Abstract
Variant D occurs frequently in Africans. However, considerably lessRHD alleles have been described in this population compared with Europeans. We characterized 5 new RHD alleles, dubbedDAU-0 to DAU-4, that shared a T379M substitution and occurred in a cDe haplotype.DAU-1 to DAU-4 were detected in Africans with partial D phenotypes. They harbored one and 2 additional missense mutations, respectively, dispersed throughout the RhD protein. An anti-D immunization was found in DAU-3. DAU-0carrying T379M only was detected by screening European blood donors and expressed a normal D phenotype. Within the phylogeny of theRHD alleles, DAU formed an independent allele cluster, separate from the DIVa, weak D type 4, and Eurasian D clusters. The characterization of the RH phylogeny provided a framework for future studies on RH alleles. The identification of theDAU alleles increased the number of known partial D alleles in Africans considerably. DAU alleles may be a major cause of antigen D variability and anti-D immunization in patients of African descent.
Introduction
The D antigen of the RH blood group (ISBT 004.001; RH1; CD240D; “Rhesus D”) is the most important blood group antigen determined by a protein, because D-negative individuals are easily anti-D immunized.1 This antibody remains the leading cause for the hemolytic disease of the newborn,2 3 and antigen D–compatible transfusion is standard in modern transfusion therapy.
D-positive individuals harboring a “partial” D antigen may produce an allo–anti-D, too. Among Europeans, the population frequency of all known partial D phenotypes combined is less than 1%.4,5The molecular basis is generally a gene conversion, in which parts of the RHD gene were substituted by the respective segments of the RHCE gene, and single missense mutations.6The molecular characterization of aberrant RHD alleles was much facilitated in Europeans by the frequent occurrence of theRHD gene deletion.7,8 Transfusion strategies were devised to ensure D-negative transfusion in carriers of D category VI, which was known to be the clinically most relevant partial D occurring in Europeans.9
The situation is more intricate in Africans: The occurrence of aberrantRHD alleles and anti-D immunizations in D-positive individuals is much more frequent than in Europeans.10The serologic testing is confounded by frequent “African” alleles that almost defy serologic recognition, like D category III types.11 Molecular analysis revealed that there are often multiple missense mutations, rather than single ones.12The molecular characterization of RHD alleles was hampered in Africans by the frequent occurrence ofRHDψ,13Ccdes,14 or the concomitant presence of 2 different partial D alleles. RHDψ andCcdes do not express a D antigen, yet they harbor a grossly intact RHD allele or an RHD-CE-Dhybrid allele, respectively, that often interferes with RHDpolymerase chain reaction (PCR) and RHD-specific sequencing.
We described 5 RHD alleles that shared a T379M substitution. Four of these alleles expressed a partial D phenotype characterized by the lack of distinct D epitopes or by an anti-D immunization event. We provide a detailed RHD phylogeny in which the DAUalleles formed a previously unknown cluster.
Materials and methods
Blood samples
There were 6 ethylenediaminetetraacetic acid (EDTA)–anticoagulated blood samples referred to our laboratory for problems with D typing or anti-D production in a D-positive individual (sample RIR no. 38 of the Rhesus Immunization Registry, accessible athttp://www.uni-ulm.de/∼wflegel/RIR). In addition, EDTA-anticoagulated blood samples were collected from blood donors of ccDee phenotype in Baden-Württemberg, Germany. DNA was isolated using QiaAmp blood kit (Qiagen, Hilden, Germany) or by a modified salting-out procedure.15 In addition, 2 DNA samples with rare RhCE phenotypes typically occurring in Africans (95-012: hrs- and 95-013: Rh: -34) were obtained from the serum, cells, and fluid exchange (SCARF).
Sequencing of the 10 RHD exons from genomic DNA
Nucleotide sequencing of the 10 RHD exons was performed as previously described.16-18 The standard PCR for RHD exon 2 amplification failed with some alleles belonging to the DIVa cluster (eg, in sample 95-013); in such samples RH exon 2 was amplified using primers re12c and re2317 and sequenced in a RHD-specific manner using primer re17. Primer sequences were re12c, attagccgggcacggtggtg; and re17, ctcgtctgcttcctcctcg.
Polymerase chain reaction with sequence-specific priming
A PCR with sequence-specific priming (PCR-SSP) was devised to detect or to confirm the 1136 C > T substitution in theDAU alleles and triggered to work under similar PCR conditions as a PCR-SSP system previously developed for RHDtyping.18 19 The positive control was a 434–base-pair (bp) PCR fragment of the human growth hormone gene. Specific primers dau1b and daub as well as control primers were used at concentrations of 1 μM. Amplifications were carried out with Taq (Qiagen) in a final volume of 10 μL. Primer sequences were dau1b, ttggccatcgtgatagctcacat; and daub, ggagatggggcacatagacatc.
Population screen for DAU
To screen for DAU alleles among Europeans, 194 random ccDee donations were screened for the 1136 C > T substitution by PCR-SSP. The presence of a DAU allele was confirmed by sequencing of RHD exon 8. The DAU type was determined by sequencing the 10 RHD exons from genomic DNA.DAU allele frequencies were calculated based on phenotype and haplotype frequencies previously determined for the local donor population.9
Antigen density and Rhesus index
Flow cytometric determination of antigen density and Rhesus index was performed as described previously.16,20 The secondary antibody was goat anti–human IgG, Fab-fragment, fluorescein isothiocyanate (FITC)–conjugated (Jackson Immunoresearch, supplied by Dianova, Hamburg, Germany). For the DAU-3 sample, which had a positive direct agglutination test, the background fluorescence was determined by incubating the sample with secondary antibody only. Monoclonal anti-D antibodies were provided by the 3rd International Workshop on Monoclonal Antibodies against Human Red Blood Cells and Related Antigens.21 The following IgG anti-D antibodies were used as primary antibodies: D-89/47 (workshop no. III-1-29); HG/92 (III-30); D-90/7 (III-31); D-90/17 (III-32); D-90/12 (III-33); 17010C9 (III-36); AUB-2F7/Fiss (III-41); LOR11-2D9 (III-43); LOR12-E2 (III-44); LOR17-6C7 (III-45); LOR17-8D3 (III-46); LOR28-21D3 (III-47); LOR28-7E6 (III-48); LOR29-F7 (III-49); LORA (III-50); LORE (III-51); NAU3-2E8 (III-53); NAU6-4D5 (III-55); NOI (III-56); SAL17-4E8 (III-58); SAL20-12D5 (III-59); SALSA-12 (III-60); 822 (III-68); BTSN4 (III-71); BTSN6 (III-72); BTSN10 (III-73); LHM76/58 (III-74); LHM76/55 (III-75); LHM76/59 (III-76); LHM77/64 (III-77); LHM59/19 (III-78); LHM50/2B (III-80); LHM169/80 (III-81); LHM169/81 (III-82); C205-29 (III-88); CLAS1-126 (III-89); F5S (III-90); H2D5D2F5 (III-93); RAB.B15 (III-94); BIRMA-DG3 (III-95); BIRMA-D6 (III-96); BIRMA-D56 (III-97); P3G6 (III-101); P3AF6 (III-102); BRAD3 (III-105); L87.1G7 (III-108); MS26 (III-112); D10 (III-114); HIRO-3 (III-117); HIRO-4 (III-118); ID6-H8 (III-119); HIRO-7 (III-120); HIRO-8 (III-121); HIRO-2 (III-122); D6DO2 (III-123); and MCAD-6 (III-124).
Epitope patterns
Agglutination was tested in a gel matrix test (LISS-Coombs 37°C, DiaMed-ID Micro Typing System; DiaMed, Cressier sur Morat, Switzerland) using the following antibodies in addition to those tested in flow cytometry: B9A4B2 (workshop no. III-1-28); HeM-92 (III-34); 175-2 (III-35); NaTH28-3C11 (III-37); NaTH87-4A5 (III-38); NaTH53-2A7 (III-39); CAZ7-4C5 (III-42); MAR-1F8 (III-52); NAU6-1G6 (III-54); NOU (III-57); VOL-3F6 (III-61); ZIG-189 (III-62); 819 (III-69); LHM70/45 (III-79); LHM174/102 (III-83); LHM50/3.5 (III-84); LHM59/25 (III-85); LHM59/20 (III-86); T3D2F7 (III-87); P3187 (III-98); P3F17 (III-99); P3F20 (III-100); MS201 (III-113); HIRO-1 (III-115); HIRO-6 (III-116); HS114 (III-134); and BS87 (III-180).
Routine D typing
Reactions of the DAU phenotypes in routine D-typing conditions were established using commercial anti-D BS226 (Biotest, Dreieich, Germany), BS232 (Biotest), RUM1 (Immucor, Norcross, GA), and D14E11 (Immucor) in tube technique. In addition, P3×61 (Diagast, Loos, France) was tested in a gel matrix test.
Phylogeny of RHD alleles
A possible phylogenetic tree for RHD alleles was developed which was based on the RHD coding sequence and the presence of a C or E allele. Nucleotide substitutions, gene conversions, recombinations, and mutations in the accompanying RHCE alleles were counted as equivalent single events. The tree was devised manually to minimize the required number of events. Within RHCE, only standard C alleles caused by the gene conversion around exon 222 were counted as C positive. Because of insufficient data, further intricacies of RHCE alleles like the 16 Trp/Cys, 245 Leu/Val, and 336 Gly/Cys polymorphisms were disregarded. Sequences from chimpanzee (Pan troglodytes Rh-like protein IIR, nucleic acid accession number L3705023) were used for external rooting. The RHD alleles shown in the phylogeny tree were published previously13,14,16-18 24-31 or described in this study for DAU-0 to DAU-4 and DIII type 5.
Nomenclature
The name DAU derived from “D of African origin” (in German: D afrikanischen Ursprungs) and is pronounced like in “now.”
Results
DAU alleles
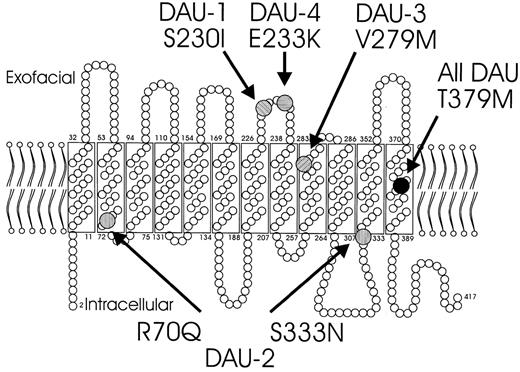
There were 5 RHD alleles identified (Table1). These alleles constituted a cluster, because they shared a 1136C > T single nucleotide polymorphism (SNP) causing a T379M substitution. T379M only was found in DAU-0 which represented the primordial allele of theDAU allele cluster. The other 4 DAU alleles harbored one or 2 additional substitutions dispersed in the various segments of the protein (Figure 1).
RHD alleles described in this study
| Trivial name . | Allele . | Nucleotide change . | Effect on protein sequence . | Exon involved . | Number of probands . | Ethnic origin . |
|---|---|---|---|---|---|---|
| DAU-0 | RHD(T379M) | 1136 C > T | Thr to Met at 379 | 8 | 3 | European* |
| DAU-1 | RHD (S230I,T379M) | 689 G > T | Ser to Ile at 230 | 5 | 3 | African |
| 1136 C > T | Thr to Met at 379 | 8 | ||||
| DAU-2 | RHD (R70Q,S333N,T379M) | 209 G > A | Arg to Gln at 70 | 2 | 1 | African† |
| 998 G > A | Ser to Asn at 333 | 7 | ||||
| 1136 C > T | Thr to Met at 379 | 8 | ||||
| DAU-3 | RHD (V279M,T379M) | 835 G > A | Val to Met at 279 | 6 | 1 | African |
| 1136 C > T | Thr to Met at 379 | 8 | ||||
| DAU-4 | RHD(E233K,T379M) | 697 G > A | Glu to Lys at 233 | 5 | 1 | African† |
| 1136 C > T | Thr to Met at 379 | 8 |
| Trivial name . | Allele . | Nucleotide change . | Effect on protein sequence . | Exon involved . | Number of probands . | Ethnic origin . |
|---|---|---|---|---|---|---|
| DAU-0 | RHD(T379M) | 1136 C > T | Thr to Met at 379 | 8 | 3 | European* |
| DAU-1 | RHD (S230I,T379M) | 689 G > T | Ser to Ile at 230 | 5 | 3 | African |
| 1136 C > T | Thr to Met at 379 | 8 | ||||
| DAU-2 | RHD (R70Q,S333N,T379M) | 209 G > A | Arg to Gln at 70 | 2 | 1 | African† |
| 998 G > A | Ser to Asn at 333 | 7 | ||||
| 1136 C > T | Thr to Met at 379 | 8 | ||||
| DAU-3 | RHD (V279M,T379M) | 835 G > A | Val to Met at 279 | 6 | 1 | African |
| 1136 C > T | Thr to Met at 379 | 8 | ||||
| DAU-4 | RHD(E233K,T379M) | 697 G > A | Glu to Lys at 233 | 5 | 1 | African† |
| 1136 C > T | Thr to Met at 379 | 8 |
Detected in the local donor population of southwestern Germany; in addition, sequencing of the 95-012 DNA sample revealed heterozygosities for 509 T/C, 667 T/G, and 1136 C/T, suggestive of aDOL/DAU-0 genotype.
Samples referred without explicit information on ethnic origin; African descent inferred from African names.
Schematic representation of the RhD proteins observed in the 5 DAU phenotypes.
All DAU types share a T379M substitution (black disk) that is located in the twelfth transmembrane protein segment. DAU-1 to DAU-4 have additional substitutions: the S230I substitution of DAU-1 and the E233Q substitution of DAU-4 are both located in exofacial loop 4; the R70Q and S333N substitutions of DAU-2 position near the border of intramembrane and intracellular protein segments; the V279M substitution of DAU-3 locates at an intramembraneous protein segment proximate to exofacial loop 5.
Schematic representation of the RhD proteins observed in the 5 DAU phenotypes.
All DAU types share a T379M substitution (black disk) that is located in the twelfth transmembrane protein segment. DAU-1 to DAU-4 have additional substitutions: the S230I substitution of DAU-1 and the E233Q substitution of DAU-4 are both located in exofacial loop 4; the R70Q and S333N substitutions of DAU-2 position near the border of intramembrane and intracellular protein segments; the V279M substitution of DAU-3 locates at an intramembraneous protein segment proximate to exofacial loop 5.
Population frequencies
DAU-1 to DAU-4 samples were referred to our laboratory because of typing problems or, in the case of DAU-3 (sample RIR no. 38), of an anti-D immunization. All probands carrying these alleles were, if known, of African descent (Table 1). To determine the possible presence of such alleles in Europeans, we screened for the common 1136C > T SNP by PCR-SSP (Figure 2). Among 194 random samples of ccDee phenotype, 3 samples (1.5%) were positive for the 1136C > T SNP. These samples, however, lacked any additional SNP in the coding sequence and represented DAU-0. The haplotype frequency of the cDe haplotype withDAU-0 was 1:3159 (95% confidence interval: 1:1170-1:11 587). The frequency of the DAU-0 phenotype in the German population was calculated to be 1:3843. All 4 other DAUalleles were infrequent in whites (cumulative frequency < 1:3164, upper limit of 95% confidence interval according to the Poisson distribution).
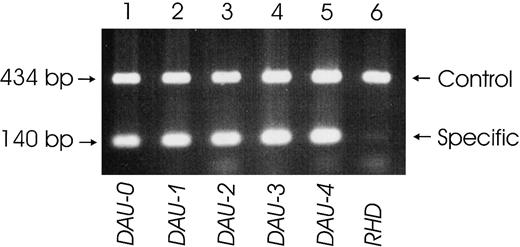
Specific detection of the T379M substitution by PCR-SSP.
A PCR-SSP was devised that amplified a specific 140-bp product from the aberrant RHD exon 8 in all DAU alleles tested (lane 1: DAU-0; lane 2: DAU-1; lane 3:DAU-2; lane 4: DAU-3; lane 5: DAU-4). In a normal D-positive sample (lane 6), only the 434-bp control product deriving from the human growth hormone gene is amplified.
Specific detection of the T379M substitution by PCR-SSP.
A PCR-SSP was devised that amplified a specific 140-bp product from the aberrant RHD exon 8 in all DAU alleles tested (lane 1: DAU-0; lane 2: DAU-1; lane 3:DAU-2; lane 4: DAU-3; lane 5: DAU-4). In a normal D-positive sample (lane 6), only the 434-bp control product deriving from the human growth hormone gene is amplified.
Phenotypes of DAU alleles
Antigen density and Rhesus index.
As previously noted for weak D samples,16 there was no simple relation of the type of substitution (Figure 1) to the antigen density or to the Rhesus index (Table 2). Both the antigen density and the Rhesus index of the DAU-0 phenotype were about normal, rendering it indistinguishable from the normal antigen D–positive phenotype. However, the extracellular substitutions in the DAU-1 and DAU-4 phenotypes correlated well with their much diminished Rhesus index, which is typical for partial D. The DAU-2 with its low antigen density was reminiscent of weak D because its 2 unique substitutions were located at the inner boundary of the red cell membrane. The Rhesus index of DAU-3 indicated its propensity to anti-D immunization, while its antigen D density at the lower end of the normal range would render DAU-3 carriers being transfused with D-positive blood units.
Antigen D density and Rhesus D similarity index
| RH phenotype . | Antigen density . | Rhesus index . | Percentiles of D epitopes detected . | Antibodies . | % of reference† . | |||
|---|---|---|---|---|---|---|---|---|
| 10% . | 50% . | 90% . | Tested . | > Cutoff* . | ||||
| DAU-0 | 15 285 | 0.74 | 12 412 | 15 285 | 16 883 | 56 | 56 | 76 |
| DAU-1 | 2 113 | 0.06 | 260 | 1 152 | 4 091 | 55 | 41 | 10 |
| DAU-2‡ | 373 | 0.17 | 276 | 373 | 1 626 | 55 | 55 | 2 |
| DAU-3 | 10 879 | 0.11 | 2 542 | 9 289 | 23 850 | 44 | 39 | 54 |
| DAU-4 | 1 909 | 0.01 | 55 | 358 | 5 595 | 56 | 25 | 9 |
| ccDee sample | 20 166 | 0.74 | 17 188 | 20 166 | 23 302 | 55 | 55 | NA2-153 |
| RH phenotype . | Antigen density . | Rhesus index . | Percentiles of D epitopes detected . | Antibodies . | % of reference† . | |||
|---|---|---|---|---|---|---|---|---|
| 10% . | 50% . | 90% . | Tested . | > Cutoff* . | ||||
| DAU-0 | 15 285 | 0.74 | 12 412 | 15 285 | 16 883 | 56 | 56 | 76 |
| DAU-1 | 2 113 | 0.06 | 260 | 1 152 | 4 091 | 55 | 41 | 10 |
| DAU-2‡ | 373 | 0.17 | 276 | 373 | 1 626 | 55 | 55 | 2 |
| DAU-3 | 10 879 | 0.11 | 2 542 | 9 289 | 23 850 | 44 | 39 | 54 |
| DAU-4 | 1 909 | 0.01 | 55 | 358 | 5 595 | 56 | 25 | 9 |
| ccDee sample | 20 166 | 0.74 | 17 188 | 20 166 | 23 302 | 55 | 55 | NA2-153 |
Number of monoclonal anti-D antibodies detecting at least 1/10 of the 90th percentile of the epitope density detected by all anti-D antibodies.
Antigen D density as percentage of control cells with ccDee phenotype.
The determination of Rhesus index and antigen density may be confounded by the very low overall epitope densities.
NA indicates not applicable because sample is reference control.
Epitope patterns.
The D epitope patterns of the DAU phenotypes were distinct (Table3). Despite DAU-1 having a much higher antigen density than DAU-2, more anti-D antibodies agglutinated DAU-2 than DAU-1 red blood cells. The profile of DAU-4 was almost identical to that reported for DHK,32 alias DYO,33which shared the E233K substitution. DAU-0 had a normal D-positive epitope pattern.
Reactivity patterns of antibodies with DAU samples
| Pattern3-150 . | Anti-D tested3-151 . | DAU-phenotype3-152 . | |||||
|---|---|---|---|---|---|---|---|
| 1-9 . | 1-37 . | IgG . | IgM . | DAU-0 . | DAU-1 . | DAU-2 . | DAU-4 . |
| 1 | 1 | 82, 123 | + | − | + | − | |
| 1 | 2 | 79 | + | − | + | − | |
| 83 | + | − | − | − | |||
| 2 | 3 | 44, 51 | + | + | + | − | |
| 2 | 4 | 48 | + | − | − | − | |
| 3 | 5 | 43, 49, 75 | + | + | + | + | |
| 4 | 6 | 45 | + | + | + | + | |
| 5 | 7 | 42, 116 | + | − | − | − | |
| 5 | 10 | 88, 89 | + | + | + | + | |
| 41 | + | − | + | − | |||
| 52 | + | − | − | − | |||
| 5 | 11 | 69 | + | − | − | − | |
| 6/7 | 12 | 102 | + | − | + | + | |
| 46 | + | − | + | − | |||
| 113 | + | − | − | + | |||
| 35 | + | − | − | − | |||
| 6/7 | 13 | 29, 36, 47, 90, 93, 105 | + | + | + | + | |
| 6/7 | 15/16 | 31, 32, 71, 80, | + | + | + | + | |
| 56, 58, 81, 95, 97, 114 | + | + | + | − | |||
| 108 | + | − | + | + | |||
| 59 | + | − | + | − | |||
| 6/7 | 17 | 37 | + | − | − | + | |
| 28, 34, 39, 98, 99, 115, 134 | + | − | − | − | |||
| 6/7 | 18 | 30 | + | + | + | + | |
| 119 | + | + | − | + | |||
| 84, 85, 86, 87, 100 | + | − | − | − | |||
| 6/7 | 20/21 | 33 | + | + | + | + | |
| 94 | + | + | + | − | |||
| 122 | + | − | + | − | |||
| 61 | + | − | − | − | |||
| 8 | 22 | 74, 78 | + | + | + | + | |
| 9 | 23 | 77, 96, 101, 112, 118, 120, 121 | + | + | + | + | |
| 97 | + | + | + | − | |||
| N/A | 31 | 54, 57 | + | − | − | − | |
| N/A | 32 | 62 | + | − | + | − | |
| N/A | 33 | 38 | + | − | − | − | |
| N/A | 34 | 50 | + | − | + | − | |
| N/A | 35 | 60 | + | − | + | − | |
| N/A | 36 | 55, 72, 76 | + | + | + | + | |
| 53 | + | − | + | − | |||
| N/A | 37 | 68, 73, 117, 124, 180 | + | + | + | + | |
| Pattern3-150 . | Anti-D tested3-151 . | DAU-phenotype3-152 . | |||||
|---|---|---|---|---|---|---|---|
| 1-9 . | 1-37 . | IgG . | IgM . | DAU-0 . | DAU-1 . | DAU-2 . | DAU-4 . |
| 1 | 1 | 82, 123 | + | − | + | − | |
| 1 | 2 | 79 | + | − | + | − | |
| 83 | + | − | − | − | |||
| 2 | 3 | 44, 51 | + | + | + | − | |
| 2 | 4 | 48 | + | − | − | − | |
| 3 | 5 | 43, 49, 75 | + | + | + | + | |
| 4 | 6 | 45 | + | + | + | + | |
| 5 | 7 | 42, 116 | + | − | − | − | |
| 5 | 10 | 88, 89 | + | + | + | + | |
| 41 | + | − | + | − | |||
| 52 | + | − | − | − | |||
| 5 | 11 | 69 | + | − | − | − | |
| 6/7 | 12 | 102 | + | − | + | + | |
| 46 | + | − | + | − | |||
| 113 | + | − | − | + | |||
| 35 | + | − | − | − | |||
| 6/7 | 13 | 29, 36, 47, 90, 93, 105 | + | + | + | + | |
| 6/7 | 15/16 | 31, 32, 71, 80, | + | + | + | + | |
| 56, 58, 81, 95, 97, 114 | + | + | + | − | |||
| 108 | + | − | + | + | |||
| 59 | + | − | + | − | |||
| 6/7 | 17 | 37 | + | − | − | + | |
| 28, 34, 39, 98, 99, 115, 134 | + | − | − | − | |||
| 6/7 | 18 | 30 | + | + | + | + | |
| 119 | + | + | − | + | |||
| 84, 85, 86, 87, 100 | + | − | − | − | |||
| 6/7 | 20/21 | 33 | + | + | + | + | |
| 94 | + | + | + | − | |||
| 122 | + | − | + | − | |||
| 61 | + | − | − | − | |||
| 8 | 22 | 74, 78 | + | + | + | + | |
| 9 | 23 | 77, 96, 101, 112, 118, 120, 121 | + | + | + | + | |
| 97 | + | + | + | − | |||
| N/A | 31 | 54, 57 | + | − | − | − | |
| N/A | 32 | 62 | + | − | + | − | |
| N/A | 33 | 38 | + | − | − | − | |
| N/A | 34 | 50 | + | − | + | − | |
| N/A | 35 | 60 | + | − | + | − | |
| N/A | 36 | 55, 72, 76 | + | + | + | + | |
| 53 | + | − | + | − | |||
| N/A | 37 | 68, 73, 117, 124, 180 | + | + | + | + | |
+ indicates a positive result, − a negative result.
Pattern as described previously by Lomas et al42 (1-9) and Scott11 (1-37), who did not differentiate epitopes 15 from 16 and epitopes 20 from 2111Tab1.
Antibody numbers as defined in “Materials and methods.” Reactivity was tested in a gel matrix test.
DAU-3 was not tested because of a positive direct antiglobulin test (DAT).
Routine D-typing issues.
Applying routine methods for D typing,34 DAU-0 typed D positive, whereas DAU-1, DAU-2, and DAU-4 were not agglutinated by most commercial monoclonal IgM anti-D antibodies (Table4). Hence, DAU-1, DAU-2, and DAU-4 would usually be typed as D-negative, triggering D-negative transfusions, which is the clinically favored management.
DAU phenotypes in routine D typing
| Phenotype . | IgM anti-D monoclonal antibody . | ||||
|---|---|---|---|---|---|
| BS226 . | BS232 . | RUM1 . | D14E11 . | P3X61 . | |
| DAU-0 | Positive | Positive | Positive | Positive | Positive |
| DAU-1 | Negative | Negative | Negative | Negative | Negative |
| DAU-2 | Negative | Negative | Negative | Negative | Negative |
| DAU-4 | Positive | Positive | Negative | Negative | Negative |
| Phenotype . | IgM anti-D monoclonal antibody . | ||||
|---|---|---|---|---|---|
| BS226 . | BS232 . | RUM1 . | D14E11 . | P3X61 . | |
| DAU-0 | Positive | Positive | Positive | Positive | Positive |
| DAU-1 | Negative | Negative | Negative | Negative | Negative |
| DAU-2 | Negative | Negative | Negative | Negative | Negative |
| DAU-4 | Positive | Positive | Negative | Negative | Negative |
DAU-3 could not be tested because of a positive DAT.
DIII type 5
Sequencing of DNA sample 95-013 revealed 6 nucleotide substitutions, 186G > T, 410C > T, 455A > C, 692C > G, 667T > G, 819G > A. This result predicted the homozygous or hemizygous presence of an RHD (L62F, A137V, N152T, T201R, F223V) allele that was dubbed DIII type 5, because of its similarity to the molecular structure described for mditDIIIa.12
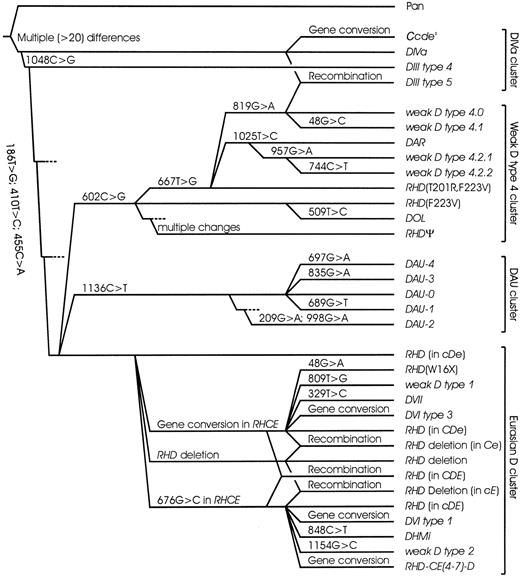
Phylogeny of RHD alleles in humans
Based on the molecular structure of the DAU alleles, the phylogenetic models35,36 of the RHD alleles were extended (Figure 3). TheDAU alleles, which were characterized by a cDehaplotype and a T379M substitution, formed a separate cluster ofRHD alleles. There were 2 other allele clusters also associated with the cDe haplotype: the weak D type 4 cluster was characterized by a common F223V substitution. The DIVa cluster comprising DIII type 4, DIVa,Ccdes, and DIII type 5 was characterized by a common N152T substitution. In addition, DIII type 4 and type 5 as well as allDIVa24 andCcdes14 37 samples investigated by us carried L62F and A137V. The remaining RHD alleles could be derived from the RHD allele prevalent in Eurasians by a single event (point mutation, gene conversion, or deletion) and formed the Eurasian D cluster.
Phylogeny of
RHD in humans. A phylogenetic tree ofRHD is shown for most “African” alleles and representative “Eurasian” alleles. There are 4 main clusters that may be discerned. The DIVa cluster encompasses the DIVa,DIII type 4, and Ccdesalleles. Most samples harboring these alleles share 3 characteristic amino acids (62F, 137V, 152T) that are ancestral, because they are also observed in chimpanzee RH (Pan troglodytesRh-like protein IIR). The weak D type 4 cluster encompassesDAR, DOL, and RHDΨ, too. For this cluster, the RHD (F223V) allele is postulated36but has yet to be shown extant. DIII type 5, a newRHD allele resembling DIIIa, evolved by a recombination between alleles of the DIVa and the weak D type 4 clusters. For the DAU cluster, its primitive type DAU-0 has been found and was shown to be the most frequent DAU allele in Europeans. All enumerated alleles occurred in a cDehaplotype and were predominantly observed in Africans. In contrast, most other RHD alleles were typical for Eurasians, derived from standard Eurasian RHD by a single event, occurred in a CDe or cDE haplotype, and formed the Eurasian D cluster. The tree was mainly based onRHD allelic variability, and dismisses the largely unknownRHCE variability beyond the C and E polymorphism. For each evolutionary step, the event is indicated; the depicted distances of the alleles are arbitrary.
Phylogeny of
RHD in humans. A phylogenetic tree ofRHD is shown for most “African” alleles and representative “Eurasian” alleles. There are 4 main clusters that may be discerned. The DIVa cluster encompasses the DIVa,DIII type 4, and Ccdesalleles. Most samples harboring these alleles share 3 characteristic amino acids (62F, 137V, 152T) that are ancestral, because they are also observed in chimpanzee RH (Pan troglodytesRh-like protein IIR). The weak D type 4 cluster encompassesDAR, DOL, and RHDΨ, too. For this cluster, the RHD (F223V) allele is postulated36but has yet to be shown extant. DIII type 5, a newRHD allele resembling DIIIa, evolved by a recombination between alleles of the DIVa and the weak D type 4 clusters. For the DAU cluster, its primitive type DAU-0 has been found and was shown to be the most frequent DAU allele in Europeans. All enumerated alleles occurred in a cDehaplotype and were predominantly observed in Africans. In contrast, most other RHD alleles were typical for Eurasians, derived from standard Eurasian RHD by a single event, occurred in a CDe or cDE haplotype, and formed the Eurasian D cluster. The tree was mainly based onRHD allelic variability, and dismisses the largely unknownRHCE variability beyond the C and E polymorphism. For each evolutionary step, the event is indicated; the depicted distances of the alleles are arbitrary.
Discussion
We found 5 RHD alleles that shared a T379M missense mutation and formed a previously unknown cluster. There were 4 of these alleles that had one or 2 additional mutations and were observed in individuals of African descent. The fifth allele, RHD(T379M), was detected in Europeans by screening blood donors.DAU alleles may be a major cause of antigen D variability and anti-D immunization in patients of African descent.
Anti-D immunization in transfusion recipients and pregnant women harboring “African” partial D is a continuing problem. For example, 11% of anti-D in pregnancies in the Cape Town area, South Africa, occurred in D-positive women.10 Current D-typing strategies are tuned to detect partial D phenotypes that are typical for white populations.9,34 Although carriers of partial D are more frequent in African populations,25 more than 25 partial D alleles are predominantly observed in Europeans and to date only 5 partial D alleles were typical for Africans.12,16 24-26 Thus, the identification of theDAU cluster increased the number of “African” partial D considerably. Because anti-D immunization may occur in carriers ofDAU alleles, our molecular characterization is instrumental for evaluating the clinical relevance in transfusion recipients.
All of the more than 50 known aberrant RHD alleles expressed variant D antigens.6 36 Unexpectedly, DAU-0encoded a normal phenotype despite its intramembraneous T379M substitution. DAU-0 may be the first example of a host of alleles harboring an amino acid substitution that does not affect their antigen D. The other 4 DAU phenotypes, however, had a low Rhesus index and qualify as partial D. The partial D phenotype was most obvious for DAU-3, in which an anti-D immunization was documented. If such immunizations were frequently occurring in populations with African admixture, the specific detection of the involved DAUalleles might be warranted.
A phylogeny model for the RH haplotypes was originally presented by Carritt et al35 in 1997 which explained the mechanisms shaping RH heterogeneity in Eurasian populations. More recently, these haplotypes were recognized to represent just one branch separated from 2 different clusters of RHD alleles that are primarily observed in African populations.36Alleles of the DAU cluster added to this diversity and represented a third “African” cluster (Figure 3). Each of these 3 “African” allele clusters was characterized by a specific amino acid substitution relative to the “Eurasian” RHD allele: (1) T379M in the DAU cluster, (2) F223V in the weak D type 4 cluster, which includedRHDΨ, DOL, and many alleles sharing F223V and T201R, and (3) N152T in the DIVa cluster, which included DIII type 4, DIVa, and Ccdes.
Recently, Rh-related proteins, including RhAG, have been shown to transport ammonia.38 It is tempting to speculate that amino acid substitutions located in transmembraneous Rh protein segments, like T379M in DAU, F223V and T201R in weak D type 4, and L245V and G336C in Ccdes, may affect the function of the Rh protein. Even a substitution that does not alter the D antigen, like T379M in DAU-0, may still be functionally effective. Malaria and other blood-borne diseases endemic in Africa may favor functional and antigenic variability, as exemplified by glucose-6-phosphate dehydrogenase deficiency39 and lack of Duffy protein expression in red cells,40respectively. Similar processes might confer evolutionary advantages to carriers of aberrant RH alleles.
The RHD alleles of the 3 “African” clusters generally occurred in a cDe haplotype, which indicated that thecE and Ce alleles of RHCE evolved in the “Eurasian” branch after its divergence from the other branches. However, haplotypes of the “Eurasian” cluster represented a sizeable fraction of haplotypes extant in Africans. For example, the frequency of antigen E–encoding “Eurasian” haplotypes is 9.01% among Barotse in Zambia.41 In contrast, even the most frequent alleles of the “African” clusters are very rare among Europeans. This was shown for DAU-0 in the present study (population frequency of 1:3159) and previously determined for weak D type 4 (1:15 000).17 The knowledge ofRH phylogeny is of practical importance because it defines the framework for determining the clinically relevantRH alleles in any population.
In populations without African admixture, including whites, Asians, Arabs, and probably American Indians, partial D phenotypes are likely to be rare and to derive from the limited and serologically well-characterized set of alleles of the Eurasian D cluster. For these populations, the current D-typing strategies applied in Europe9 appear to be appropriate and sufficient. Typing strategies for African populations and those with African admixture may take account of the various frequently occurring alleles of the “African” clusters. Several of these alleles characterized by multiple dispersed amino acid substitutions are difficult to discern by serologic means and may in the future warrant genotyping approaches for detection in patients and donors.
We are greatly indebted to all contributors of the 3rd International Workshop on Monoclonal Antibodies against Human Red Blood Cells and Related Antigens in Nantes, France, 1996, who provided most other monoclonal anti-D antibodies. We thank John J. Moulds and Joann M. Moulds, Philadelphia, PA, for rare DNA samples from the SCARF Exchange program. We acknowledge the expert technical assistance of Marianne Lotsch, Anita Hacker, Sabine Kaiser, and Sabine Zahn.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2002-01-0320.
Supported by the DRK-Blutspendedienst Baden-Württemberg–Hessen, Mannheim; by the University of Ulm (Forschungsförderungsprojekt P. 531); and by the Deutsche Gesellschaft für Transfusionsmedizin und Immunhämatologie (project DGTI/fle/00-01).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Willy A. Flegel, Abteilung Transfusionsmedizin, Universitätsklinikum Ulm, and DRK Blutspendedienst Baden-Württemberg–Hessen, Institut Ulm, Helmholtzstrasse 10, D-89081 Ulm, Germany; e-mail: waf@ucsd.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal