Deficient von Willebrand factor (VWF) degradation has been associated with thrombotic thrombocytopenic purpura (TTP). In hereditary TTP, the specific VWF-cleaving protease (VWF-cp) is absent or functionally defective, whereas in the nonfamilial, acquired form of TTP, an autoantibody inhibiting VWF-cp activity is found transiently in most patients. The gene encoding for VWF-cp has recently been identified as a member of the metalloprotease family and designatedADAMTS13, but the functional activity of the ADAMTS13 gene product has not been verified. To establish the functional activity of recombinant VWF-cp, we cloned the complete cDNA sequence in a eukaryotic expression vector and transiently expressed the encoded recombinant ADAMTS13 in HEK 293 cells. The expressed protein degraded VWF multimers and proteolytically cleaved VWF to the same fragments as those generated by plasma VWF-cp. Furthermore, recombinant ADAMTS13-mediated degradation of VWF multimers was entirely inhibited in the presence of plasma from a patient with acquired TTP. These data show that ADAMTS13 is responsible for the physiologic proteolytic degradation of VWF multimers.
Introduction
Von Willebrand factor (VWF) is a blood glycoprotein that has 2 hemostatic functions.1-3 It is required for platelet adhesion to sites of vascular damage and acts as a carrier protein for blood-clotting factor VIII in the circulation. The VWF molecule is synthesized by vascular endothelial cells and megakaryocytes, with a large propeptide at its N-terminal end. After translocation into the endoplasmic reticulum, dimers of VWF precursor subunits are formed by disulfide bonding near their C-termini. These tail-to-tail dimers are transported to the Golgi complex, where additional head-to-head disulfide bonds are formed near the N-termini of the dimers. After cleavage of the propeptide, the multimers are stored in the endothelial Weibel-Palade bodies or are secreted into the plasma.
The largest multimers of circulating VWF contain more than 40 dimers and can exceed a molecular weight of 20 000 kDa. The largest VWF multimers are hemostatically most active and effectively mediate platelet adhesion. Multimeric VWF is degraded by a plasma protease,4,5 and, if this process is impaired, unusually large VWF (ULvWF) multimers are found in plasma. These ULvWF multimers have the propensity to agglutinate circulating platelets at sites with high levels of shear stress. Increased ULvWF plasma concentrations have been correlated with the risk for platelet clumping in the microvasculature in chronic relapsing thrombotic thrombocytopenic purpura (TTP).6 This disease, originally described by Moschcowitz,7 is characterized by microangiopathic hemolytic anemia, thrombocytopenia, fluctuating neurologic impairment, renal dysfunction, and fever.8-11 There are 3 clinical subcategories of TTP: an acute idiopathic or sporadic form, an intermittent form with eventual relapse, and a chronic relapsing form, often but not always occurring in siblings. In the familial or hereditary form of TTP, VWF-cleaving protease (VWF-cp) is not produced in sufficient quantity, whereas in acute idiopathic TTP, an autoantibody-inhibiting VWF-cp activity is found transiently in most patients.12-14 The reappearance of the inhibitor is associated with intermittent relapsing TTP.13 In children, a phenotypically similar disorder termed hemolytic uremic syndrome (HUS), in which renal dysfunction predominates, can be found. However, this disease is not associated with deficiency or inhibition of VWF-cp.12
From 1988 to 1991, the annual TTP incidence in the United States was an estimated 3.7 cases per 1 million residents.15 Most of these cases were acute idiopathic TTP. For unknown reasons, but probably not simply because of improved recognition, the incidence of the disease seems to have increased in recent years.9 10
A treatment regimen of fresh-frozen plasma (FFP) or cryosupernatant plasma results in the disappearance of ULvWF multimers in patients with chronic relapsing TTP.16 From normal human plasma,5 a specific VWF-cp of high molecular weight has been partially purified.4 This metalloprotease was activated by Ca2+/Ba2+, and it metabolized large VWF multimers to smaller forms. It was not inhibited by inhibitors of serine and cysteine proteases. The peptide bond between residues Tyr842 and Met843 of VWF, which is cleaved in vivo,17 has also been shown to be cleaved by the partially purified protease.4
Because the protease is only present at low concentrations in plasma, making it difficult to purify therapeutic quantities, the preferred treatment strategy for TTP remains therapeutic plasma exchange with FFP or cryosupernatant.18 Such treatment has been repeatedly found to be effective in several large patient series.9,18,19 Patients with hereditary TTP and severe constitutional VWF-cp deficiency can be treated and kept in remission by simple FFP infusion.8,20 This treatment is, however, not only inconvenient but also associated with complications.21 Furthermore, FFP carries a risk for transmission of infectious diseases.
The gene encoding VWF-cp was recently identified as a member of the ADAMTS metalloprotease family22,23 and was designatedADAMTS13.24-27 Analysis of the genomic DNA of patients with hereditary TTP showed that mutations ofADAMTS13, putatively leading to severe VWF-cp deficiency, are the molecular mechanisms responsible for constitutional TTP.24 Identification of the gene is an important step toward the production of recombinant VWF-cp. The recombinant protein holds the promise of being a new treatment modality for patients with TTP and will improve our understanding of the pathogenesis of the disease. ADAMTS13 data available to date have fallen short of establishing the functional activity of the gene product. We therefore set out to clone and express the gene of ADAMTS13and to conclusively characterize the function of the protein.
Materials and methods
Database searches
Nucleic acid sequence information was obtained by searching the NCBI GenBank (http://www.ncbi.nlm.nih.gov/). Exon searches were conducted with the Grail program (http://compbio.ornl.gov/Grail-1.3). The SMART program (http://smart.embl-heidelberg.de) was used for analyzing domain structures.28 Information about the ADAMTS proteins can be found in (http://www.gene.ucl.ac.uk/nomenclature/genefamily/adamts.html). Amino acid sequences were compared by use of the EMBL protein database (http://www.ebi.ac.uk).
Reverse transcription of poly-A RNA
To obtain long transcripts, 3 μg poly-A RNA from human liver (Stratagene, San Diego, CA) was heated for 5 minutes at 64°C and reverse transcribed for 1.5 hours at 42°C with the Expand RT kit from Roche Molecular Biochemicals (Mannheim, Germany) in a final volume of 120 μL with concentrations of reagents according to the instructions of the manufacturer using 100 pmol primer spdT consisting of a poly-dT part, a randomly chosen specific sequence (MC18), and 2 undefined nucleotides (NN) at the 3′ end for specific fixation of the primer (Table 1).
Sequences of primers
| Name . | Sequence . | Remark . |
|---|---|---|
| spdT | 5′-GAGCAAATTCCTGTACTGAC (T)30 NN-3′ | Production of cDNA |
| MC18 | 5′-GAGCAAATTCCTGTACTGAC-3′ | Specific part of spdT |
| 3113 | 5′-CGGATAACAATTTCACACAGG-3′ | UniversallacZ reversed |
| 6142 | 5′-CGGGCTGCAGGCGGGATCCTACACCTGGAG-3′ | Exon 3 forward |
| 6277 | 5′-AATGGTGACTCCCAGGTCGAG-3′ | Exon 6 reversed |
| 6278 | 5′-CGCTCCCTGGTGGAGCTGACC-3′ | Exon 11 forward |
| 6346 | 5′-ATCATGAAGCGTGGAGACAGC-3′ | Exon 13 forward |
| 6351 | 5′-GAGTTGCCTGATGGTAACCG-3′ | Exon 6 forward |
| 6395 | 5′-CCTGGAGGGGTCCCCAGATG-3′ | Exon 16 reversed |
| 6406 | 5′-TGCAGCCCACGGAAGGGCTC-3′ | Exon 14 forward |
| 6407 | 5′-GAGCCCTTCCGTGGGCTGCA-3′ | Exon 14 reversed |
| 6480 | 5′-CATCCTGCGTCTGGACCTGGCTG-3′ | β-Actin forward |
| 6482 | 5′-GACCTGGCCGTCAGGCAGCTCG-3′ | β-Actin reversed |
| 6506 | 5′-CAGGGCTCCAGGCTGCAGGC-3′ | Exon 23 reversed |
| 6546 | 5′-TGGAGGTCAGCACCAACACA-3′ | Exon 11 reversed |
| 6548 | 5′-AGGAAGAGCTGTGTGGCCTG-3′ | Exon 22 forward |
| 6601 | 5′-AGCGAATTCGCTGCAGGCGGCATCCTACACC-3′ | Exon 3 forwardEcoRl |
| 6617 | 5′-AGCCTCGAGCTGGCCAGACACGGAACAAAT-3′ | 3′ untranslated reversed Xhol |
| 6650 | 5′-AGCCTCGAGGGTTCCTTCCTTTCCCTTCCAGGA-3′ | Exon 29 reversed |
| 6727 | 5′-CTGCTGGAAATGGGAGGGTC-3′ | Exon 1 reversed |
| 6728 | 5′-CCGCCTCTGCCTCTGCCTCT-3′ | Exon 2 reversed |
| 6753 | 5′-GACATGTTGCTGCTTTGGGGC-3′ | Exon 27 forward |
| 6839 | 5′-GATCGAATTCGCCGGCCACCATGCACCAGCGTCACCCCCG-3′ | Exon 1 forward EcoRl |
| Name . | Sequence . | Remark . |
|---|---|---|
| spdT | 5′-GAGCAAATTCCTGTACTGAC (T)30 NN-3′ | Production of cDNA |
| MC18 | 5′-GAGCAAATTCCTGTACTGAC-3′ | Specific part of spdT |
| 3113 | 5′-CGGATAACAATTTCACACAGG-3′ | UniversallacZ reversed |
| 6142 | 5′-CGGGCTGCAGGCGGGATCCTACACCTGGAG-3′ | Exon 3 forward |
| 6277 | 5′-AATGGTGACTCCCAGGTCGAG-3′ | Exon 6 reversed |
| 6278 | 5′-CGCTCCCTGGTGGAGCTGACC-3′ | Exon 11 forward |
| 6346 | 5′-ATCATGAAGCGTGGAGACAGC-3′ | Exon 13 forward |
| 6351 | 5′-GAGTTGCCTGATGGTAACCG-3′ | Exon 6 forward |
| 6395 | 5′-CCTGGAGGGGTCCCCAGATG-3′ | Exon 16 reversed |
| 6406 | 5′-TGCAGCCCACGGAAGGGCTC-3′ | Exon 14 forward |
| 6407 | 5′-GAGCCCTTCCGTGGGCTGCA-3′ | Exon 14 reversed |
| 6480 | 5′-CATCCTGCGTCTGGACCTGGCTG-3′ | β-Actin forward |
| 6482 | 5′-GACCTGGCCGTCAGGCAGCTCG-3′ | β-Actin reversed |
| 6506 | 5′-CAGGGCTCCAGGCTGCAGGC-3′ | Exon 23 reversed |
| 6546 | 5′-TGGAGGTCAGCACCAACACA-3′ | Exon 11 reversed |
| 6548 | 5′-AGGAAGAGCTGTGTGGCCTG-3′ | Exon 22 forward |
| 6601 | 5′-AGCGAATTCGCTGCAGGCGGCATCCTACACC-3′ | Exon 3 forwardEcoRl |
| 6617 | 5′-AGCCTCGAGCTGGCCAGACACGGAACAAAT-3′ | 3′ untranslated reversed Xhol |
| 6650 | 5′-AGCCTCGAGGGTTCCTTCCTTTCCCTTCCAGGA-3′ | Exon 29 reversed |
| 6727 | 5′-CTGCTGGAAATGGGAGGGTC-3′ | Exon 1 reversed |
| 6728 | 5′-CCGCCTCTGCCTCTGCCTCT-3′ | Exon 2 reversed |
| 6753 | 5′-GACATGTTGCTGCTTTGGGGC-3′ | Exon 27 forward |
| 6839 | 5′-GATCGAATTCGCCGGCCACCATGCACCAGCGTCACCCCCG-3′ | Exon 1 forward EcoRl |
Polymerase chain reaction
Genomic DNA was purified with the Qiagen Blood kit and polymerase chain reaction (PCR) products with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). When multiple PCR fragments were obtained, the PCR products were subjected to agarose gel electrophoresis and purified by the CONCERT Rapid Gel Extraction System (Life Technologies, Lofer, Austria). All purifications were conducted according to the supplier's protocols. The cDNA solution or genomic DNA (0.5 μL) was subjected to PCR using thermally activated DNA polymerases (FastStart Taq DNA polymerase, Roche Molecular Biochemicals or HotStar Taq polymerase, Qiagen). PCR was carried out in a total volume of 50 μL containing 1 U polymerase in the buffer supplied by the manufacturer (with the addition of GC solution [Roche Molecular Biochemicals] as recommended), 200 μM dNTP, and 50 pmol each forward and reverse primer (Table 1). Samples were overlaid with mineral oil, incubated for 4 minutes at 95°C (14 minutes for HotStar polymerase), and amplified for 45 cycles in a TRIO-Thermoblock (Biometra, Göttingen, Germany) with the following cycle profile: 50 seconds at 95°C, 50 seconds at 62°C, and 1 to 6 minutes at 72°C (1-minute extension for approximately 1 kb DNA). Samples were then fractionated on a 1% agarose gel stained with ethidium bromide.
Amplification of the full-length 5′ cDNA end
The GeneRacer Kit (Invitrogen, Carlsbad, CA) was used to obtain a full-length 5′ end of the cDNA. Truncated mRNA is eliminated from the amplification process by treatment with calf intestinal phosphatase, removal of the 5′ cap structure from intact mRNA with tobacco acid pyrophosphatase, and ligation of the GeneRacer RNA oligo (supplied by the kit) to the decapped RNA. Two hundred fifty nanograms poly-A RNA from human liver (Stratagene) was treated according to the instructions of the supplier. Subsequent reverse transcription (RT) was performed using the specific reversed primer 6728 located on exon 2. PCR using the GeneRacer 5′ primer, primer 6728, nested GeneRacer 5′ primer, and nested primer 6727 was carried out as described above.
Sequencing of PCR products
For each primer pair, multiple independently created PCR products were sequenced on an Applied Biosystems (Foster City, CA) model 373A sequencer using the cycle-sequencing method with dye terminators (BIG Dye Terminator Cycle Sequencing Ready Reaction; Applied Biosystems) and the same primers used in the PCR. Chromatograms were aligned and assembled to the complete cDNA sequence using the Sequence Editor (SeqEd; Applied Biosystems).
Gene expression panel
The mRNA accumulation in various human tissues and developmental stage was determined semiquantitatively with the Rapid-Scan Gene Expression Panel Kit (OriGene Technologies, Rockville, MD). The kit contains a panel of normalized first-strand cDNAs of tissues from brain, heart, kidney, spleen, liver, colon, lung, small intestine, muscle, stomach, testis, placenta, salivary gland, thyroid, adrenal, pancreas, ovary, uterus, prostate, skin, peripheral blood leukocyte (PBL), bone marrow, fetal brain, and fetal liver. In contrast to the instructions of the supplier, the lyophilized cDNAs were dissolved in 10 μL H20. Then either 3.3 or 1 μL of the respective solutions (resulting in 330, 100, 33, 10, 3, 1, 0.33, and 0.1 pg) were subjected to PCR as described above using primers 6753/6650 and the primers specific for the β-actin (6480 and 6428) control.
Construction of an ADAMTS13 cDNA clone
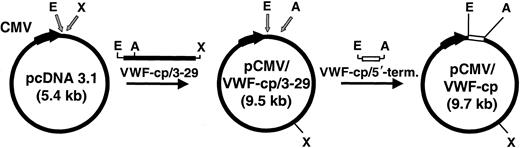
For construction of a cDNA clone, we started with the information of the N-terminal 15–amino acid residues AAGGILHLELLVAVG of the purified plasmatic human VWF-cp.29 Database searching yielded the corresponding nucleic acid sequence, GCT GCA GGC GGC ATC CTA CAC CTG GAG CTG CTG GTG GCC GTG GGC, located on chromosome 9, clone RP11-244N20 (AL158826, GI11544459). Database and exon search programs were then combined with PCR methods to obtain a full-length cDNA clone. Poly-A RNA from liver was reverse transcribed, and the cDNA was PCR amplified with primers specific for some of the putative exons (Table 1). The PCR products of primers 6142 to 6277, 6142 to 6546, 6351 to 6546, 6278 to 6407, 6346 to 6395, and 6406 to 6506 (derived from exons 3-6, 3-11, 6-11, 11-14, 13-16, 14-23, respectively) revealed bands of the expected size. The 3′ end of the cDNA sequence was obtained by PCR amplification of the cDNA using forward primer 6548 of exon 22 and the specific MC18 sequence of the RT primer spdT. Knowing the complete sequence of ADAMTS13, the cDNA from exons 3 to 29 was then amplified with primers 6601 and 6617, digested with EcoRI and XhoI, and cloned intoEcoRI/XhoI sites of pcDNA 3.1 (Invitrogen) resulting in pCMV/VWF-cp/3-29. The 5′-terminal sequence upstream of exon 3 was amplified with primer 6839 containing the 5′-terminal nucleotide sequence of exon 1 (including the start codon), a Kozak consensus sequence, and an EcoRI recognition site and primer 3113. This PCR product was digested with EcoRI andAscI (the amplification product contains an internalAscI site) and was cloned into theEcoRI/AscI sites of pCMV/VWF-cp/3-29, resulting in pCMV/VWF-cp containing the complete VWF-cp cDNA under the control of the cytomegalovirus (CMV) immediate-early promoter/enhancer. A schematic drawing of the cloning of the expression plasmid is shown in Figure 1.
Schematic drawing of the cloning of the recombinant ADAMTS13 expression plasmid.
The 4.12-kb ADAMTS13 cDNA encompassing exons 3 to 29 (black bar) was PCR amplified and cloned into pcDNA3.1 digested withEcoRI/XhoI (gray arrows). In pcDNA3.1, the CMV promoter drives the eukaryotic expression (bold black arrow indicating direction of transcription). The resultant vector pCMV/VWF-cp/3-29 was digested with EcoRI/AscI (gray arrows) and ligated to the 0.7-kb VWF-cp/5′-terminal cDNA fragment (white bar). The final plasmid pCMV/VWF-cp (9.7 kb) was used for the expression of the complete rADAMTS13 gene. A indicates AscI; E,EcoRI; X, XhoI.
Schematic drawing of the cloning of the recombinant ADAMTS13 expression plasmid.
The 4.12-kb ADAMTS13 cDNA encompassing exons 3 to 29 (black bar) was PCR amplified and cloned into pcDNA3.1 digested withEcoRI/XhoI (gray arrows). In pcDNA3.1, the CMV promoter drives the eukaryotic expression (bold black arrow indicating direction of transcription). The resultant vector pCMV/VWF-cp/3-29 was digested with EcoRI/AscI (gray arrows) and ligated to the 0.7-kb VWF-cp/5′-terminal cDNA fragment (white bar). The final plasmid pCMV/VWF-cp (9.7 kb) was used for the expression of the complete rADAMTS13 gene. A indicates AscI; E,EcoRI; X, XhoI.
Transient transfection
The VWF-cp constructs were transiently expressed in HEK 293 (human embryonic kidney fibroblasts; ATCC CRL-1573), Chang liver cells (ATCC CCL-13), and CHO (Chinese hamster ovary; ATCC CRL-9096) cells recombinantly expressing the human endoprotease furin.30The cells were routinely grown in Dulbecco modified Eagle medium/Ham F12 (1:1) medium (Life Technologies) with 10% fetal calf serum (full medium). Transfection was carried out at 85% to 95% confluence using lipofectamine 2000 (Life Technologies) according to the supplier's protocol. Twenty-four hours after transfection, the medium was changed and serum-free full medium was applied to the confluent cells for 24 hours. Conditioned medium was collected, clarified by centrifugation, and concentrated 20-fold using Amicon Centriprep YM-30 (Millipore, Bedford, MA) at 4°C. Cells were harvested with No-zyme (JRH Biosciences, Lenexa, KS), counted, and lysed in 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA (ethylenediaminetetraacetic acid) containing 0.5% Triton X-100 and 1 mM Pefabloc SC at a concentration of 5 × 107 cells/mL as described.31
Antibodies against VWF-cp and Western blot analysis
Protein samples were reduced, denatured, and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on 4% stacking/6% or 8% separation gels and were visualized by Western blotting as described previously.32 For the generation of antibodies directed against the catalytic domain of VWF-cp, a PCR fragment corresponding to nucleotides 221 to 744 in the VWF-cp cDNA was cloned into the NdhI/EcoRI sites of plasmid pRSET-B (Invitrogen) and was used for prokaryotic expression according to the supplier's instructions. The catalytic domain fragment was purified by SDS-PAGE and designated rvWF-cp/cat. Monoclonal antibodies were generated by standard hybridoma techniques. Spleen cell donor mice were immunized subcutaneously twice at a 2-week interval with purified rVWF-cp/cat using Al(OH)3 as an adjuvant. Antibodies from clone 242/H1 were used for immunostaining of recombinant VWF-cp together with alkaline phosphatase–conjugated goat antimouse immunoglobulin G (IgG) antibodies (Sigma, St Louis, MO). For the detection of VWF cleavage products, rabbit anti-VWF antiserum (Dakopatts, Copenhagen, Denmark) and alkaline phosphatase–conjugated goat antirabbit IgG antibodies (Promega, Madison, WI) were used. As a substrate, 5-bromo-4-chloro-3-indolyl-phosphate/p-nitroblue-tetrazolium (BCIP/NBT; Promega) was used.
Assay of VWF-cp activity
The activity of rADAMTS13 was determined at low ionic strength and in the presence of urea as described previously.4Briefly, cell lysate (equivalent to 5 × 107 cells/mL) and conditioned medium (20-fold concentrated) from transfected cells were diluted 1:10, 1:20, and 1:40 in 0.15 M NaCl and 10 mM Tris, pH 7.4, containing 10 mM BaCl2 and were incubated for 5 minutes at 37°C. The incubation mixture (100 μL) was immediately added to 50 μL plasma-derived VWF (50 μg/mL; Stago, Parsippany, NJ) and was transferred to circular dialysis membranes (VSWP, 25-mm diameter; Millipore) floating on 50 mL 1.5 M urea/5 mM Tris, pH 8.0. Overnight incubation at 37°C was stopped by the addition of 10 μL 0.2 M EDTA, pH 7.4. Dilutions of normal human plasma pool (NHP; Baxter AG, Vienna, Austria) were used for assay calibration.
Inhibition of VWF-cp activity
For inhibition of VWF-cp activity, equal volumes of NHP or conditioned medium from cells transfected with the pCMV/VWF-cp vector or the parental vector were mixed with the inhibitory plasma from a patient with acquired TTP and were incubated for 10 minutes at 37°C before dilution and activation with BaCl2. Accordingly, these mixtures contained a 1:20 dilution of each sample and inhibitory plasma. In addition, inhibitory TTP plasma (and NHP as a negative control) was fractionated using Protein G Sepharose (Amersham Pharmacia Biotech, Uppsala, Sweden). Briefly, 150 μL plasma diluted with an equal volume of binding buffer (0.1 M Tris/0.15 M NaCl, pH 7.5) was incubated for 2 hours at 4°C with 75 μL (bed-volume) Protein G Sepharose that had been equilibrated with binding buffer. Nonbound plasma was separated by centrifugation. After washing the Sepharose extensively with binding buffer, IgG was eluted in 0.1 M glycine/HCl, pH 2.8, and was immediately neutralized with 1 M Tris, pH 9.0. Quantitative estimation and analysis of IgG in the individual fractions were achieved by UV absorption, reducing SDS-PAGE, followed by silver staining and by immunostaining with alkaline phosphatase–conjugated antihuman IgG antibody (Sigma). Conditioned medium containing rADAMTS13 and NHP were incubated either concentrated or in serial dilutions as indicated with a constant volume (5 μL) of IgG-depleted plasma or purified IgG before dilution and activation; the VWF-cp activity assay was performed as described earlier.
N-terminal amino acid sequence analysis
The VWF cleavage fragments generated by incubation with rADAMTS13 were separated by reducing SDS-PAGE and were electrotransferred to a polyvinylidene difluoride (PVDF) membrane using 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11, in 10% methanol as transfer buffer. After transfer, the blot was stained with 0.1% Coomassie blue-R250 in 40% methanol/1% acetic acid and was destained with 50% methanol. N-terminal amino acid sequence analysis of the excised protein bands was carried out using standard Edman chemistry with a Procise 491A Protein Sequencer (Perkin Elmer, Shelton, CT).
Results
Cloning of the ADAMTS13 cDNA and semiquantitative determination of mRNA expression levels in various tissues
The N-terminal amino acid sequence AAGGILHLELLVAVG was obtained from plasma-derived VWF-cp29 and was used to construct a full-length cDNA clone (Figure 1). Comparison of the nucleotide sequence of our clone with the published sequence revealed no difference on the amino acid level. However, using a methodology specifically designed to obtain the 5′-end of an mRNA (GeneRacer Kit), we found the nucleotide sequence 5′-CCCATTCCATACTGACCAG-3′ located upstream of the published cDNA sequence (NCBI GenBank AF414401). Additional sequences did not result in an additional exon upstream of exon 1.
The expression pattern of the VWF-cleaving protease was examined semiquantitatively using PCR techniques and a multitissue panel consisting of first-strand cDNA from a variety of human tissue samples. The method is based on the assumption that once a specific RNA becomes rare at a certain dilution, PCR will not be able to pick up a signal consistently. At the limiting concentration, a series of identical PCRs yields positive and negative results. Therefore, titration of a specific cDNA by running the identical PCR reactions with different sample dilutions allows semiquantitative determination of the relative concentration of a given cDNA in a tissue-specific cDNA pool. To screen for the complete transcribed gene, we used primers located at the 3′-terminal end of ADAMTS13. Through this methodology, moderate levels of ADAMTS13 mRNA expression could be found in tissues such as liver, brain, heart, kidney, placenta, muscle, testis, and fetal liver (Table 2). Expression of VWF-cp–specific mRNA was compared against β-actin and was found to be at least 3000-fold less.
Semiquantitative analysis of ADAMTS13 expression in human tissues
| Tissue . | 333 pg . | 100 pg . | 33 pg . | 10 pg . | 3.3 pg . | 1 pg . | 0.33 pg . |
|---|---|---|---|---|---|---|---|
| Brain | + | + | + | + | ± | ± | − |
| Heart | + | + | + | ± | − | − | − |
| Kidney | + | + | + | − | − | − | − |
| Spleen | + | + | − | − | − | − | − |
| Liver | + | + | + | + | + | − | − |
| Colon | − | − | − | − | − | − | − |
| Lung | + | − | − | − | − | − | − |
| Intestine | + | − | − | − | − | − | − |
| Muscle | + | + | + | − | − | − | − |
| Stomach | + | − | − | − | − | − | − |
| Testis | + | + | + | ± | − | − | − |
| Placenta | + | + | + | ± | − | − | − |
| Salivary | + | − | − | − | − | − | − |
| Thyroid | + | ± | − | − | − | − | − |
| Adrenal gland | + | + | − | − | − | − | − |
| Pancreas | + | + | − | − | − | − | − |
| Ovary | + | − | − | − | − | − | − |
| Uterus | + | + | − | − | − | − | − |
| Prostate | + | ± | − | − | − | − | − |
| Skin | + | − | − | − | − | − | − |
| PBL | − | − | − | − | − | − | − |
| Bone marrow | + | − | − | − | − | − | − |
| Fetal brain | + | + | − | − | − | − | − |
| Fetal liver | + | + | + | − | − | − | − |
| Tissue . | 333 pg . | 100 pg . | 33 pg . | 10 pg . | 3.3 pg . | 1 pg . | 0.33 pg . |
|---|---|---|---|---|---|---|---|
| Brain | + | + | + | + | ± | ± | − |
| Heart | + | + | + | ± | − | − | − |
| Kidney | + | + | + | − | − | − | − |
| Spleen | + | + | − | − | − | − | − |
| Liver | + | + | + | + | + | − | − |
| Colon | − | − | − | − | − | − | − |
| Lung | + | − | − | − | − | − | − |
| Intestine | + | − | − | − | − | − | − |
| Muscle | + | + | + | − | − | − | − |
| Stomach | + | − | − | − | − | − | − |
| Testis | + | + | + | ± | − | − | − |
| Placenta | + | + | + | ± | − | − | − |
| Salivary | + | − | − | − | − | − | − |
| Thyroid | + | ± | − | − | − | − | − |
| Adrenal gland | + | + | − | − | − | − | − |
| Pancreas | + | + | − | − | − | − | − |
| Ovary | + | − | − | − | − | − | − |
| Uterus | + | + | − | − | − | − | − |
| Prostate | + | ± | − | − | − | − | − |
| Skin | + | − | − | − | − | − | − |
| PBL | − | − | − | − | − | − | − |
| Bone marrow | + | − | − | − | − | − | − |
| Fetal brain | + | + | − | − | − | − | − |
| Fetal liver | + | + | + | − | − | − | − |
Semiquantitative PCR analysis of ADAMTS13 mRNA expression levels in a panel of human tissues is shown. A commercially available kit containing different concentrations of normalized first-strand cDNAs was used, and the indicated amounts of cDNA from the tissues were PCR amplified. + indicates PCRs always positive; ±, PCRs with mixed results (positive and negative); −, PCRs always negative.
Transient expression of rADAMTS13
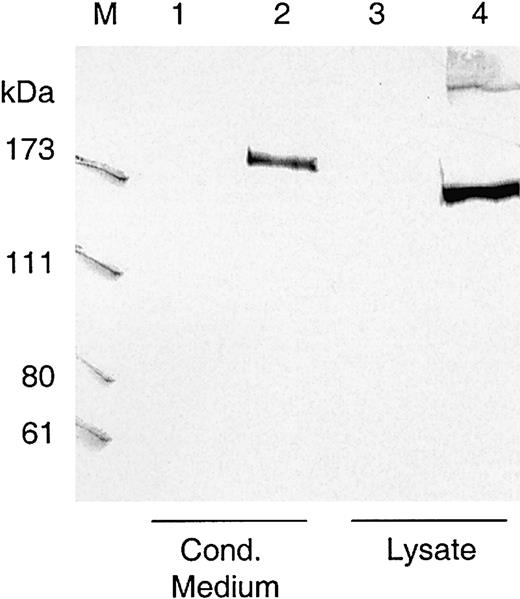
The complete cDNA sequence of ADAMTS13 was cloned in a eukaryotic expression plasmid in front of the CMV promoter, and the encoded VWF-cp was transiently expressed in HEK 293, Chang liver, and CHO/furin cells. Conditioned medium was collected and 20-fold concentrated, and nuclei-free lysate was recovered from the cells. Amounts equivalent to approximately 1 × 106 cells were subjected to Western blot analysis. In lysates of HEK 293 cells transfected with the ADAMTS13 expression construct, the monoclonal antibody 242/H1 directed against the catalytic domain of VWF-cp detected a band at approximately 175 kDa, which was not present in the lysates of HEK 293 cells transfected with the control vector (Figure 2). An immunoreactive VWF-cp band slightly larger than 175 kDa was also detectable in the conditioned medium. There was no obvious difference in the expression pattern, whether solely endogenous cellular (Chang liver cells) or recombinant furin (CHO/furin cells) was present (data not shown).
Western blot of transiently expressed ADAMTS13.
Lysates and conditioned media from transfected HEK 293 cells transiently expressing ADAMTS13 were loaded on SDS-polyacrylamide gels, blotted to nitrocellulose, and visualized by murine monoclonal antibody 242/H1 directed against the catalytic domain of VWF-cp. The amount of lysate and conditioned medium loaded onto the gel was equivalent to approximately 1 × 106 cells. Lanes 1 and 3, conditioned medium and lysate from HEK 293 cells transfected with control vector pcDNA3.1; lanes 2 and 4, conditioned medium and lysate from cells transfected with VWF-cp expression vector pCMV/VWF-cp. M indicates protein standard in kDa.
Western blot of transiently expressed ADAMTS13.
Lysates and conditioned media from transfected HEK 293 cells transiently expressing ADAMTS13 were loaded on SDS-polyacrylamide gels, blotted to nitrocellulose, and visualized by murine monoclonal antibody 242/H1 directed against the catalytic domain of VWF-cp. The amount of lysate and conditioned medium loaded onto the gel was equivalent to approximately 1 × 106 cells. Lanes 1 and 3, conditioned medium and lysate from HEK 293 cells transfected with control vector pcDNA3.1; lanes 2 and 4, conditioned medium and lysate from cells transfected with VWF-cp expression vector pCMV/VWF-cp. M indicates protein standard in kDa.
Degradation of VWF multimers by transiently expressed rADAMTS13
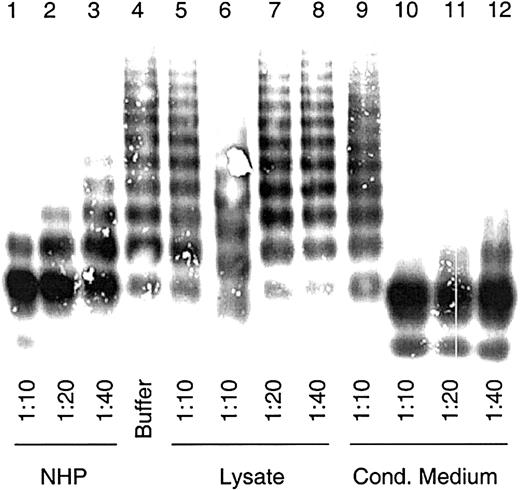
To examine the VWF-cp activity, cell lysate and conditioned medium were assayed for multimer degradation capability as described for plasma-derived VWF-cp.4 A modest proteolytic activity of intracellular recombinant ADAMTS13 (rADAMTS13), similar to that of the 1:40 diluted NHP, was observed in 1:10 diluted lysates of HEK 293 transfections (Figure 3). No VWF degradation was found in the vector controls. However, VWF-cp activity was observed in the cell culture supernatant. In the 1:40 diluted conditioned medium, which had been previously 20-fold concentrated, VWF-cp activity similar to that of NHP at 1:10 dilution was detected. The degradation of VWF multimers was concentration dependent.
VWF-cleaving protease activity in lysates and conditioned media of cells transiently expressing ADAMTS13.
Diluted lysates and conditioned media from transfected HEK 293 cells were activated with BaCl2 and incubated with 2.5 μg VWF. NHP dilutions were used for assay calibration. Multimeric analysis of VWF was carried out by SDS gel electrophoresis in 1% agarose gels, and VWF was detected by immunostaining. Sample dilutions are indicated. VWF-cp assay was carried out with cell lysates (lanes 6-8) and conditioned medium (lanes 10-12) from transfections with plasmid pCMV/VWF-cp and the control vector (lanes 5 and 9) and with NHP (lanes 1-3) and buffer (lane 4) as probes.
VWF-cleaving protease activity in lysates and conditioned media of cells transiently expressing ADAMTS13.
Diluted lysates and conditioned media from transfected HEK 293 cells were activated with BaCl2 and incubated with 2.5 μg VWF. NHP dilutions were used for assay calibration. Multimeric analysis of VWF was carried out by SDS gel electrophoresis in 1% agarose gels, and VWF was detected by immunostaining. Sample dilutions are indicated. VWF-cp assay was carried out with cell lysates (lanes 6-8) and conditioned medium (lanes 10-12) from transfections with plasmid pCMV/VWF-cp and the control vector (lanes 5 and 9) and with NHP (lanes 1-3) and buffer (lane 4) as probes.
VWF-cp activity was estimated by the determination of the collagen-binding activity of degraded VWF substrate after digestion with VWF-cp. We found approximately 4 U/mL rADAMTS13 in the 20-fold concentrated conditioned medium and less than 0.1 U/mL in the lysate of cells transfected with plasmid pCMV/VWF-cp. No activity was obtained in the conditioned medium or lysate from cells transfected with the parental vector. The discrepancy between the amount of intracellular rADAMTS13 detected by immunoblotting and the activity actually found can, at least in part, be explained by the substantial inhibitory activity observed in the cell lysates. When the assay was carried out with conditioned medium containing functional VWF-cp, which was mixed with the lysate from cells transfected with the parental vector, rADAMTS13 activity was strongly impaired (data not shown).
Cleavage of VWF subunits into proteolytic fragments by rADAMTS13
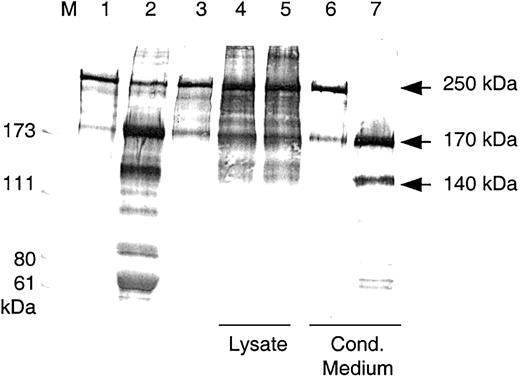
Purified plasma-derived VWF was incubated with diluted conditioned medium that had been activated with BaCl2, and the mixture was dialyzed overnight against 1.5 M urea/5 mM Tris, pH 8.0. The reaction mixture was then subjected to SDS-PAGE under reducing conditions and was immunoblotted. In contrast to lysates and to the buffer control, digestion mixtures containing rADAMTS13 from culture medium degraded the VWF 250 kDa subunit completely into proteolytic fragments of 170 kDa and 140 kDa (Figure4). These fragments were also found with VWF digested by BaCl2- activated NHP.
Cleavage of VWF subunits by rADAMTS13.
Western blot analysis of VWF fragments after VWF-cp digestion. VWF cleavage fragments were separated by SDS-PAGE under reducing conditions. VWF substrate (lane 1) was incubated with NHP (lane 2), buffer (lane 3), lysates, and conditioned media from HEK 293 transfections with the control vector (lanes 4 and 6) and with the VWF-cp expression plasmid (lanes 5 and 7). Lysates, conditioned media, and NHP were diluted 1:10 in the VWF-cp assay. M indicates protein standard in kilodaltons.
Cleavage of VWF subunits by rADAMTS13.
Western blot analysis of VWF fragments after VWF-cp digestion. VWF cleavage fragments were separated by SDS-PAGE under reducing conditions. VWF substrate (lane 1) was incubated with NHP (lane 2), buffer (lane 3), lysates, and conditioned media from HEK 293 transfections with the control vector (lanes 4 and 6) and with the VWF-cp expression plasmid (lanes 5 and 7). Lysates, conditioned media, and NHP were diluted 1:10 in the VWF-cp assay. M indicates protein standard in kilodaltons.
To verify the specificity of the cleavage site, we analyzed the N-terminal amino acid sequence of the cleavage products generated by rADAMTS13. Analysis of the 170-kDa cleavage product yielded the sequence MVTGNPASDEIKRLP, corresponding to amino acid residues 843 to 857 in the intact mature VWF. The N-terminal sequence of the 140-kDa fragment was SLS(C)RPPMVKLV(C)PA, which coincides with the first amino acid residues of the mature VWF. These data indicate that rADAMTS13 cleaved the peptide bond between Tyr842 and Met843, identical to the in vivo cleavage site of plasma-derived VWF-cp.
Inhibition of rADAMTS13-mediated VWF degradation by plasma from a patient with acquired TTP
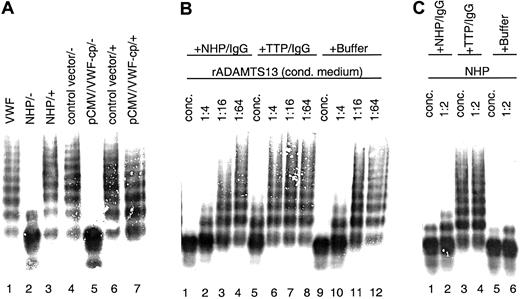
Conditioned medium samples with rADAMTS13 activity were mixed with plasma from a TTP patient containing antibodies against VWF-cp before activation and incubation with the VWF substrate. Assay mixtures were then subjected to VWF multimer analysis. The rADAMTS13-mediated VWF degradation was entirely prevented by the addition of this plasma, and the full multimeric pattern of the VWF substrate was preserved (Figure5A). No change in the multimeric pattern was observed when conditioned medium derived from transfections with the vector control was used. Furthermore, NHP and TTP patient plasma from which the IgG fraction was depleted by incubation with Protein G Sepharose failed to inhibit VWF-cp activity (data not shown). In contrast, when purified IgG (3 mg/mL) from TTP patient plasma was added to serial dilutions of conditioned medium containing rADAMTS13, the VWF-cp activity was diminished with progressive dilution of the conditioned medium (Figure 5B). Similar results were obtained with plasma-derived VWF-cp in NHP (Figure 5C). Neither purified IgG (3.3 mg/mL) from NHP nor buffer contained any VWF-cp inhibitory activity. Taken together, these results clearly show that inhibitors present in TTP patient plasma are IgG antibodies specifically directed toward the ADAMTS13 protein.
Inhibition of rADAMTS13 activity by TTP patient plasma containing antibodies neutralizing VWF-cp activity.
VWF-cp assay mixtures were loaded onto an SDS-1% agarose gel, and the VWF multimer pattern was detected by immunostaining. (A) Inhibitory TTP patient plasma was added (+) to the samples before dilution (1:20) activation and incubation with VWF. Buffer was used instead of TTP plasma in some samples (−). Samples used were pooled NHP (lanes 2 and 3), conditioned medium derived from transfections using the vector control (lanes 4 and 6), and VWF-cp expression vector (lanes 5 and 7). VWF-substrate is shown in lane 1. (B) Purified IgG derived from pooled NHP or acquired TTP patient plasma and buffer as control was mixed with conditioned medium containing rADAMTS13 either concentrated (lanes 1, 5, 9) or diluted 1:4 (lanes 2, 6, 10), 1:16 (lanes 3, 7, 11), or 1:64 (lanes 4, 8, 12). (C) NHP was incubated directly (lanes 1, 3, 5) or diluted 1:2 (lanes 2, 4, 6) with purified IgG from NHP or inhibiting TTP patient plasma and buffer.
Inhibition of rADAMTS13 activity by TTP patient plasma containing antibodies neutralizing VWF-cp activity.
VWF-cp assay mixtures were loaded onto an SDS-1% agarose gel, and the VWF multimer pattern was detected by immunostaining. (A) Inhibitory TTP patient plasma was added (+) to the samples before dilution (1:20) activation and incubation with VWF. Buffer was used instead of TTP plasma in some samples (−). Samples used were pooled NHP (lanes 2 and 3), conditioned medium derived from transfections using the vector control (lanes 4 and 6), and VWF-cp expression vector (lanes 5 and 7). VWF-substrate is shown in lane 1. (B) Purified IgG derived from pooled NHP or acquired TTP patient plasma and buffer as control was mixed with conditioned medium containing rADAMTS13 either concentrated (lanes 1, 5, 9) or diluted 1:4 (lanes 2, 6, 10), 1:16 (lanes 3, 7, 11), or 1:64 (lanes 4, 8, 12). (C) NHP was incubated directly (lanes 1, 3, 5) or diluted 1:2 (lanes 2, 4, 6) with purified IgG from NHP or inhibiting TTP patient plasma and buffer.
Discussion
The nucleotide and predicted amino acid sequence of the VWF-cp has recently been published.24,25 27 The protease was identified as a new member of the ADAMTS family and was designated ADAMTS13. However, experiments verifying the expected functional property—that is, a VWF-cleaving activity of a recombinant ADAMTS13—have not been published so far.
Choice of an appropriate cell line derived from a certain tissue is critical for the production of substantial amounts of recombinant ADAMTS13. Using Northern blot analysis, Zheng et al,25Levy et al,24 and Soejima et al27 found strong mRNA expression of the gene in liver. With a sensitive RT-PCR, the specific mRNA was detected in trace amounts in a number of different tissues. Nonquantitative RT-PCR does not, however, measure the expression levels in tissue. Using a semiquantitative approach, we have provided a more detailed description of the expression pattern in a large number of different tissues. Moderate expression of ADAMTS13 is not restricted to liver. We detected similar amounts of mRNA in, for example, brain, heart, and kidney. However, the expression levels were considerably lower than those of β-actin. The overall low expression of ADAMTS13 is in accordance with that of most mammalian ADAMTS genes in adult tissues.35-37
We transiently expressed the full-length cDNA sequence of theADAMTS13 gene in HEK 293 cells derived from human kidney. We found that most rADAMTS13 protein in the cellular extracts had an estimated molecular weight of 175 kDa, which is in agreement with the molecular weight of VWF-cp purified from human plasma.26 29 Secreted VWF-cp was slightly larger than the corresponding intracellular form. Posttranslational modification, such as glycosylation, might explain the migration differences. The only modest VWF-cp activity observed for intracellular rADAMTS13, at least under the assay conditions used, might also reflect incomplete modification. A lack of chaperonelike proteins essential for adequate secretion might additionally explain the accumulation of inactive rADAMTS13 in the cell. Low concentrations of VWF-cp found in plasma might reflect a fine-tuned and regulated expression of the enzyme, making it difficult to express active VWF-cp constitutively in high amounts. Although the apparent VWF-cp concentration in the conditioned medium was lower than in the cell lysate, VWF-cp activity in conditioned medium was much higher than in the cell lysate, obviously because of inhibitory activity observed in the cell lysates. It remains to be elucidated whether the lysis conditions used in this study or a putative intracellular inhibitor blocks VWF-cp activity. The degradation of VWF multimers by rADAMTS13 was completely inhibited by the addition of plasma derived from a TTP patient and by the purified IgG antibodies. These results confirm that idiopathic TTP patient plasma contains antibodies capable of directly neutralizing VWF-cp activity. They also confirm clinical data that inhibitors present in idiopathic TTP patient plasma are directed against the VWF-cleaving protease encoded by the gene ofADAMTS13. Moreover, the specificity of the cleavage site recognized by rADAMTS13 was verified by N-terminal sequencing of the generated cleavage products. These data clearly confirm that rADAMTS13 cleaves VWF between Tyr842 and Met843, identical to the plasma VWF-cp.
In conclusion, our experimental data provide convincing evidence that ADAMTS13 is the enzyme responsible for proteolytic degradation of VWF multimers. Verification of this functionality, added to the cloning of the gene of ADAMTS13, opens the way to production of a recombinant protein in commercially viable amounts. This protein potentially provides an improved treatment strategy for hereditary forms of TTP to replace the current inconvenient FFP and cryosupernatant treatment, which have been associated with complications.21 The recombinant protein might also be useful for developing an immune adsorption system for removal of the inhibiting antibody. Furthermore, availability of recombinant ADAMTS13 will be helpful in gaining further insight into the pathogenesis of TTP.
We thank Gabriele Mohr, Waltraud Wernhart, Sabrina Pable, Monika Grillowitzer, Luigina Tagliavacca, and Thomas Lindner for excellent technical assistance, Ewald Kapeller for N-terminal sequencing, Peter L. Turecek for helpful discussions, Katalin Varadi for the collagen-binding assay, and Elise Langdon-Neuner for editing.
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2002-05-1397.
B.P. and K.Z. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Friedrich Scheiflinger, Baxter BioScience, Biomedical Research Center, Uferstr. 15, 2304 Orth/Donau, Austria; e-mail: scheiff@baxter.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal