunity in control of HHV-8 infection comes from several studies on immune control of acute and persistent infections with 2 other gammaherpesviruses, specifically, Epstein-Barr virus (EBV) and murine herpesvirus 68 (MHV-68).6-10 Although EBV latency proteins can induce strong CD8+ T-cell responses,11 lytic cycle proteins are considered as major CD8+ T-cell targets in both acute and persistent EBV infections.12 Immunodominant epitopes restricted to different HLA class I alleles, which are important for our understanding of viral immunopathogenesis and development of appropriate vaccines,13 have been determined for peptides derived from EBV structural proteins gp350 and gp85, and immediate-early (IE) and early proteins such as BZLF1, BRLF1, and BMLF1.6-9 Compared to latency proteins, the frequency of CD8+ T lymphocytes specific for peptides derived from lytic cycle proteins is higher in PBMCs of healthy virus carriers.14 CD8+ CTLs have also been demonstrated to secrete IFN-γ in response to EBV lytic cycle antigens in seropositive individuals.15 During primary EBV infectious mononucleosis, up to 60% of CD8+ T cells circulating in blood are EBV-specific.16 Furthermore, by use of a peptide-tetramer complex, Tan et al17 have directly visualized from 0.5% to 6.6% of CD8+ T blood cells in virus carriers that are specific for a single BMLF-1 epitope.
In MHV-68 infection, lytic cycle proteins also induce CD8+T cell responses.10,18,19 T cells specific for peptides derived from lytic cycle proteins dominate the acute phase of MHV-68 infection.10,19 Moreover, vaccination with immunodominant CD8+ T-cell epitopes derived from lytic cycle proteins of MHV-68 significantly reduces viral titers and the level of infected cells during acute infection.20
We have tested CD8+ T-cell reactivity to peptides derived from 5 HHV-8 lytic cycle proteins based on a prediction model for the HLA A*0201 motif in HHV-8–infected subjects to determine immunodominant epitopes to the virus. We chose HLA A*0201 because it is prevalent in a large percentage of the white population and it is the most extensively studied HLA class I antigen.21 We found that one peptide derived from the glycoprotein B (gB) homolog was immunogenic for CD8+ T cells from HHV-8+, HLA A*0201 individuals, particularly when presented by autologous, mature dendritic cells (DCs). CD8+ T-cell responses to this HHV-8 lytic cycle peptide may be important in the control of acute and persistent HHV-8 infection and in vaccine design.
Materials and methods
Study subjects and detection of HHV-8 infection
The participants were volunteers from the Pittsburgh portion of Multicenter AIDS Cohort Study,22 laboratory volunteers, and commercial blood donors (Central Blood Bank, Pittsburgh, PA) based on their HHV-8 antibody or DNA status and HLA class I genotype. The volunteers gave written informed consent. None of these subjects were infected with HIV-1. The subjects were classified into 3 groups based on HHV-8 serum antibody or DNA in PBMCs, and HLA A*0201 genotype: group A (n = 14) was HHV-8+, HLA A*0201+; group B (n = 5) was HHV-8−, HLA A*0201+; and group C (n = 7) was HHV-8+, HLA A*0201−.
Detection of HHV-8 serum antibody specific for lytic antigens and HLA molecular typing were done as described elsewhere.2,3,23HHV-8 DNA was detected in PBMCs by DNA hybridization to HHV-8–specific polymerase chain reaction (PCR) products. PCR analyses were performed using primers that amplified a 233–base pair (bp) fragment of the minor capsid gene encoded by open reading frame (ORF) 2624,25 as described previously.3 PCR products were separated on 2% agarose gels, transferred to Nytran membranes and hybridized with an internal oligonucleotide probe labeled with 32P. The sequence of oligonucleotide probe was TGCAGCAGCTGTTGGTGTACCACAT. PCR products that hybridized to the probe were detected by image analysis with an ImageQuant PhosphorImager (Molecular Dynamics, Sunnyvale, CA). This procedure has a sensitivity of detection of 10 or more copies of HHV-8 DNA from 1 × 106 PBMCs. We have also found that PBMCs from HHV-8 seronegative controls are negative for HHV-8 DNA, whereas a cell line persistently infected with HHV-8 is consistently positive by this procedure.
Synthetic peptides
The 9-mer and 10-mer peptides were derived from the Los Alamos GenBank sequence for HHV-826 using a prediction model that ranks peptides based on a predicted half-time of dissociation to HLA A*020127 28 (Table1). The peptides were synthesized by Fmoc chemistry and the purity was determined to average approximately 90% by analytical high-performance liquid chromatography profile and mass spectrometry in the Peptide Synthesis Facility of the University of Pittsburgh Cancer Institute and by Dr Robert Lee of the University of Illinois.
T2 cell line HLA A*0201 stabilization assay
A peptide-induced stabilization assay of the HLA A*0201 molecule expressed by the T2 cell line was performed using a method described elsewhere.29,30 T2 cells are a transporter associated with antigen processing (TAP)-deficient cell line that has low levels of HLA A*0201 molecules expressed on the cell surface and impaired endogenous peptide presentation.31 The T2 stabilization assay determines the conformation of HLA A*0201 molecules when exogenous synthetic peptides and β2-microglobulin (β2M) are delivered to the T2 cells. Two control peptides in addition to the 25 HHV-8 lytic cycle peptides were tested. A peptide derived from HIV-1 p24 protein (Gag p24263-272; KRWIILGLNK; single-letter amino acid codes), which is HLA B27–restricted,32 served as a negative control; a peptide representing an HLA A*0201 epitope from the influenza A virus matrix protein (M158-66; GILGFVFTL)33 served as a positive control. The 5 × 105 T2 cells (gift from Dr Russell Salter, University of Pittsburgh) were incubated at 37°C and 5% CO2 overnight in the presence of various concentrations of peptide in RPMI 1640 medium (Life Technologies, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (Hyclone, Logan, UT) and 1 μM β2M (Sigma, St Louis, MO). The cells were washed twice with cold phosphate-buffered saline (PBS) and incubated for 30 minutes at 4°C with monoclonal antibody (mAb) specific for the conformationally changed HLA class I molecule (BB7.2; American Type Culture Collection, Manassas, VA). The cells were then washed twice with PBS and incubated with fluorescein isothiocyanate–conjugated IgG F(ab′)2 (Organon-Teknika, Durham, NC) for 30 minutes at 4°C. The cells were washed twice with cold PBS and fixed with 1% paraformaldehyde (Polysciences, Warrington, PA). The labeled cells were analyzed by flow cytometry (Elite; Beckman Coulter, Fullerton, CA). The positive binding capacity cutoff was determined by the mean + 3 SD of the ratio for HIV-1 p24263-272, which was calculated as mean fluorescence intensity (MFI) of the test peptides compared to the MFI of T2 cells without peptides added. The mean ± SD value for HIV-1 p24263-272 binding was 0.93 ± 0.18, giving a cutoff value for positive binding of 1.5.
Single-cell IFN-γ production
A single-cell enzyme immunoassay for IFN-γ production was done using standard methods described previously,3 with minor modifications. Frozen PBMCs were thawed and incubated overnight at 1 × 105 cells with peptides at a concentration of 50 μg/mL in nitrocellulose-bottom, 96-well plates (Millipore, Bedford, MA). An immunodominant peptide derived from an EBV lytic cycle protein (BMLF1280-288; GLCTLVAML),6 the influenza matrix peptide M158-66, and HIV-1 p24 Gag peptide (p24151-159; TLNAWVKVV)34 and phorbol 12-myristrate 13-acetate (1 ng/mL)–ionomycin (1 μM; PMA-iono) were used as positive controls. A plasmid expressing the ORF for gB (ORF8) was cloned into the vaccinia virus shuttle vector, pSC11, as previously described,2 3 and used for expression of the native form of gB protein (designated VgB). VSC11 not containing the HHV-8 plasmid was used as the control for background, vaccinia virus gene expression.
After overnight stimulation with the antigens, the plates were washed and processed, and the number of spots counted with a dissecting microscope.3 The results are given as the number of spots per 106 PBMCs stimulated by the viral peptides or autologous B lymphocyte cell lines (BLCLs) infected with VgB, minus the number of spots per 106 PBMCs stimulated by the medium control or BLCLs infected with the VSC11 vector, respectively. The mean (± SE) number of IFN-γ–producing cells induced by medium alone was 3 (± 1) per 106 PBMCs (n = 26). The cutoff value for positive IFN-γ production to the HHV-8 peptides was defined as more than 40 spot-forming cells per 106 PBMCs based the range of values for IFN-γ production to the 25 HHV-8 peptides in the group 2 and 3 control participants (ie, median, 0; range, 0-20; spot-forming cells per 106 PBMCs; n = 12).
In certain experiments, the peptides were presented to PBMCs by autologous DCs derived from CD14+ blood monocytes that were positively selected using CD14 immunomagnet microbeads (Miltenyi Biotec, Auburn, CA). The CD14+ cells were resuspended at 1 × 106 cells/mL in AIM-V medium containing 1000 U/mL each of recombinant human interleukin (IL)-4 and human granulocyte-monocyte colony-stimulating factor (GM-CSF). The cells were cultured for 7 days with fresh IL-4 and GM-CSF added at 2-day intervals as previously described.34 The cells were then treated with human CD40 ligand trimer (CD40L) at 1 μg/mL (Immunex, Seattle, WA) for 2 days to augment phenotypic and functional maturation. These DCs have a mature phenotype as determined by staining with fluorescent dye-conjugated mAb and analyzed by FACS as previously described.34 The mature DCs were loaded with peptide at 10 μg/mL for 2 hours in AIM V at room temperature, washed 3 times and counted with a light microscope. The peptide-loaded mature DCs were then used as stimulators (5 × 103) for autologous PBMCs (5 × 104) in triplicate in the single-cell enzyme immunoassay overnight at 37°C (termed zero week stimulation, or S0). To examine longer-term effects of stimulation of T cells by DCs, parallel DC-PBMC mixtures were cultured with IL-2 at 100 U/mL for 7 days, harvested, and restimulated (1 × 105 cells) overnight at 37°C with either untreated or peptide loaded, autologous PBMCs (1 × 104) in triplicate in nitrocellulose-bottom microwells in the single-cell enzyme immunoassay (termed 1-week stimulation, or S1). Stimulation of PBMCs with DCs without peptide was included as a control for the S0 and S1 cultures.
CTL assay
The bulk lysis CTL assay was performed as described previously using 3 effector-to-target (E/T) ratios,2,3 except for the preparation of target cells. On day 13, autologous BLCLs infected with the VgB or VSC11 control vectors, or T2 cells pulsed with the viral peptides (50 μg/mL) and B2M, were labeled with51Cr (100 μCi/mL [3.7 MBq] Na2[51Cr]O4, Dupont NEN, Boston, MA) and incubated overnight. On day 14, 51Cr release was determined in a gamma counter. The data are presented as percent virus-specific lysis, which equals percent specific lysis against the HHV-8 antigen-expressing targets minus percent specific lysis against the VSC11 control antigen-expressing targets. Mean (± SE) background lysis of the uninfected and VSC11-infected targets was 0 (± 1%) for lysis at 3 E/T ratios (n = 9). The cutoff value for positive lysis was 10% based on anti-HHV-8 CTL lysis in HHV-8–seronegative individuals.2
Establishment of CD8+ T-cell lines
For the establishment of CD8+ T-cell lines, T2 cells pulsed with the gB492-500 peptide and β2M were used as stimulators and plated into 96-well U-bottom plates at a concentration of 5 × 104 in RPMI 1640 medium, supplemented with 15% fetal calf serum, IL-2 (100 U/mL; Chiron, Emeryville, CA), and anti-CD3 mAb (12-F6, 10 ng/mL; gift from Dr Johnson Wang, Massachusetts General Hospital, Boston, MA). Frozen PBMCs were thawed and plated at 50 cells/well in 96-microwell U-bottom plates. Then, 50 μL gamma-irradiated (4000 rad) allogeneic PBMCs were plated at a concentration of 1 × 106/mL as feeder cells to enhance T-cell growth. The cultures were incubated at 37°C in 5% CO2 for 2 to 3 weeks. Primary screening was performed when large cell pellets were visually evident. The cell lines were approximately 70% CD8+ by mAb staining. Radioactively labeled T2 cells loaded with the corresponding peptides and B2M were used as targets. Wells positive for CTL activity were then transferred to 48-well plates and used for the single-cell IFN-γ production assay and T-cell phenotyping. T-cell subsets were phenotyped by flow cytometry (XL; Beckman Coulter) after staining with mAbs specific for CD3, CD4, and CD8 (Becton Dickinson; Mountain View, CA).
Results
Selection of potential CD8+ T-cell epitopes of HHV-8 lytic cycle proteins for binding to HLA A*0201
To identify potential HLA A*0201–restricted epitopes within HHV-8 lytic cycle proteins, amino acid sequences of the 5 HHV-8 proteins were analyzed based on a prediction model for the HLA A*0201 binding motif.27 28 This prediction model allows location and ranking of 9-mer peptides that contain putative peptide-binding motifs for HLA class I molecules, based on an estimation of the half-time dissociation (T1/2 dis) of the HLA-peptide complex. The 5 highest predicted binding scores for 9-mer peptides derived from each of the 5 HHV-8 lytic cycle proteins are listed in Table 1.
We tested the 25 peptides for actual HLA A*0201 binding capacity using the T2 cell-binding assay. Thirteen of the 25 peptides bound to HLA A*0201 in a concentration-dependent manner (Table 1). The positive control M158-66 peptide also bound, whereas no binding was associated with the negative control p24 Gag263-272 peptide at any concentration tested.
Characterization of HHV-8 peptides for stimulation of peripheral blood, CD8+ T cells from HHV-8 seropositive, healthy persons
We next characterized CD8+ T-cell reactivity to the HHV-8 peptides using a single-cell IFN-γ production assay in the 3 groups of HIV-1−, healthy subjects. Even though 12 of the peptides did not bind to HLA A*0201 based on our T2 cell-binding assay, we chose to screen all 25 peptides for T-cell activation in the event that this assay was more sensitive than the binding assay for detecting peptide antigenicity. We found that PBMCs from 2 of the 14 group A persons, who were HHV-8+ and HLA A*0201–genotypic, produced IFN-γ (subject A27 = 50 and subject A29 = 240 spots/106 PBMCs) in response to one of the 13 HHV-8 peptides that bound to HLA A*0201, that is, gB492-500(LMWYELSKI), and to none of the 12 peptides that did not bind (data not shown). No HHV-8 peptide-specific, IFN-γ production was noted in PBMCs from either the 5 group B subjects, who were HHV-8−and HLA A*0201–genotypic, or the 7 group C subjects, who were HHV-8+ but HLA A*0201− (median, 0; range, 0-20 spots/106 PBMCs). These results support that IFN-γ production induced by gB492-500 was HHV-8–immune specific and HLA A*0201–restricted.
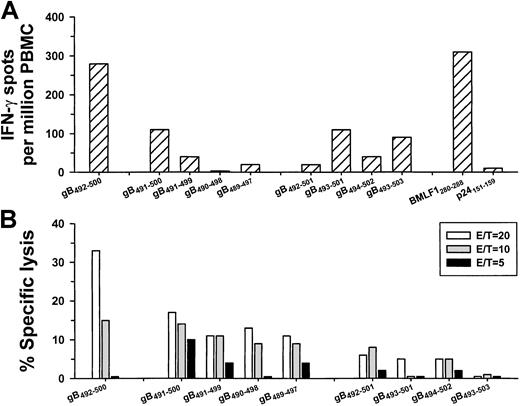
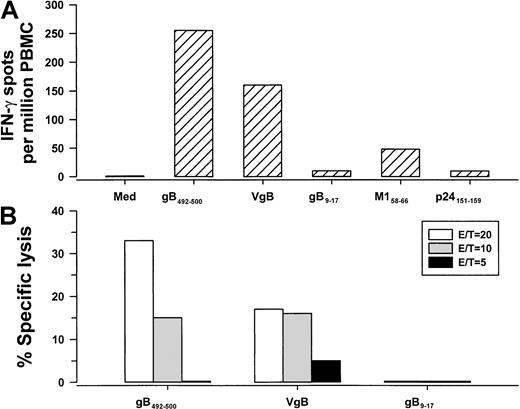
Next, we determined that both PBMCs and CD8+ T cells, but not CD4+ T cells, produced IFN-γ in response to gB492-500 in a concentration-dependent manner, confirming that this 9-mer peptide was specific for CD8+ T cells (Figure 1). We then compared single-cell IFN-γ production and CTL activity stimulated by the HHV-8 synthetic peptide that was loaded directly in PBMCs with that induced by BLCLs expressing naturally processed peptides produced by VgB (which we have previously reported induces HHV-8–specific IFN-γ production in CD8+ T cells2 3). The results show IFN-γ production and CTL reactivity to both gB492-500 and cells infected with the vaccinia virus gB expression vector (Figure2), indicating that this HHV-8 peptide induced similar T-cell reactivity as the corresponding, naturally processed HHV-8 protein. In contrast, there were very low IFN-γ and CTL responses to gB9-17 in the VgB-stimulated cultures (Figure 2), which bound to HLA A*0201 (Table 1) but did not induce IFN-γ in any of the HLA A*0201, HHV-8+, group A persons (data not shown).
Production of IFN-γ.
Concentration-dependent production of IFN-γ in response to HLA A*0201 HHV-8 gB492-500, influenza A virus M158-66, and HIV-1 p24 Gag151-159 by PBMCs (A), CD8+ T cells (B), and CD4+ T cells (C) of group A subject 29 (HLA A*0201, HHV-8–seropositive, HIV-1–seronegative).
Production of IFN-γ.
Concentration-dependent production of IFN-γ in response to HLA A*0201 HHV-8 gB492-500, influenza A virus M158-66, and HIV-1 p24 Gag151-159 by PBMCs (A), CD8+ T cells (B), and CD4+ T cells (C) of group A subject 29 (HLA A*0201, HHV-8–seropositive, HIV-1–seronegative).
IFN-γ production and CTL reactivity.
IFN-γ production (A) and CTL reactivity (B) at 3 E/T ratios in PBMCs from group A subject 29 (HLA A*0201, HHV-8 seropositive) specific for gB492-500 peptide and the corresponding gB protein produced by VgB. The controls for the IFN-γ assay were HLA A*0201 peptides for HHV-8 gB9-17, influenza A virus M158-66, and HIV-1 p24 Gag151-159. Med indicates medium control.
IFN-γ production and CTL reactivity.
IFN-γ production (A) and CTL reactivity (B) at 3 E/T ratios in PBMCs from group A subject 29 (HLA A*0201, HHV-8 seropositive) specific for gB492-500 peptide and the corresponding gB protein produced by VgB. The controls for the IFN-γ assay were HLA A*0201 peptides for HHV-8 gB9-17, influenza A virus M158-66, and HIV-1 p24 Gag151-159. Med indicates medium control.
We examined whether presence of HHV-8 DNA in the blood was associated with the ability of CD8+ T cells to respond to HHV-8 peptides. We tested PBMCs from a subset of 7 of the 14 group A persons for HHV-8 DNA, that is, subjects A25 through A30 and A34. We found that PBMCs from only 2 of these 7 subjects (A25 and A30) were positive for HHV-8 DNA. Thus, there was no association between HHV-8 DNAemia and ability of the CD8+ T cells to respond to these 25 HHV-8 peptides.
Determination of the minimal length of the gB epitope
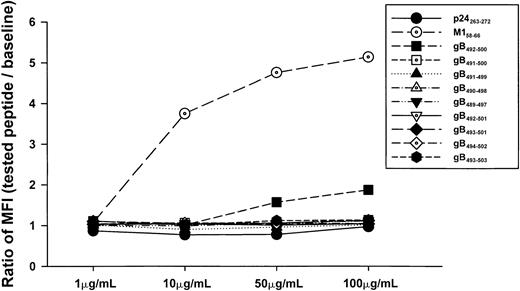
Because the gB492-500 peptide was derived from a model of predicted binding of a 9-mer to an HLA motif, we next defined the true minimal epitope of this gB peptide by binding to HLA A*0201 and T-cell reactivity using six 9-mer peptides and two 10-mer peptides derived from overlapping sequences (Table2). None of these overlapping peptides bound to the T2 cells (Figure 3). Nevertheless, we tested all 8 of these peptides for IFN-γ and CTL reactivity in the event that these assays were more sensitive measures of a peptide's T-cell stimulatory capacity. Our results show that greater IFN-γ production and CTL lysis was induced by the predicted, 9-mer gB492-500 peptide than by the overlapping 9-mer and 10-mer peptides in PBMCs of HHV-8+, HLA A*0201 individuals from group A (Figure 4A,B). Some of the peptides (eg, gB491-500) induced low CTL and IFN-γ reactivity even though they did not bind to HLA A*0201 in the T2 cell assay (Figure 3).
Binding capacity of overlapping 9-mer and 10-mer peptides.
Concentration-dependent binding capacity of HHV-8 peptides gB492-500 and overlapping 9-mer and 10-mer peptides for gB492-500. The HLA B27 p24 Gag263-272 peptide and the HLA A*0201 M158-66 peptide served as negative and positive controls, respectively.
Binding capacity of overlapping 9-mer and 10-mer peptides.
Concentration-dependent binding capacity of HHV-8 peptides gB492-500 and overlapping 9-mer and 10-mer peptides for gB492-500. The HLA B27 p24 Gag263-272 peptide and the HLA A*0201 M158-66 peptide served as negative and positive controls, respectively.
IFN-γ production and CTL lysis induced by 9-mer gB492-500.
IFN-γ production (A) and CTL activity (B) to overlapping 9-mer and 10-mer peptides for gB492-500 in PBMC from group A subject 29 (HLA A*0201, HHV-8–seropositive). HLA A*0201 p24 Gag151-159 and BMLF1280-288 peptides served as negative and positive controls, respectively.
IFN-γ production and CTL lysis induced by 9-mer gB492-500.
IFN-γ production (A) and CTL activity (B) to overlapping 9-mer and 10-mer peptides for gB492-500 in PBMC from group A subject 29 (HLA A*0201, HHV-8–seropositive). HLA A*0201 p24 Gag151-159 and BMLF1280-288 peptides served as negative and positive controls, respectively.
We next established CD8+ T-cell lines to test T-cell reactivity to these overlapping HHV-8 peptides. Two cell lines generated against gB492-500 had higher IFN-γ production in response to restimulation by the predicted 9-mer gB492-500 peptide than by the overlapping 9-mer and 10-mer peptides (Table 2).
T-cell response to gB492-500 in HLA A*0201 HHV-8 seropositive persons using gB492-500 peptide-loaded DCs
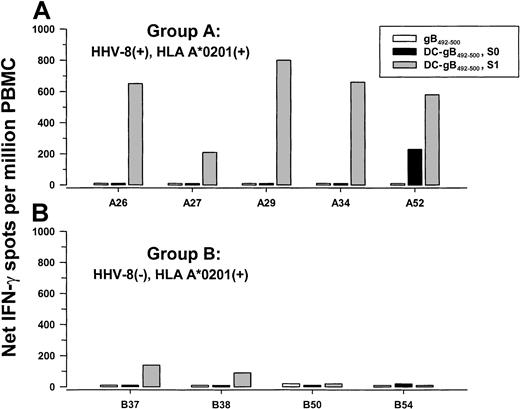
We and others have previously shown that human blood monocyte-derived DCs loaded with HIV-1,35EBV,36,37 and human cytomegalovirus (CMV)38 39peptides are potent inducers of antiviral CD8+ T-cell responses in PBMCs of individuals seropositive for these viruses. Therefore, we determined if autologous DCs loaded with gB492-500 peptide were able to induce peptide-specific T-cell reactivity as compared to the conventional method of stimulation with peptide alone. Peptide-loaded and nonloaded, CD40L-matured DCs were added to autologous PBMCs derived from 5 group A and 4 group B subjects for 1 day (S0) and 7 days (S1), and tested in a single-cell IFN-γ assay. As shown in Figure 5A, PBMCs from all 5 HHV-8 seropositive, group A subjects responded to gB492-500 when the peptide was presented by autologous DCs and cocultured for 1 week (S1; P = .004, 2-tailedt test, as compared to group B subjects). Coculture of the PBMCs overnight with peptide alone, or with peptide-loaded, autologous DCs (S0), was insufficient to induce anti-gB492-500 T-cell responses except in PBMCs from subject A52. There was little or no response of PBMCs from the 4 HHV-8 group B seronegative donors after stimulation with peptide-loaded, autologous DCs either overnight or for 7 days (Figure 5B). PBMCs from all 9 subjects responded strongly to stimulation with PMA-iono (median, 2225; range, 1150-5520 spots/106 cells). None of these PBMCs were positive for HHV-8 DNA.
IFN-γ production to gB492-500 presented by autologous DCs.
PBMCs from 5 group A HHV-8 seropositive (A) and 4 group B HHV-8 seronegative (B), HLA A*0201 subjects were stimulated for 1 day with gB492-500 alone, or for 1 day (S0) or 1 week (S1) with autologous DCs loaded with gB492-500 and matured with CD40L. Median (range) of PBMC responses in the single-cell enzyme immunoassays to medium alone after 1 day = 0 (0-10) and to DCs alone for S0 = 0 (0-10) and S1 = 0 (0-250).
IFN-γ production to gB492-500 presented by autologous DCs.
PBMCs from 5 group A HHV-8 seropositive (A) and 4 group B HHV-8 seronegative (B), HLA A*0201 subjects were stimulated for 1 day with gB492-500 alone, or for 1 day (S0) or 1 week (S1) with autologous DCs loaded with gB492-500 and matured with CD40L. Median (range) of PBMC responses in the single-cell enzyme immunoassays to medium alone after 1 day = 0 (0-10) and to DCs alone for S0 = 0 (0-10) and S1 = 0 (0-250).
IFN-γ production to peptide gB492-500 at sequential time points
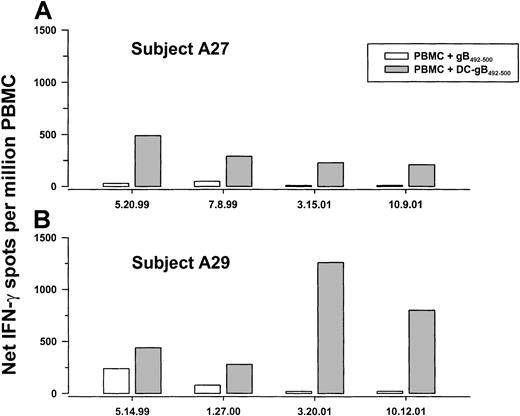
Notably, PBMCs obtained from group A subjects 27 and 29 at a later time did not produce IFN-γ in response to the gB492-500peptide alone, but did produce IFN-γ in response to 1 week of stimulation with gB492-500 peptide–labeled DCs (Figure 5). Further assessment using the DC-peptide system was therefore done to determine the persistence of anti-gB492-500 T-cell responses over time in 2 HHV-8+, HLA A*0201 individuals (A27 and A29). Fresh DCs were obtained from the blood of each subject and used to stimulate PBMCs that had been cryopreserved at approximately 6-month intervals over more than 2 years. The data show that IFN-γ production to gB492-500-loaded, autologous DCs in the 7-day assay was consistent over time (Figure6A,B). Moreover, the numbers of IFN-γ–producing cells were higher at all time points compared to stimulation of PBMCs directly with the free peptide, supporting the higher efficiency of T-cell activation by the antigen-expressing DCs. The subjects' PBMCs also had consistently high responses to PMA-iono over this time period (median, 2220; range, 1900-2500 spots/106 cells).
IFN-γ production in PBMCs obtained at sequential time points from 2 HHV-8–seropositive subjects.
PBMCs cryopreserved at the indicated dates from group A subjects A27 and A29 were thawed and cocultured for 1 week with gB492-500 alone or with freshly obtained, autologous, mature DCs loaded with gB492-500.
IFN-γ production in PBMCs obtained at sequential time points from 2 HHV-8–seropositive subjects.
PBMCs cryopreserved at the indicated dates from group A subjects A27 and A29 were thawed and cocultured for 1 week with gB492-500 alone or with freshly obtained, autologous, mature DCs loaded with gB492-500.
We examined whether HHV-8 DNA in blood was related to persistence of anti-HHV-8 CD8+ T-cell responses. The results indicate that all sequential PBMC samples from subjects A27 and A29 were negative for HHV-8 DNA (data not shown).
Discussion
We provide evidence for an HLA A*0201–restricted, CD8+ T-cell epitope specific for an HHV-8 lytic cycle protein, gB. Twenty-five 9-mer peptides derived from 5 lytic cycle proteins were selected for screening based on an HLA motif, peptide-binding prediction model.27,28 One of the 25 HHV-8 peptides, gB492-500 (LMWYELSKI), was clearly identified as immunogenic based on positive T-cell reactivity in HHV-8+, HLA A*0201 individuals. This response was shown to be HHV-8–immune specific and HLA A*0201–restricted in that there was no IFN-γ induced by the HHV-8 peptide in either HHV-8−, HLA A*0201 individuals or HHV-8+, non-HLA A*0201 controls. By use of overlapping 9-mer and 10-mer peptides with PBMCs and CD8+T-cell lines derived from the HHV-8 seropositive individuals, we confirmed that gB492-500 was the minimal epitope for HLA A*0201–restricted CD8+ T cells. The amino acid sequence of gB492-500 also corresponds well to the published motif for HLA A*0201 binding peptides,28 with preferred amino acids at anchor positions 2 and 9, that is, methionine and isoleucine. As expected, CD8+ and not CD4+ T cells produced HHV-8–specific IFN-γ in response to gB492-500. This HHV-8 peptide also was immunogenic for the native form of the HHV-8 gB protein produced during active viral replication, in that IFN-γ production and CTL activity were comparable when stimulated by the synthetic peptide or autologous BLCLs infected with recombinant vaccinia viruses expressing gB.
Of importance is that our data support gB492-500 being a major immunogenic HLA A*0201 epitope for HHV-8–specific, CD8+ T cells. This was demonstrated by induction of high numbers of IFN-γ–producing cells in PBMCs from all 5 HHV-8+, HLA A*0201 subjects tested that were stimulated for 1 week with peptide-loaded, CD40L matured, autologous DCs. This response was persistent over more than 2 years in 2 HHV-8–seropositive persons. In contrast, PBMCs from 4 HHV-8 seronegative HLA A*0201 subjects had low or negative responses to their peptide-loaded DCs even after 1 week of stimulation. Stimulation for 1 day with peptide alone or peptide-loaded DCs was not as efficient at activating peptide-specific T-cell responses. These data support the enhanced ability to detect epitopes for memory CD8+ T cells by use of peptide-loaded, CD40L-matured DCs as antigen-presenting cells. Similarly, DCs matured with monocyte-conditioned medium and loaded with peptides from influenza A virus matrix protein40 or EBV latency or lytic cycle proteins36,37 are most proficient at inducing peptide-specific, memory CD8+ T-cell responses. Other studies have revealed new epitopes for HIV-1 proteins using HIV-1 peptide-loaded, mature DCs.41 Activation of T cells in vitro in our study may require presentation of antigen in the context of high expression of T-cell coreceptors42 and production of IL-1243 and IL-1544 by the CD40L-matured DCs. This could also explain the recent failure to detect T-cell responses to gB492-500 in PBMCs from HHV-8 seropositive, HIV-1–infected persons that were stimulated with peptide-loaded PBMCs.45
We postulated that presence of latent HHV-8 infection in the blood may be related to the ability of the host's CD8+ T cells to respond to HHV-8 peptides. We did not find, however, a relationship of HHV-8 DNA in PBMCs with reactivity of T cells to these HHV-8 peptides. Indeed, most of our HHV-8–seropositive, healthy subjects were negative for HHV-8 DNA in blood by our PCR assay that can detect as few as 10 copies of HHV-8 DNA per 1 × 106 cells. This suggests that an infrequent, low level of latent HHV-8 infection occurs in the blood of healthy seropositive individuals, similar to other reports.46 This is in contrast to frequent detection of EBV47 and CMV48 DNA in the blood of asymptomatic carriers by highly sensitive PCR assays. Such a model of HHV-8 latency would predict a lower frequency and level of viral reactivation. This could account at least in part for the lower anti–HHV-8 T-cell responses detected in healthy, HHV-8–seropositive individuals,2-4 as compared to T-cell responses to EBV and CMV peptides in healthy, EBV- and CMV-seropositive persons.36-39
Although the precise function of HHV-8 gB is unknown, a recent report shows that it is part of the virion and infected cell membrane,49 in contrast to gB of EBV and MHV-68.50,51 Indeed, gB may be important for HHV-8 infectivity and pathogenesis in that it binds to cell surface heparan sulfate molecules.49 Regardless of its function in HHV-8, it is likely that the immune response to gB is related to the level of HHV-8 lytic replication. Several studies have shown an association of DNA positivity and higher levels of HHV-8 DNA in blood and tissues with development of KS and other HHV-8–related diseases.52-56We hypothesize that such viral replication may boost CD8+T-cell responses against HHV-8 lytic cycle proteins. This T-cell reactivity in turn acts to quell viral replication and prevent development of disease. Similarly, CD8+ T cells specific for peptides of EBV lytic cycle antigens have been observed during primary and latent EBV infection,7,16,17,57 suggesting that this immune response is involved in containment of EBV replication and related diseases. CTL reactivity to lytic cycle proteins is also postulated to be important in control of viral replication during acute, primary infection with MHV-68.10
Our study demonstrates that there is a novel HLA A*0201–restricted, immunogenic CD8+ T-cell epitope for HHV-8 lytic cycle protein gB. The identification of such CD8+ T-cell epitopes for HHV-8 proteins will be useful to our understanding of the role of HLA class I-restricted immunity in development of KS and other HHV-8–related diseases. We2,3 and others4 have previously shown that HHV-8 lytic cycle proteins gB, gH, MCP, MiCP, IE, and K8.1 elicit CD8+ T-cell responses during primary and persistent HHV-8 infections. A vaccine approach based on such T-cell–specific targets of the virus could potentially prevent HHV-8 infection, reactivation, and the development of associated diseases.
We thank Dr Xiao-Qing Zhao, Aki Hoji, Christine Kalinyak, Luann Borowski and Susan McQuiston for their research suggestions and technical assistance, and Judy Malenka for secretarial assistance. We also thank Bill Buchanan for clinical assistance and the Pitt Men's Study MACS staff and volunteers for their dedication and support. This work was done as part of the requirements for the doctorate degree by Q.J.W. in the Department of Infectious Diseases and Microbiology of the University of Pittsburgh Graduate School of Public Health.
Supported in part by National Institutes of Health grants P30 CA47904, R01 CA82053, R01 CA75957, and R03 CA81600.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Charles R. Rinaldo, Jr, Department of Infectious Diseases and Microbiology, Graduate School of Public Health, University of Pittsburgh, A427 Crabtree Hall, 130 DeSoto St, Pittsburgh, PA 15261; e-mail: rinaldo+@pitt.edu.