Enhancing the pharmacologic activity of all-transretinoic acid (ATRA) is potentially useful in the management of acute promyelocytic leukemia (APL) and other types of myeloid leukemia. In this report, we identify a novel class of experimental agents selectively potentiating the cytodifferentiating activity of ATRA and synthetic retinoic acid receptor α agonists in APL and other myeloid leukemia cell lines. These agents have a bis-indolic structure (BISINDS), and ST1346 is the prototypical compound of the series. Gene-profiling experiments and determination of the level of expression of myeloid-associated markers indicate that ST1346 stimulates many aspects of the granulocytic maturation process set in motion by ATRA. Stimulation of the cytodifferentiating activity of ATRA by ST1346 enhances the efficacy of the retinoid in vivo, as demonstrated in the APL model of the severe combined immunodeficiency (SCID) mouse receiving transplants of NB4 cells. Although the molecular mechanisms underlying the ATRA-potentiating action of ST1346 and congeners have not been completely clarified, bis-indols are not ligands and do not exert any direct effect on the ATRA-dependent transactivation of nuclear receptors. However, ST1346 inhibits the down-regulation of cyclic adenosine monophosphate (cAMP)–dependent CREB transcriptional complexes and enhances the level of expression of signal transducers and activators of transcription-1 (STAT1), 2 putative molecular determinants of the differentiation process activated by ATRA in APL cells. More importantly, ST1346 relieves the down-regulation of Jun N-terminal kinases (JNK) afforded by ATRA. In addition, a specific JNK inhibitor blocks the enhancing effect of ST1346 on ATRA-induced maturation of NB4 cells. This demonstrates an important role for the mitogen-activated protein kinase in the molecular mechanisms underlying the pharmacologic activity of the bis-indol.
Introduction
Acute promyelocytic leukemia (APL) is a form of acute myelogenous leukemia with typical chromosomal translocations leading to the expression of abnormal fusion proteins involving the nuclear retinoic acid receptor (RAR) α.1 These fusion proteins act as oncogenes and are responsible for the differentiation block and the expansion of the leukemic clone.1 In the majority of APL patients, the translocation involves chromosomes 15 and 17 and leads to the synthesis of promyelocytic leukemia (PML)–RARα.1 APL is the object of intense study, as it represents the only example of neoplastic disease that can be treated with a cytodifferentiating approach.2 APL patients are induced into clinical remission with all-transretinoic acid (ATRA),3 which forces the leukemic blast to acquire many of the characteristics of the terminally differentiated neutrophil. These include a short lifespan and the propensity to undergo a natural process of programmed cell death or apoptosis.4
Although the success obtained with APL patients has raised enthusiasm for the clinical use of ATRA in the treatment of leukemia and other neoplastic diseases, the therapeutic efficacy of this compound is still burdened by problems such as resistance5 and toxicity.6 7 One possible strategy to increase the therapeutic index of ATRA is the development of ATRA-based pharmacologic combinations that are more powerful and easily tolerated than the individual components.
Relevant aspects of the differentiation program set in motion by ATRA in APL cells can be reproduced in primary cultures of leukemic blasts and in the derived NB4 cell line,8,9 which is a unique model for the study of the pharmacologic activity of ATRA and derivatives.10-12 Pharmacologic concentrations of ATRA arrest the growth of NB4 blasts and differentiate them into cells that resemble mature neutrophils.13,14 This is followed by a slow process of apoptosis.15 We and others used NB4 cells to demonstrate that such diverse compounds as cytokines, cyclic adenosine monophosphate (cAMP) analogs, and STI57116-20enhance the pharmacologic activity of ATRA and other retinoids.
In this report, we identify a novel class of compounds capable of potentiating the cytodifferentiating activity of ATRA in NB4 cells. These molecules have a bis-indol structure and enhance the efficacy of ATRA not only in vitro but also in vivo in the APL model of the severe combined immunodeficiency (SCID) mouse receiving transplants of NB4.21 22
Materials and methods
Chemicals
We used ATRA (Sigma, St Louis, MO), AM580,23 CD2915, and CD280915 (CIRD-Galderma Research and Development, Sophia Antipolis, France), U0126, PD169316, and SP600125 (Calbiochem, La Jolla, CA). ST1707, ST1346, ST1422, ST1860, ST1381, ST1730, ST1350, ST1866, ST1371, ST1385, ST1345, and ST1750 were synthesized by Sigma-Tau Industrie Farmaceutiche Riunite S.p.A.
Cell culture, treatments, and transfections
NB4,9 NB4.306,24 U937, Kazumi, HL-60, KG1 (American Type Culture Collection [ATCC], Rockville, MD), and PR9 (a U937-derived cell clone expressing PML-RARα upon induction with zinc sulfate25) cells were cultured in the presence of RPMI 1640 with 10% fetal calf serum (FCS) (20% FCS in the case of Kazumi). COS-7 cells (ATCC) were grown in Dulbecco modified Eagle medium containing 10% FCS. Cells were free from mycoplasma contamination (Hoechst 33258 staining method).
Cellular proliferation, viability, and cytodifferentiation
Cell number and viability were determined following staining with erythrosin (Sigma).27 CD11b, CD38, CD14, ICAM-1, and integrin α-5 were measured by flow cytometry using specific monoclonal antibodies (Becton Dickinson, Franklin Lakes, NJ).28 The nitrobluetetrazolium (NBT) reduction assay was performed on extracts of phorbol myristate acetate–stimulated cells,20 and the percentage of NBT+ cells was determined as described.20
Western blot analysis, JNK kinase activity, and determination of cytokine secretion and gel retardation assays
Extracts of NB4 cells (from 3 × 106cells) were subjected to Western blot analysis as reported.20 The experiments were performed with antibodies against RARα (RPαF'),29 RXRα, 4RX1F6,30 CEBPε (C-22; Santa Cruz Biotechnology, Santa Cruz, CA), signal transducers and activators of transcription-1 (STAT1) p91 (C-24; Santa Cruz Biotechnology), STAT1 p91 and p84 (C-136; Santa Cruz Biotechnology), Tyr701 phosphorylated STAT1 p91 and p84 (Upstate Biotechnology, Lake Placid, NY), cathepsin D (C-20; Santa Cruz Biotechnology), tyk-2 (C-20; Santa Cruz Biotechnology), Mlh1 (C-20; Santa Cruz Biotechnology), or βactin (C-11; Santa Cruz Biotechnology). The antibodies directed against the phosphorylated forms of extracellular signal-regulated kinase-1 (ERK-1), ERK-2, Jun N-terminal kinase (JNK), and p38, and against histone H3 and its acetylated form were from Cell Signaling Technology (Beverly, MA). Protein bands were visualized with the enhanced chemiluminescence detection kit (Amersham, Little Chalfont, United Kingdom). JNK kinase activity was determined on JNK immunoprecipitates with c-JUN as a substrate, using a commercial kit (Cell Signaling Technology).
Enzyme-linked immunosorbent assays (ELISAs) (Endogen, Woburn, MA31) were used for the determination of interleukin-1 (IL-1), tumor necrosis factor (TNF), and monocyte chemoattractant protein 1 (MCP-1). Gel retardation assays were performed on nuclear extracts of NB4 cells, as already described.27 The following double-strand oligonucleotide probes (Santa Cruz) were used:CREB = 5′GAGAGATTGCCTGACGTCAGAGAGCTAG 3′; and Ets-1 = 5′GGATCTCGAGCAGGAAGTTCGA 3′.
RNA extraction and gene profiling
RNA was extracted with the TRIzol reagent (Invitrogen SRL, S. Giuliano Milanese, Italy) and the polyadenylated fraction selected with the Atlas Pure kit (Clontech, Palo Alto, CA). The human 1.2 ATLAS nylon filters (Clontech) were used for the microarray experiments. Single-strand 32P-labeled cDNA probes were synthesized from poly(A+) RNA as suggested by the manufacturer. Video imaging was performed with the Storm 860 Phosphorimager (Molecular Dynamics, Sunnyvale, CA). Microarray data were quantitated with the ATLAS IMAGE 2.0 analysis software (Clontech).
In vivo experiments
The in vivo experiments were carried out as described.22 Cells were suspended in 199 Hanks Medium, and 0.1 mL (1 × 106 cells per mouse) were intraperitoneally inoculated in SCID mice. ST1346 was dissolved in Cremophor/ethanol 1:1 solution, diluted 1:4 in phosphate-buffered saline at the concentrations of 15 and 30 mg/kg, respectively. ATRA (Sigma) was dissolved in carboxy methyl cellulose (CMC) 0.5% + 0.01% Tween 80 solution at the concentration of 5 mg/kg. Drugs were administered intraperitoneally in a volume of 10 mL/kg one day after the inoculation, and the treatment continued for 3 weeks (5 daily injections per week.
Results
BISINDs as a novel class of retinoid sensitizers
A screening process based on the APL-derived NB4 cell line and aimed to select compounds that potentiate the cytodifferentiating activity of ATRA resulted in the identification of a novel class of agents with bis-indolic structures (BISINDs). As exemplified for the prototypical compounds ST1346 and ST1707 in Figure1, when NB4 cells are exposed to micromolar concentrations of BISINDs for 3 days, no significant effect on the growth, viability, or differentiation of the leukemic blasts is observed. However, when cells are simultaneously exposed to suboptimal concentrations of ATRA (0.1 μM) and ST1346 or ST1707, NBT-reducing activity (NBT-R), a myeloid differentiation marker,32 is enhanced in a dose-dependent manner. Combinations of ATRA and ST1346 or ST1707 are no more effective than the retinoid alone in terms of growth inhibition. In our experimental conditions, cell viability is always above 80% and is not affected by any treatment.
Effect of ST1346 and ST1707 on the ATRA-dependent induction of NBT-reducing activity and inhibition of NB4 cell growth.
NB4 cells were seeded (200 000/mL) and treated for 3 days with the indicated concentrations of ST1346 and ST1707 in the presence of vehicle (○) or ATRA (0.1μM) (●) for 3 days. Aliquots of cells were used for the determination of NBT-reducing activity (A,D), total number of cells (B,E), and cell viability (C,F). The results shown are the means ± SD of 3 separate cultures. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Student t test). °Significantly higher than the sum of the effects observed in the corresponding ST1346-, ST1707-, and ATRA-treated groups, following 2-way analysis of variance and measurement of the F score of the interaction (P < .01 according to Tukey test).
Effect of ST1346 and ST1707 on the ATRA-dependent induction of NBT-reducing activity and inhibition of NB4 cell growth.
NB4 cells were seeded (200 000/mL) and treated for 3 days with the indicated concentrations of ST1346 and ST1707 in the presence of vehicle (○) or ATRA (0.1μM) (●) for 3 days. Aliquots of cells were used for the determination of NBT-reducing activity (A,D), total number of cells (B,E), and cell viability (C,F). The results shown are the means ± SD of 3 separate cultures. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Student t test). °Significantly higher than the sum of the effects observed in the corresponding ST1346-, ST1707-, and ATRA-treated groups, following 2-way analysis of variance and measurement of the F score of the interaction (P < .01 according to Tukey test).
To get insight into the structural determinants responsible for the pharmacologic activity of BISINDs, we evaluated 12 congeners (Table1). No molecule in the series increases NBT-R, regardless of the concentration used. However, ST1707, ST1346, ST1422, and ST1860 significantly enhance the induction of NBT-R afforded by ATRA (0.1μM). ST1381 causes a much lower effect, whereas ST1730, ST1350, ST1866, and ST1371 are devoid of any potentiating action. Significantly, ST1385, ST1345, and ST1750 inhibit ATRA-dependent induction of NBT-R. In the presence of a fixed concentration of ATRA (0.1μM), the AC50 values for the potentiating effect vary from 0.3 μM in the case of ST1381 to 8.2 μM in the case of ST1422. The relative potency of each BISIND was calculated at 10 μM (except for ST1730) and results in the rank order: ST1707 > ST1346 > ST1422 > ST1860 > ST1381. In the absence of ATRA, all the compounds are devoid of significant antiproliferative effects. With the exception of ST1730, all the BISINDs are cytotoxic only at concentrations that are significantly above their corresponding AC50. A first analysis of structure/activity relationships indicates that the bis-indol is strictly required for the observed pharmacologic activity (Table 1; data not shown). Attempts to identify other determinants of the potentiating activity of BISINDs suggest that the alkyl chain bound to the carbon bridging the 2 indol rings and the terminal carboxylic group are important.
Effect of BISINDs on NBT-R activity in NB4 cells
| Compound . | Chemical structure . | Activity . | Differentiation AC50 or IC50 (μM) . | Differentiation Relative potency, (10 μM) . | Cytotoxicity (μM) . | Cell growth (10 μM) % T/C . |
|---|---|---|---|---|---|---|
| ST1707 | potentiating | 2.7 ± 2.0 | +108 (±10) | >10 | 98 ± 6 | |
| ST1346 | potentiating | 2.8 ± 1.6 | +95 (±11) | >50 | 81 ± 5 | |
| ST1422 | potentiating | 8.2 ± 1.8 | +76 (±7) | >50 | 97 ± 16 | |
| ST1860 | potentiating | 3.2 ± 0.6 | +76 (±6) | >10 | 88 ± 7 | |
| ST1381 | potentiating | 0.3 ± 0.2 | +55 (±2) | >10 | 83 ± 3 | |
| ST1730 | nil | ND | +18 (±7)* | >1 | ND | |
| ST1350 | nil | 21.8 ± 0.1 | +3 (±1) | >50 | 98 ± 3 | |
| ST1866 | nil | >50 | −2 (±1) | >50 | 122 ± 6 | |
| ST1371 | nil | 27.7 ± 0.3 | −3 (±1) | >50 | 100 ± 4 | |
| ST1385 | inhibitory | 0.5 ± 0.1 | −43 (±1) | >5 | ND | |
| ST1345 | inhibitory | 0.7 ± 0.1 | −56 (±1) | >5 | ND | |
| ST1750 | inhibitory | 3.2 ± 0.6 | −67 (±4) | >50 | 74 ± 1 | |
| Compound . | Chemical structure . | Activity . | Differentiation AC50 or IC50 (μM) . | Differentiation Relative potency, (10 μM) . | Cytotoxicity (μM) . | Cell growth (10 μM) % T/C . |
|---|---|---|---|---|---|---|
| ST1707 | potentiating | 2.7 ± 2.0 | +108 (±10) | >10 | 98 ± 6 | |
| ST1346 | potentiating | 2.8 ± 1.6 | +95 (±11) | >50 | 81 ± 5 | |
| ST1422 | potentiating | 8.2 ± 1.8 | +76 (±7) | >50 | 97 ± 16 | |
| ST1860 | potentiating | 3.2 ± 0.6 | +76 (±6) | >10 | 88 ± 7 | |
| ST1381 | potentiating | 0.3 ± 0.2 | +55 (±2) | >10 | 83 ± 3 | |
| ST1730 | nil | ND | +18 (±7)* | >1 | ND | |
| ST1350 | nil | 21.8 ± 0.1 | +3 (±1) | >50 | 98 ± 3 | |
| ST1866 | nil | >50 | −2 (±1) | >50 | 122 ± 6 | |
| ST1371 | nil | 27.7 ± 0.3 | −3 (±1) | >50 | 100 ± 4 | |
| ST1385 | inhibitory | 0.5 ± 0.1 | −43 (±1) | >5 | ND | |
| ST1345 | inhibitory | 0.7 ± 0.1 | −56 (±1) | >5 | ND | |
| ST1750 | inhibitory | 3.2 ± 0.6 | −67 (±4) | >50 | 74 ± 1 | |
NB4 cells were treated with increasing concentrations of the indicated BISIND in the presence or absence of ATRA (0.1 μM), and dose-response curves for NBT-reducing activity and cell growth similar to those reported in Figure 1 were constructed. Differentiation AC50 = the concentration of the compound giving 50% of the maximal induction of NBT-reducing activity in the presence of ATRA. Differentiation relative potency is the percent increase in NBT-reducing activity observed for a given combination of BISIND (at the concentration of 10 μM) and ATRA over that observed with ATRA alone. A “+” symbol indicates potentiation and a “−” symbol indicates inhibition. A value below 30% is considered negative, and the test compound is labeled as inactive (nil activity). In the case of ST1730, the value was calculated at the concentration of 1 μM. The value under the column labeled “Cytotoxicity” indicates the concentration of test compound above which we observed a decrease in cell viability of at least 40% following a standard treatment period of 3 days. The effect on cell growth was evaluated following 3 days of treatment with the test compound alone (10 μM). The value is expressed as the percentage of the total number of viable cells in treated (T) versus control (C) conditions. Each value is the mean ± SD of 3 separate culture dishes.
We focused our attention on ST1346. Figure2 demonstrates that treatment of NB4 cells with ST1346 enhances ATRA-induced NBT-R time dependently (panel A). Potentiation is observed also in terms of the number of NBT-R+ cells, which is significantly higher following 3 days of treatment with ATRA + ST1346 than with ATRA alone (panel B). This demonstrates that the BISIND accelerates the process of granulocytic maturation activated by ATRA. The growth curve of NB4 blasts observed in control conditions is not altered by ST1346 (panel C). Similarly, the growth arrest of the leukemic cells triggered by ATRA is not significantly affected by the BISIND. The level of cell viability is higher than 85% in all the experimental conditions considered.
Time-dependent potentiation of the ATRA-dependent induction of NBT-reducing activity by ST1346 in NB4 cells.
NB4 cells were seeded (200 000/mL) and treated with vehicle, ST1346 (5 μM), ATRA (0.1 μM), or the combination of ATRA + ST1346 for 3 days (panel B) for various lengths of time, as indicated (panels A,C). Aliquots of cells were used for the determination of NBT-reducing activity (panel A), the number of cells scoring positive for NBT-reducing activity (panel B), and the total number of viable cells (panel C). In all cases the cellular viability was above 90%, as assessed by the erythrosin exclusion test. The results are the means ± SD of 3 separate cultures. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Student t test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and ATRA-treated groups, following 2-way analysis of variance and measurement of the F of interaction (P < .01 according to Tukey test).
Time-dependent potentiation of the ATRA-dependent induction of NBT-reducing activity by ST1346 in NB4 cells.
NB4 cells were seeded (200 000/mL) and treated with vehicle, ST1346 (5 μM), ATRA (0.1 μM), or the combination of ATRA + ST1346 for 3 days (panel B) for various lengths of time, as indicated (panels A,C). Aliquots of cells were used for the determination of NBT-reducing activity (panel A), the number of cells scoring positive for NBT-reducing activity (panel B), and the total number of viable cells (panel C). In all cases the cellular viability was above 90%, as assessed by the erythrosin exclusion test. The results are the means ± SD of 3 separate cultures. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Student t test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and ATRA-treated groups, following 2-way analysis of variance and measurement of the F of interaction (P < .01 according to Tukey test).
Figure 3A shows that the enhancing effect of ST1346 on ATRA-induced stimulation of NBT-R is already visible at the retinoid concentration of 0.01μM and is maintained at higher concentrations. Potentiation is not the result of a priming effect of ATRA or ST1346 on NB4 cells because continuous treatment with both compounds is necessary. As shown in Figure 3B, pretreatment of NB4 cells with ST1346 for one day followed by washing with fresh medium and incubation with the retinoid for 3 days causes an induction of NBT-R that is significantly lower than that observed following continuous treatment for 4 days with the combination of ST1346 + ATRA. Changing the order of addition of ST1346 and ATRA or extending the pretreatment period to 3 days results in similar effects. Figure 3C shows that ST1346-dependent potentiation is observed in the retinoid-sensitive NB4 parental cell line, but not in the retinoid-resistant NB4.306 subline.24 Figure 3D demonstrates that the cytodifferentiating effect of ST1346 + ATRA is evident also in the myeloid leukemia cell line, PR9, which is engineered to allow conditional expression of the APL oncogene PML-RARα.25 However, induction of the oncogene by zinc sulfate (data not shown) in PR925 does not result in an increase of NBT+ cells, which is significantly different from that observed in the absence of the inducing agent. These data suggest that PML-RARα is not strictly necessary for the ST1346 + ATRA–mediated cytodifferentiating response. Nevertheless, we cannot formally exclude the possibility that PML-RARα has some effect on the interaction, given a certain level of leaky expression of the oncogene in uninduced PR9 cells. As shown in Figure 3E-H, we extended our analysis to 4 other PML-RARα− myeloid leukemia cell types, that is, the myelocytic HL-60, the myelomonoblastic U937, and Kazumi, as well as the myeloblastic KG1 cell lines. Treatment of the ATRA-sensitive HL-60 blasts with increasing concentrations of ST1346 and a suboptimal concentration of ATRA leads to significant induction of NBT-R. In this case, maximal induction of NBT-R is observed following addition of 50 μM ST1346 to ATRA. In the experimental conditions used, U937 and Kazumi cells are refractory to challenge with ATRA, while the combination of ST1346 + ATRA causes a variable proportion of both cell populations to become NBT positive. It is worth noticing that maximal induction of NBT positivity in Kazumi cells by ST1346 + ATRA is observed at 2 different concentrations of the BISIND (10 and 50 μM) and takes longer than in other cell types. KG1 is totally unresponsive to both ATRA and the combination of the bis-indol and the retinoid.
Influence of the cellular context and exposure schedule on the potentiation of the ATRA-dependent induction of NBT-reducing activity by ST1346.
(A) NB4 cells were treated for 3 days with vehicle (○) or ST1346 (5 μM) (●) in the presence of the indicated concentrations of ATRA. (B) NB4 cells were treated with vehicle, ST1346 (5 μM), ATRA (0.1 μM), or the combination of ATRA + ST1346 (ST1346/ATRA) for the indicated amount of time in days (d). Pretreatment means that ST1346 or ATRA (+) is added to the culture medium for the indicated amount of time. This is followed by a cell-washing step in complete culture medium and subsequent addition of fresh medium containing the complementary compound not present in the pretreatment phase. In the case of ATRA + ST1346, we added a control group in which a washing step was introduced after 3 days of pretreatment (rightmost data bar). (D) PR9 cells were treated with ATRA (1 μM), ST1346 (10 μM), or the combination of the 2 compounds for 3 days in the absence or presence of the PML-RARα transgene inducer zinc sulfate (Zn++; 50 μM). (C,E-H) NB4, NB4.306, HL-60, U937, Kazumi, or KG1 cells were treated with vehicle (control), ST1346 (at the indicated concentrations), ATRA (0.1 or 1 μM), or the combination of ATRA + ST1346 for 3 days (panels C, E, F, and H) or the indicated number of days (panel G). For all the experiments, cells were seeded at a concentration of 200 000/mL and aliquots used for the determination of NBT-reducing activity (panels A, B, C, and E) or the number of NBT+ cells (panels D, F, G, and H). In all cases the cellular viability was above 90%, as assessed by the erythrosin exclusion test. The results are the means ± SD of 3 separate cultures. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Studentt test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and ATRA-treated groups, following 2-way analysis of variance and measurement of the F of interaction (P < .01 according to Tukey test).
Influence of the cellular context and exposure schedule on the potentiation of the ATRA-dependent induction of NBT-reducing activity by ST1346.
(A) NB4 cells were treated for 3 days with vehicle (○) or ST1346 (5 μM) (●) in the presence of the indicated concentrations of ATRA. (B) NB4 cells were treated with vehicle, ST1346 (5 μM), ATRA (0.1 μM), or the combination of ATRA + ST1346 (ST1346/ATRA) for the indicated amount of time in days (d). Pretreatment means that ST1346 or ATRA (+) is added to the culture medium for the indicated amount of time. This is followed by a cell-washing step in complete culture medium and subsequent addition of fresh medium containing the complementary compound not present in the pretreatment phase. In the case of ATRA + ST1346, we added a control group in which a washing step was introduced after 3 days of pretreatment (rightmost data bar). (D) PR9 cells were treated with ATRA (1 μM), ST1346 (10 μM), or the combination of the 2 compounds for 3 days in the absence or presence of the PML-RARα transgene inducer zinc sulfate (Zn++; 50 μM). (C,E-H) NB4, NB4.306, HL-60, U937, Kazumi, or KG1 cells were treated with vehicle (control), ST1346 (at the indicated concentrations), ATRA (0.1 or 1 μM), or the combination of ATRA + ST1346 for 3 days (panels C, E, F, and H) or the indicated number of days (panel G). For all the experiments, cells were seeded at a concentration of 200 000/mL and aliquots used for the determination of NBT-reducing activity (panels A, B, C, and E) or the number of NBT+ cells (panels D, F, G, and H). In all cases the cellular viability was above 90%, as assessed by the erythrosin exclusion test. The results are the means ± SD of 3 separate cultures. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Studentt test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and ATRA-treated groups, following 2-way analysis of variance and measurement of the F of interaction (P < .01 according to Tukey test).
In NB4 cells, ST1346 enhances the cytodifferentiating activity not only of ATRA but also of AM580, a synthetic and selective RARα/PML-RARα agonist.23 As illustrated in Figure4, the BISIND (5 μM) augments the level of NBT-R induced by AM580 (0.001 and 0.01 μM) significantly. ATRA cannot be substituted by the RXR agonists CD2915 and CD2809. The small NBT-R–inducing effect caused by ST1346 and the highest concentrations of CD2915 (1 μM) is due to the residual RAR agonistic activity associated with the retinoid. In fact, a similar phenomenon is not observed in the case of the combination ST1346 + CD2809.
Effect of combinations of ST1346 and RARα agonist or RXR agonists on NBT-reducing activity in NB4 cells.
NB4 cells were seeded at an initial concentration of 200 000/mL, and aliquots of the cell cultures were used for the determination of NBT-reducing activity. In all cases the cellular viability was above 80%, as assessed by the erythrosin exclusion test. (A) Cells were treated with the RARα agonist AM580 or the RXR agonist CD2915 at the indicated concentrations in the presence or absence of ST1346 (10 μM). (B) Cells were treated for 3 days with vehicle, ST1346 (5 μM), the RXR agonist CD2809 (1 μM), or the combination of the 2 compounds. Results are expressed as the means ± SD of 3 separate culture dishes and are representative of at least 2 independent experiments. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Studentt test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and RARα- or RXR-agonist–treated groups, following 2-way analysis of variance and measurement of the F of interaction (P < .01 according to Tukey test).
Effect of combinations of ST1346 and RARα agonist or RXR agonists on NBT-reducing activity in NB4 cells.
NB4 cells were seeded at an initial concentration of 200 000/mL, and aliquots of the cell cultures were used for the determination of NBT-reducing activity. In all cases the cellular viability was above 80%, as assessed by the erythrosin exclusion test. (A) Cells were treated with the RARα agonist AM580 or the RXR agonist CD2915 at the indicated concentrations in the presence or absence of ST1346 (10 μM). (B) Cells were treated for 3 days with vehicle, ST1346 (5 μM), the RXR agonist CD2809 (1 μM), or the combination of the 2 compounds. Results are expressed as the means ± SD of 3 separate culture dishes and are representative of at least 2 independent experiments. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Studentt test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and RARα- or RXR-agonist–treated groups, following 2-way analysis of variance and measurement of the F of interaction (P < .01 according to Tukey test).
Influence of ST1346 on ATRA-dependent myeloid differentiation markers and the gene expression profile of undifferentiated and retinoid-differentiated NB4 cells
We established whether the potentiating activity of ST1346 affects the levels of the myeloid CD11b and the monocytic CD14 markers. A typical fluorescence-activated cell-sorter profile of CD11b in NB4 cells treated for 3 days with ATRA (0.01 μM), ST1346 (5 μM), or the combination of the 2 compounds is shown in Figure5A. A small proportion of NB4 cells is positive for CD11b in basal conditions and following challenge with the BISIND alone. ATRA increases both the number of CD11b+cells and CD11b cell–associated fluorescence, and these effects are enhanced by ATRA + ST1346. As illustrated in Figure 5B, an increase in the number of CD11b+ cells is evident at ATRA concentrations of 0.001 μM and 0.01 μM and is lost at 0.1 μM (left panel). This is expected, as 0.1 μM ATRA induces expression of CD11b practically on almost all the NB4 cell population. Significantly, however, even at the highest concentration of the retinoid, the amount of CD11b cell–associated fluorescence is higher following challenge with ATRA + ST1346 than with ATRA alone (right panel). None of these effects is evident in the case of CD38, which is maximally induced by ATRA and not further enhanced by ST1346. Treatment of NB4 cells with ATRA or ATRA + ST1346 does not result in the appearance of CD14+cells.
Influence of ST1346 on ATRA-induced surface expression of the NB4 differentiation markers.
(A) NB4 cells were treated for 3 days with vehicle alone, ST1346 (5 μM), ATRA (0.01μM), or the combination of the 2 compounds. Typical flow cytometric profiles following staining of cells with a fluoresceinated anti-CD11b antibody (right graph) and an isotype-matched negative control (left graph) are shown. (B) NB4 cells were treated for 3 days with vehicle alone, ST1346 (5 μM), the indicated concentrations of ATRA (open symbols), or the combination of the 2 compounds (closed symbols). The level of expression of the indicated phenotypic markers was determined by flow cytometry using fluorescein-labeled monoclonal antibodies against CD11b, CD38, and CD14 or irrelevant isotype-matched antibodies (neg). The percentage of marker-positive cells is shown in the left graph, and the amount of cell-associated fluorescence in arbitrary units is illustrated in the right graph. In all cases, cells were seeded at an initial concentration of 200 000/mL. Results are representative of 2 independent experiments.
Influence of ST1346 on ATRA-induced surface expression of the NB4 differentiation markers.
(A) NB4 cells were treated for 3 days with vehicle alone, ST1346 (5 μM), ATRA (0.01μM), or the combination of the 2 compounds. Typical flow cytometric profiles following staining of cells with a fluoresceinated anti-CD11b antibody (right graph) and an isotype-matched negative control (left graph) are shown. (B) NB4 cells were treated for 3 days with vehicle alone, ST1346 (5 μM), the indicated concentrations of ATRA (open symbols), or the combination of the 2 compounds (closed symbols). The level of expression of the indicated phenotypic markers was determined by flow cytometry using fluorescein-labeled monoclonal antibodies against CD11b, CD38, and CD14 or irrelevant isotype-matched antibodies (neg). The percentage of marker-positive cells is shown in the left graph, and the amount of cell-associated fluorescence in arbitrary units is illustrated in the right graph. In all cases, cells were seeded at an initial concentration of 200 000/mL. Results are representative of 2 independent experiments.
To identify the genes modulated in NB4 cells by ST1346 alone or in combination with ATRA, we analyzed the expression of approximately 1200 genes with cDNA microarrays. We focused on genes expressed either early (8 hours) or relatively late (3 days) along the granulocytic maturation process, and the results are summarized in Table2. We observed 38 genes induced by ATRA. The expression of 20 of these genes is enhanced by cotreatment with ST1346 for either 8 or 72 hours (group A). Among the products of these genes, the myeloid nuclear cell differentiation antigen, intercellular adhesion molecule-1 (ICAM-1), MCP-1, migration inhibitory factor–related protein (MRP)–14, MRP-8, and granulocyte colony-stimulating factor (G-CSF) receptor are known myeloid differentiation markers. For some genes in group A (PRK1, G-CSF receptor, LIM-kinase, LD-78, c-fgr, and Tyk-2), the combination of ATRA + ST1346 maintains increased expression for a longer time than ATRA alone. For other genes (MAPKAP-K2 and cathepsin D), treatment of NB4 cells with the retinoid and the BISIND accelerates the up-regulation observed in the presence of ATRA. The remainder of genes whose expression is induced by ATRA is not further up-regulated by the addition of ST1346 to the medium (group B). Indeed, in some cases, the BISIND inhibits the induction of gene transcription afforded by the retinoid (P19INK, TNF receptor, RALB, etc). Group C consists of 10 genes that are activated only following treatment of NB4 cells with ATRA + ST1346. The majority of these genes are turned on at 8 hours, and increased activity is either maintained or turned off at 72 hours. Paradoxically, however, the steady-state levels of the transcripts encoding human DNA replication licensing factor (hRlf) beta subunit, vascular endothelial growth factor (VEGF), and classical late-infantile neuronal ceroid lipofuscinosis (CLN2) show a biphasic pattern of expression upon treatment with ATRA + ST1346, that is, an increase at 8 hours and a decrease at 72 hours.
Gene expression profile in NB4 cells treated with ATRA, ST1346, or the combination of the two compounds
| . | 8 hours . | 72 hours . | ||||||
|---|---|---|---|---|---|---|---|---|
| Control . | ATRA . | ST1346 . | ATRA + ST1346 . | Control . | ATRA . | ST1346 . | ATRA + ST1346 . | |
| Group A | ||||||||
| ICAM-1 (J03132) | 1 368 | 7 530* | 1 491 | 16 649* | <1 000 | 7 909* | <1 000 | 20 326* |
| MCP-1 (M24545) | <1 000 | 4 734* | 1 062 | 7 668* | <1 000 | 23 911* | <1 000 | 51 927* |
| Id-2 (M97796) | <1 000 | 1 773* | <1 000 | 3 023* | <1 000 | 2 357* | <1 000 | 5 518* |
| Mlh1 (U07418) | 16 128 | 9 100 | 14 897 | 9 913 | <1 000 | 2 261* | <1 000 | 4 713* |
| MAPKAP-K2 (U12779) | 2 368 | 1 836 | 1 826 | 4 552* | <1 000 | 2 253* | <1 000 | 7 329* |
| Cathepsin D (M11233) | 1 712 | 1 753 | 2 331 | 3 963* | 3 604 | 16 269* | 3 606 | 6 787* |
| FLI-1 (M93255) | <1 000 | <1 000 | <1 000 | 1 308* | <1 000 | 5 949* | 8 474* | 9 568* |
| MRP14 (X06233) | <1 000 | 3 399* | <1 000 | 3 889* | <1 000 | 6 933* | <1 000 | 12 376* |
| MRP8 (X06234) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 11 299* | <1 000 | 23 344* |
| MNDA (M81750) | <1 000 | 1 607* | <1 000 | 1 583* | <1 000 | 2 234* | <1 000 | 6 609* |
| IRF7 (U73036) | <1 000 | 2 412* | <1 000 | 1 726* | <1 000 | 2 510* | <1 000 | 4 216* |
| Net (Z36715) | <1 000 | 1 038 | <1 000 | 1 612* | <1 000 | 2 095* | 1 656 | 6 806* |
| PRK1 (U33053) | <1 000 | 3 182* | <1 000 | 1 700* | 1 364 | <1 000 | 1 097 | 6 899* |
| G-CSF receptor (M59818) | 1 072 | 3 370* | <1 000 | 3 483* | 2 428 | 1 782 | 3 095 | 8 136* |
| LIM-kinase (D26309) | <1 000 | 3 374* | <1 000 | 1 912* | 1 084 | 1 186 | 1 641 | 5 484* |
| LD-78 (D10924) | <1 000 | 2 570* | <1 000 | 1 951* | <1 000 | <1 000 | 1 710 | 3 730* |
| c-fgr (M19722) | 1 116 | 2 884* | <1 000 | 4 025* | 1 296 | 1 323 | <1 000 | 3 963* |
| PKC delta (D10495) | 3 132 | 5 548* | 3 062 | 5 568* | <1 000 | <1 000 | <1 000 | 3 080* |
| Tyk-2 (X54637) | 2 784 | 3 842 | 2 288 | 2 362 | 2 104 | 3 109 | 2 524 | 5 180* |
| EBI (U24166) | 1 948 | 2 286 | 1 418 | 3 518* | 2 752 | 7 422* | 2 553 | 8 378* |
| Group B | ||||||||
| Cyclin-B (M25753) | 2 260 | 4 746* | 3 414 | 4 814* | 4 696 | 9 553* | 8 794 | 5 577 |
| P19 INK (U40343) | <1 000 | 7 222* | 1 019 | 6 065* | 2 016 | 2 800 | 1 698 | 1 493 |
| Integrin alpha-4 (L12002) | 2 308 | 2 870 | 2 521 | 3 127 | 4 379 | 8 720* | 5 548 | 7 428 |
| TNF receptor (M32315) | <1 000 | 2 728* | <1 000 | 1 654* | <1 000 | 2 453* | <1 000 | <1 000 |
| GST pi (M24485) | 18 472 | 13 777 | 13 877 | 14 985 | <1 000 | 3 392* | 2 829 | 3 957* |
| Neutrophil elastase inhibitor (M93056) | <1 000 | 1 492* | <1 000 | 1 794* | <1 000 | 5 640* | <1 000 | <1 000 |
| G0S19-1 (M23452) | <1 000 | 2 298* | 1 541 | 1 514* | <1 000 | <1 000 | <1 000 | <1 000 |
| Glutharedoxin (X76648) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 4 096* | <1 000 | 2 280* |
| RALB (M35416) | <1 000 | 1 687 | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 |
| Glial growth factor (L12261) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 3 581* | <1 000 | <1 000 |
| c-sis (X02744) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 3 253* | <1 000 | <1 000 |
| Estrogen sulfotransferase (U08098) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 4 736* | <1 000 | <1 000 |
| NKSF (M65291) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 3 912* | <1 000 | <1 000 |
| S19 ribosomal protein (M81757) | 4 772 | 12 426* | 8 619 | 7 603 | 3 360 | 3 759 | 2 511 | 4 177 |
| Cathepsin H (X07549) | 3 996 | 2 076† | 4 241 | 3 384 | <1 000 | 4 589* | <1 000 | <1 000 |
| Stromelysin-3 (X57766) | 2 992 | 5 591* | 2 969 | 2 912 | 4 116 | 1 880† | 3 519 | 3 053 |
| DNA-binding protein A (M24069) | <1 000 | 1 414* | 2 530* | 1 305* | 2 556 | <1 000† | 2 634 | <1 000† |
| Nucleobindin (M96824) | <1 000 | 2 098* | 2 342* | 1 623* | 1 608 | 1 924 | 3 594* | 1 944 |
| Group C | ||||||||
| c-raf (X03484) | <1 000 | 1 026 | <1 000 | 1 439* | <1 000 | <1 000 | <1 000 | <1 000 |
| APNH1 (M81768) | <1 000 | <1 000 | 1 072 | 1 452* | <1 000 | <1 000 | <1 000 | <1 000 |
| Lnk (AF055581) | <1 000 | <1 000 | <1 000 | 1 334* | <1 000 | <1 000 | <1 000 | 2 262* |
| hRlf beta subunit (D38073) | <1 000 | <1 000 | <1 000 | 2 117* | 2 792 | 2 131 | 1 626 | <1 000† |
| RbAp46 (U35143) | <1 000 | <1 000 | <1 000 | 1 368* | 1 300 | <1 000 | <1 000 | 5 810* |
| CD19 (M21097) | <1 000 | <1 000 | <1 000 | 1 563* | <1 000 | <1 000 | <1 000 | <1 000 |
| VEGF (M32977) | <1 000 | <1 000 | <1 000 | 1 321* | 6 228 | 4 326 | 6 453 | 1 116† |
| CLN2 protease (AF017456) | <1 000 | <1 000 | <1 000 | 1 597* | 6 812 | 6 433 | 1 928 | <1 000† |
| MAPKAP (U09578) | 2 484 | 1 019† | 1 949 | 1 422 | 3 084 | 1 433† | 2 416 | 7 880* |
| RAB7 GTPase (X93499) | 3 080 | 1 212† | 1 929 | 2 518 | <1 000 | <1 000 | <1 000 | 2 237* |
| Group D | ||||||||
| Cathepsin C (X87212) | 2 628 | <1 000 | 2 693 | 2 052 | <1 000 | <1 000 | 3 361* | 2 979* |
| Heat shock factor 1 (M64673) | <1 000 | <1 000 | 3 067* | 1 912* | <1 000 | <1 000 | 5 753* | 1 723* |
| Repl. factor C (M87339) | <1 000 | <1 000 | 1 822* | 2 240* | 4 044 | 4 664 | 4 500 | <1 000† |
| Group E | ||||||||
| RanGTPase act. Protein 1 (X82260) | 2 212 | 1 078† | 1 830 | <1 000† | <1 000 | <1 000 | <1 000 | <1 000 |
| c-myc (V00568) | 10 500 | 4 884† | 10 851 | 4 436† | 7 560 | <1 000† | 6 526 | <1 000† |
| TNF (X01394) | 8 292 | 3 265† | 7 822 | 3 801† | 2 424 | 2 345 | 2 500 | <1 000† |
| IL-2 receptor (X01057) | 5 832 | 2 716† | 5 537 | 3 039† | 15 236 | 3 749† | 14 835 | 4 269† |
| IL-2 receptor gamma (D11086) | 3 424 | 1 433† | 3 132 | 1 977† | 2 812 | 1 573 | 4 410 | 3 173 |
| HSP27 (X54079) | 15 452 | 5 319† | 14 741 | 7 652† | 5 076 | 3 226 | 3 421 | 3 161 |
| Histidine decarboxylase (X54297) | 2 080 | 1 074† | 2 398 | <1 000† | 2 512 | <1 000† | 3 920 | 1 901† |
| Repl. factor C (M87338) | 3 016 | 1 733 | 2 597 | 1 081† | 9 048 | 1 260† | 4 718 | 4 994 |
| Control | ATRA | ST1346 | ATRA + ST1346 | Control | ATRA | ST1346 | ATRA + ST1346 | |
| TRRAP (AF076974) | 3 920 | 1 903† | 4 068 | 2 316† | 1 432 | 2 371 | 8 519* | <1 000† |
| ALG-2 (AF035606) | 4 976 | 2 712† | 5 929 | 3 433 | 2 840 | 3 304 | 6 438* | <1 000 |
| HER-3 (M34309) | 1 616 | <1 000† | 1 773 | <1 000† | 3 256 | <1 000† | 3 089 | <1 000† |
| HDLCl (U32944) | 7 220 | 3 320† | 8 343 | 5 583 | 5 136 | 2 104† | 4 859 | 3 897 |
| CDC47 homolog (D55716) | 4 692 | 2 345† | 4 112 | 3 176 | 7 288 | 2 132† | 5 690 | <1 000† |
| Uracil-DNA glyc. (X15653) | 2 480 | <1 000† | 3 032 | 1 534† | 14 940 | <1 000† | 3 818† | <1 000† |
| EGR1(X52541) | 2 344 | <1 000† | 1 869 | 2 269 | 3 316 | <1 000† | 10 176* | 1 174† |
| HSE 70 (Y00371) | 4 608 | 1 891† | 5 417 | 4 537 | 10 164 | 2 288† | 11 668 | 5 663† |
| Hepatoma-derived GF (D16431) | 4 660 | 1 149† | 4 019 | 2 316† | 7 772 | 6 226 | 11 680 | 5 565 |
| IL-1 (K02770) | 4 104 | 2 092 | 3 601 | 1 676† | <1 000 | <1 000 | <1 000 | <1 000 |
| TAXREB 67 (D90209) | 7 272 | 6 709 | 8 802 | 6 375 | 5 772 | 2 754† | 7 088 | 3 241† |
| Group F | ||||||||
| ARF1 (M36340) | 2 148 | 2 483 | 1 793 | 1 166† | 2 524 | 2 525 | 4 045 | 4 717 |
| DNA pol. Delta (M80397) | 2 656 | 1 832 | 2 188 | 1 078† | 1 272 | <1 000 | 1 571 | 1 571 |
| M-CSF (M37435) | 10 608 | 6 353 | 10 423 | 5 189† | 4 184 | 3 453 | 2 073 | <1 000† |
| . | 8 hours . | 72 hours . | ||||||
|---|---|---|---|---|---|---|---|---|
| Control . | ATRA . | ST1346 . | ATRA + ST1346 . | Control . | ATRA . | ST1346 . | ATRA + ST1346 . | |
| Group A | ||||||||
| ICAM-1 (J03132) | 1 368 | 7 530* | 1 491 | 16 649* | <1 000 | 7 909* | <1 000 | 20 326* |
| MCP-1 (M24545) | <1 000 | 4 734* | 1 062 | 7 668* | <1 000 | 23 911* | <1 000 | 51 927* |
| Id-2 (M97796) | <1 000 | 1 773* | <1 000 | 3 023* | <1 000 | 2 357* | <1 000 | 5 518* |
| Mlh1 (U07418) | 16 128 | 9 100 | 14 897 | 9 913 | <1 000 | 2 261* | <1 000 | 4 713* |
| MAPKAP-K2 (U12779) | 2 368 | 1 836 | 1 826 | 4 552* | <1 000 | 2 253* | <1 000 | 7 329* |
| Cathepsin D (M11233) | 1 712 | 1 753 | 2 331 | 3 963* | 3 604 | 16 269* | 3 606 | 6 787* |
| FLI-1 (M93255) | <1 000 | <1 000 | <1 000 | 1 308* | <1 000 | 5 949* | 8 474* | 9 568* |
| MRP14 (X06233) | <1 000 | 3 399* | <1 000 | 3 889* | <1 000 | 6 933* | <1 000 | 12 376* |
| MRP8 (X06234) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 11 299* | <1 000 | 23 344* |
| MNDA (M81750) | <1 000 | 1 607* | <1 000 | 1 583* | <1 000 | 2 234* | <1 000 | 6 609* |
| IRF7 (U73036) | <1 000 | 2 412* | <1 000 | 1 726* | <1 000 | 2 510* | <1 000 | 4 216* |
| Net (Z36715) | <1 000 | 1 038 | <1 000 | 1 612* | <1 000 | 2 095* | 1 656 | 6 806* |
| PRK1 (U33053) | <1 000 | 3 182* | <1 000 | 1 700* | 1 364 | <1 000 | 1 097 | 6 899* |
| G-CSF receptor (M59818) | 1 072 | 3 370* | <1 000 | 3 483* | 2 428 | 1 782 | 3 095 | 8 136* |
| LIM-kinase (D26309) | <1 000 | 3 374* | <1 000 | 1 912* | 1 084 | 1 186 | 1 641 | 5 484* |
| LD-78 (D10924) | <1 000 | 2 570* | <1 000 | 1 951* | <1 000 | <1 000 | 1 710 | 3 730* |
| c-fgr (M19722) | 1 116 | 2 884* | <1 000 | 4 025* | 1 296 | 1 323 | <1 000 | 3 963* |
| PKC delta (D10495) | 3 132 | 5 548* | 3 062 | 5 568* | <1 000 | <1 000 | <1 000 | 3 080* |
| Tyk-2 (X54637) | 2 784 | 3 842 | 2 288 | 2 362 | 2 104 | 3 109 | 2 524 | 5 180* |
| EBI (U24166) | 1 948 | 2 286 | 1 418 | 3 518* | 2 752 | 7 422* | 2 553 | 8 378* |
| Group B | ||||||||
| Cyclin-B (M25753) | 2 260 | 4 746* | 3 414 | 4 814* | 4 696 | 9 553* | 8 794 | 5 577 |
| P19 INK (U40343) | <1 000 | 7 222* | 1 019 | 6 065* | 2 016 | 2 800 | 1 698 | 1 493 |
| Integrin alpha-4 (L12002) | 2 308 | 2 870 | 2 521 | 3 127 | 4 379 | 8 720* | 5 548 | 7 428 |
| TNF receptor (M32315) | <1 000 | 2 728* | <1 000 | 1 654* | <1 000 | 2 453* | <1 000 | <1 000 |
| GST pi (M24485) | 18 472 | 13 777 | 13 877 | 14 985 | <1 000 | 3 392* | 2 829 | 3 957* |
| Neutrophil elastase inhibitor (M93056) | <1 000 | 1 492* | <1 000 | 1 794* | <1 000 | 5 640* | <1 000 | <1 000 |
| G0S19-1 (M23452) | <1 000 | 2 298* | 1 541 | 1 514* | <1 000 | <1 000 | <1 000 | <1 000 |
| Glutharedoxin (X76648) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 4 096* | <1 000 | 2 280* |
| RALB (M35416) | <1 000 | 1 687 | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 |
| Glial growth factor (L12261) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 3 581* | <1 000 | <1 000 |
| c-sis (X02744) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 3 253* | <1 000 | <1 000 |
| Estrogen sulfotransferase (U08098) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 4 736* | <1 000 | <1 000 |
| NKSF (M65291) | <1 000 | <1 000 | <1 000 | <1 000 | <1 000 | 3 912* | <1 000 | <1 000 |
| S19 ribosomal protein (M81757) | 4 772 | 12 426* | 8 619 | 7 603 | 3 360 | 3 759 | 2 511 | 4 177 |
| Cathepsin H (X07549) | 3 996 | 2 076† | 4 241 | 3 384 | <1 000 | 4 589* | <1 000 | <1 000 |
| Stromelysin-3 (X57766) | 2 992 | 5 591* | 2 969 | 2 912 | 4 116 | 1 880† | 3 519 | 3 053 |
| DNA-binding protein A (M24069) | <1 000 | 1 414* | 2 530* | 1 305* | 2 556 | <1 000† | 2 634 | <1 000† |
| Nucleobindin (M96824) | <1 000 | 2 098* | 2 342* | 1 623* | 1 608 | 1 924 | 3 594* | 1 944 |
| Group C | ||||||||
| c-raf (X03484) | <1 000 | 1 026 | <1 000 | 1 439* | <1 000 | <1 000 | <1 000 | <1 000 |
| APNH1 (M81768) | <1 000 | <1 000 | 1 072 | 1 452* | <1 000 | <1 000 | <1 000 | <1 000 |
| Lnk (AF055581) | <1 000 | <1 000 | <1 000 | 1 334* | <1 000 | <1 000 | <1 000 | 2 262* |
| hRlf beta subunit (D38073) | <1 000 | <1 000 | <1 000 | 2 117* | 2 792 | 2 131 | 1 626 | <1 000† |
| RbAp46 (U35143) | <1 000 | <1 000 | <1 000 | 1 368* | 1 300 | <1 000 | <1 000 | 5 810* |
| CD19 (M21097) | <1 000 | <1 000 | <1 000 | 1 563* | <1 000 | <1 000 | <1 000 | <1 000 |
| VEGF (M32977) | <1 000 | <1 000 | <1 000 | 1 321* | 6 228 | 4 326 | 6 453 | 1 116† |
| CLN2 protease (AF017456) | <1 000 | <1 000 | <1 000 | 1 597* | 6 812 | 6 433 | 1 928 | <1 000† |
| MAPKAP (U09578) | 2 484 | 1 019† | 1 949 | 1 422 | 3 084 | 1 433† | 2 416 | 7 880* |
| RAB7 GTPase (X93499) | 3 080 | 1 212† | 1 929 | 2 518 | <1 000 | <1 000 | <1 000 | 2 237* |
| Group D | ||||||||
| Cathepsin C (X87212) | 2 628 | <1 000 | 2 693 | 2 052 | <1 000 | <1 000 | 3 361* | 2 979* |
| Heat shock factor 1 (M64673) | <1 000 | <1 000 | 3 067* | 1 912* | <1 000 | <1 000 | 5 753* | 1 723* |
| Repl. factor C (M87339) | <1 000 | <1 000 | 1 822* | 2 240* | 4 044 | 4 664 | 4 500 | <1 000† |
| Group E | ||||||||
| RanGTPase act. Protein 1 (X82260) | 2 212 | 1 078† | 1 830 | <1 000† | <1 000 | <1 000 | <1 000 | <1 000 |
| c-myc (V00568) | 10 500 | 4 884† | 10 851 | 4 436† | 7 560 | <1 000† | 6 526 | <1 000† |
| TNF (X01394) | 8 292 | 3 265† | 7 822 | 3 801† | 2 424 | 2 345 | 2 500 | <1 000† |
| IL-2 receptor (X01057) | 5 832 | 2 716† | 5 537 | 3 039† | 15 236 | 3 749† | 14 835 | 4 269† |
| IL-2 receptor gamma (D11086) | 3 424 | 1 433† | 3 132 | 1 977† | 2 812 | 1 573 | 4 410 | 3 173 |
| HSP27 (X54079) | 15 452 | 5 319† | 14 741 | 7 652† | 5 076 | 3 226 | 3 421 | 3 161 |
| Histidine decarboxylase (X54297) | 2 080 | 1 074† | 2 398 | <1 000† | 2 512 | <1 000† | 3 920 | 1 901† |
| Repl. factor C (M87338) | 3 016 | 1 733 | 2 597 | 1 081† | 9 048 | 1 260† | 4 718 | 4 994 |
| Control | ATRA | ST1346 | ATRA + ST1346 | Control | ATRA | ST1346 | ATRA + ST1346 | |
| TRRAP (AF076974) | 3 920 | 1 903† | 4 068 | 2 316† | 1 432 | 2 371 | 8 519* | <1 000† |
| ALG-2 (AF035606) | 4 976 | 2 712† | 5 929 | 3 433 | 2 840 | 3 304 | 6 438* | <1 000 |
| HER-3 (M34309) | 1 616 | <1 000† | 1 773 | <1 000† | 3 256 | <1 000† | 3 089 | <1 000† |
| HDLCl (U32944) | 7 220 | 3 320† | 8 343 | 5 583 | 5 136 | 2 104† | 4 859 | 3 897 |
| CDC47 homolog (D55716) | 4 692 | 2 345† | 4 112 | 3 176 | 7 288 | 2 132† | 5 690 | <1 000† |
| Uracil-DNA glyc. (X15653) | 2 480 | <1 000† | 3 032 | 1 534† | 14 940 | <1 000† | 3 818† | <1 000† |
| EGR1(X52541) | 2 344 | <1 000† | 1 869 | 2 269 | 3 316 | <1 000† | 10 176* | 1 174† |
| HSE 70 (Y00371) | 4 608 | 1 891† | 5 417 | 4 537 | 10 164 | 2 288† | 11 668 | 5 663† |
| Hepatoma-derived GF (D16431) | 4 660 | 1 149† | 4 019 | 2 316† | 7 772 | 6 226 | 11 680 | 5 565 |
| IL-1 (K02770) | 4 104 | 2 092 | 3 601 | 1 676† | <1 000 | <1 000 | <1 000 | <1 000 |
| TAXREB 67 (D90209) | 7 272 | 6 709 | 8 802 | 6 375 | 5 772 | 2 754† | 7 088 | 3 241† |
| Group F | ||||||||
| ARF1 (M36340) | 2 148 | 2 483 | 1 793 | 1 166† | 2 524 | 2 525 | 4 045 | 4 717 |
| DNA pol. Delta (M80397) | 2 656 | 1 832 | 2 188 | 1 078† | 1 272 | <1 000 | 1 571 | 1 571 |
| M-CSF (M37435) | 10 608 | 6 353 | 10 423 | 5 189† | 4 184 | 3 453 | 2 073 | <1 000† |
The numbers indicate the adjusted intensity values of the spots on the array corresponding to the genes listed on the left. They were obtained following average background subtraction and normalization for the average intensity of all the arrayed spots using the ATLAS 2.0 analysis software. The Genbank accession number for each gene is indicated in parentheses. Genes were classified in each group according to their expression profiles. Less than 1000 indicates that the intensity of the signal was equivalent to or below the background level.
An induction of the mRNA equal to or higher than 1.6 relative to the corresponding control.
A decrease of the mRNA equal to or higher than 0.4 relative to the corresponding control.
Group D is composed of the genes coding for cathepsin C, heat shock factor 1, and replication factor C, which are activated by ST1346 alone. Group E consists of 19 genes whose expression is significantly down-regulated by ATRA and includes those coding for the 2 cytokines IL-1 and TNF. Finally, group F includes 3 genes, ARF1, DNA polymerase Delta, and M-CSF, whose level of expression is significantly down-regulated only by ATRA + ST1346.
For a number of genes in Table 2, we measured the relative protein or RNA products. Flow cytometric analysis of ICAM-1 demonstrates that more than 90% of the undifferentiated NB4 blasts express the adhesion molecule. As shown in Figure 6A, ST1346 (10 μM) does not alter the levels of cell-associated ICAM-1, whereas ATRA augments it in a dose-dependent manner. Cotreatment of NB4 cells for 3 days with the BISIND and ATRA enhances the surface expression of ICAM-1 by approximately 2-fold at both the concentrations of the retinoid considered. By contrast, ATRA and ATRA + ST1346 are equally efficient in up-regulating integrin α4. As illustrated in Figure 6B, the combination of the BISIND and ATRA is more effective than the retinoid alone in inducing the mutL homolog 1 (Mlh-1) DNA repair protein, the protease cathepsin D, and the tyrosine kinase Tyk-2. A similar situation is observed in the case of the mRNAs encoding the transcription factor Id-2 and the myeloid nuclear differentiation antigen (data not shown). The most dramatic example of the positive interaction between ST1346 and ATRA is represented by the chemokine MCP-1. Following treatment with ATRA, NB4 cells secrete detectable amounts of MCP-1 (Figure 6C). At 3 days, the levels of secreted chemokines are increased approximately 6-fold by ATRA + ST1346. We also evaluated the secretion of the 2 cytokines TNF and IL-1, whose mRNAs are down-regulated by ATRA and the combination of ST1346 and ATRA (Table 2). Figure 6C demonstrates that undifferentiated NB4 cells secrete measurable amounts of TNF and IL-1. Both ST1346 and ATRA reduce the secretion of the 2 cytokines. In the case of IL-1, this effect is significant at day 1 and is maximal at day 3. Given the low levels of TNF already observed following treatment with the BISIND or ATRA, it is difficult to establish whether the combination of BISIND + ATRA is more effective than the single components in inhibiting the secretion of the 2 cytokines. By contrast, inhibition of IL-1 secretion by ATRA + ST1346 is not significantly different from that afforded by ST1346 or ATRA alone.
Influence of ST1346 on the steady-state levels of ATRA-modulated proteins and the secretion of the MCP-1 chemokine, TNF, and IL-1 proinflammatory cytokines.
NB4 cells were treated for 1 or 3 days, as indicated, with vehicle alone, ST1346 (10 μM), the indicated concentrations of ATRA, or the combination of the 2 compounds. (A) The level of expression of the indicated phenotypic markers was determined by flow cytometry using fluorescein-labeled monoclonal antibodies against ICAM-1 and integrin α4 or irrelevant isotype-matched antibodies (Neg). NB4 cells are more than 95% positive for the expression of the 2 integrins in all the experimental conditions considered. The results are expressed in mean cell-associated fluorescence. (B) The amounts of the indicated proteins were determined by Western blot analysis with the use of anti–cathepsin D, anti-Tyk2, anti-Mlh1, and antiactin polyclonal antibodies. (C) The amounts of MCP-1, TNF, and IL-1 secreted in the conditioned medium of NB4 cells treated as indicated were determined with the use of specific ELISA assays. Results are expressed as the means ± SD of 3 separate culture dishes and are representative of at least 3 independent experiments.
Influence of ST1346 on the steady-state levels of ATRA-modulated proteins and the secretion of the MCP-1 chemokine, TNF, and IL-1 proinflammatory cytokines.
NB4 cells were treated for 1 or 3 days, as indicated, with vehicle alone, ST1346 (10 μM), the indicated concentrations of ATRA, or the combination of the 2 compounds. (A) The level of expression of the indicated phenotypic markers was determined by flow cytometry using fluorescein-labeled monoclonal antibodies against ICAM-1 and integrin α4 or irrelevant isotype-matched antibodies (Neg). NB4 cells are more than 95% positive for the expression of the 2 integrins in all the experimental conditions considered. The results are expressed in mean cell-associated fluorescence. (B) The amounts of the indicated proteins were determined by Western blot analysis with the use of anti–cathepsin D, anti-Tyk2, anti-Mlh1, and antiactin polyclonal antibodies. (C) The amounts of MCP-1, TNF, and IL-1 secreted in the conditioned medium of NB4 cells treated as indicated were determined with the use of specific ELISA assays. Results are expressed as the means ± SD of 3 separate culture dishes and are representative of at least 3 independent experiments.
Effects of ST1346 on the levels, stability, and activation of retinoic acid receptors and histone acetylation
NB4 blasts express significant amounts of both RARα and PML-RARα, and transfection of this cell line is technically challenging. Thus, the effect of ST1346 on the trans-activating properties of RARα, PML-RARα, or RXRα was investigated in COS-7 cells, which are endowed with relatively low levels of nuclear retinoic acid receptors23 and easily transfected. As illustrated in Figure 7A, following cotransfection with a void plasmid (pSG5) and TRE-TK-Luc (containing a luciferase reporter gene controlled by a retinoid-dependent artificial promoter), treatment of cells with ATRA results in an approximately 5-fold increase in the levels of luciferase activity relative to what is observed in basal conditions. This effect is explained by the presence of endogenous retinoic acid receptors in COS-7 cells. ST1346 has no significant effect on either the basal or the ATRA-induced levels of luciferase activity. Forced expression of RARα RXRα RARα RXRα, and PML-RARα (data not shown) increases the ATRA-dependent induction of luciferase activity to varying degrees. Again, ST1346 has no significant effect on the ligand-dependent or independent trans-activating properties of the various receptors. Varying the concentrations of ATRA and ST1346 or changing the type of recipient cell lines and reporter constructs does not alter the results (data not shown). Although our data do not rule out possible specific effects related to the myeloid cell context, they suggest that ST1346 does not modulate the transcriptional activity of RARα or PML-RARα complexes directly.
Influence of ST1346 and ATRA on the transcriptional activity of RARα in transfected COS-7 cells and on the levels of PML-RARα, RARα, and RXRα proteins as well as histone acetylation in NB4 cells.
(A) COS-7 cells were transfected with the plasmid pSG5 (void vector) and the same plasmid containing RARα or RXRα as indicated. The transfection mixture also contained the retinoid-responsive reporter construct TRE-TK-Luc (containing the luciferase reporter gene) and the β-galactosidase expression vector pCH110. Twenty-four hours following transfection, cells were treated with vehicle (dimethyl sulfoxide [DMSO]), ST1346 (5 μM), ATRA (0.1 μM), or the combination of the 2 compounds for a further 24 hours. Cell extracts were used for the determination of luciferase enzymatic activity. The results are always normalized for the transfection efficiency as assessed by determination of the β-galactosidase enzymatic activity. Results are expressed as the means ± SD of 3 separate dishes and are representative of at least 3 experiments. (B) NB4 cells were treated for 72 hours with vehicle, ATRA (0.1 μM), and ST1346 at the indicated concentrations, or combinations of ATRA + ST1346. Aliquots of cell extracts were subjected to Western blot analysis with anti-RARα, anti-RXRα, and anti–β-actin antibodies. The positions of relevant molecular weight markers are indicated on the right. (●) indicates the undegraded form; * represents a degradation band of PML-RARα. (C) NB4 cells were treated for 72 hours with vehicle, ST1346 (5 μM), sodium butyrate (1 mM), and ATRA at the indicated concentrations or combinations of ATRA + ST1346 and ATRA + sodium butyrate. Aliquots of cell extracts were subjected to Western blot analysis with antibodies recognizing histone H3 or the acetylated form of histone H3 (Ac-H3). Results are representative of 2 independent experiments.
Influence of ST1346 and ATRA on the transcriptional activity of RARα in transfected COS-7 cells and on the levels of PML-RARα, RARα, and RXRα proteins as well as histone acetylation in NB4 cells.
(A) COS-7 cells were transfected with the plasmid pSG5 (void vector) and the same plasmid containing RARα or RXRα as indicated. The transfection mixture also contained the retinoid-responsive reporter construct TRE-TK-Luc (containing the luciferase reporter gene) and the β-galactosidase expression vector pCH110. Twenty-four hours following transfection, cells were treated with vehicle (dimethyl sulfoxide [DMSO]), ST1346 (5 μM), ATRA (0.1 μM), or the combination of the 2 compounds for a further 24 hours. Cell extracts were used for the determination of luciferase enzymatic activity. The results are always normalized for the transfection efficiency as assessed by determination of the β-galactosidase enzymatic activity. Results are expressed as the means ± SD of 3 separate dishes and are representative of at least 3 experiments. (B) NB4 cells were treated for 72 hours with vehicle, ATRA (0.1 μM), and ST1346 at the indicated concentrations, or combinations of ATRA + ST1346. Aliquots of cell extracts were subjected to Western blot analysis with anti-RARα, anti-RXRα, and anti–β-actin antibodies. The positions of relevant molecular weight markers are indicated on the right. (●) indicates the undegraded form; * represents a degradation band of PML-RARα. (C) NB4 cells were treated for 72 hours with vehicle, ST1346 (5 μM), sodium butyrate (1 mM), and ATRA at the indicated concentrations or combinations of ATRA + ST1346 and ATRA + sodium butyrate. Aliquots of cell extracts were subjected to Western blot analysis with antibodies recognizing histone H3 or the acetylated form of histone H3 (Ac-H3). Results are representative of 2 independent experiments.
Treatment of APL blasts with ATRA is associated with degradation of both PML-RARα and RARα.33 Furthermore, ATRA-triggered granulocytic maturation is accompanied by down-regulation of the transcript encoding RXRα.34 As illustrated in Figure 7B, treatment of NB4 cells with the retinoid for 72 hours results in the disappearance of the protein band corresponding to PML-RARα and in a substantial decrease in the steady-state levels of the RARα polypeptide. This phenomenon is left unaffected by addition of ST1346 to the medium. Similarly, ST1346 does not exert any effect on the RXRα protein, which is synthesized in significant amounts by undifferentiated NB4 cells and down-regulated by ATRA treatment. Finally, as shown in Figure 7C, ST1346 is not a general inducer of histone acetylation, a phenomenon associated with the transcriptional activation of retinoic acid receptors and stimulated by agents such as sodium butyrate.35 36
Influence of ST1346 on nuclear transcriptional factors and complexes involved in ATRA-dependent granulocytic maturation of APL cells
Various types of transcriptional factors, including the STAT1 and CEBPε proteins as well as the ets and cAMP-dependent CREB binding complexes, are implicated in the process of APL blast differentiation activated by ATRA.37-40 As expected37 and as illustrated in Figure 8A (left panel), treatment of NB4 cells for 3 days with ATRA causes a significant increase in the amounts of STAT1α (91 kDa) and STAT1β(84 kDa). ST1346 enhances the ATRA-dependent induction of both forms of STAT1 (left panel). A quantitative assessment of the enhancing effect of ST1346 on the ATRA-dependent induction of STAT1α demonstrates that the BISIND causes a 3-fold induction of the protein over what is observed in the presence of the retinoid alone (right panel). At least a proportion of the ATRA- and ST1346 + ATRA–induced STAT proteins must be in the transcriptionally active state, as indicated by the level of phosphorylation of Tyr701 (left panel). As shown in Figure 8B, CEBPε is not synthesized by undifferentiated NB4 cells. Following ATRA treatment, significant amounts of CEBPε are already evident at 12 hours and increase up to 72 hours. ST1346 treatment does not result in the synthesis of the CEBPε protein and has only marginal effects on the level of induction afforded by the retinoid. Figure 8C demonstrates that nuclear extracts of undifferentiated NB4 cells contain detectable amounts of CREB and Ets-1complexes, as assessed by gel retardation assays. Treatment of cells for 4 hours with ST1346, but not ATRA, results in the disappearance of the Ets-1 complex. This inhibitory effect is completely suppressed if cells are simultaneously exposed to ST1346 and ATRA. ST1346-dependent inhibition disappears following 16 hours of culturing. By this time, NB4-associated Ets-1 binding activity is similar in all the experimental conditions considered. In the case of CREB, ATRA reduces the amounts of the specifically retarded band observed in control conditions at both 4 and 16 hours. This effect is reversed by addition of ST1346 to the culture medium.
Influence of ST1346 and ATRA on the levels and state of transcriptional factors or complexes involved in ATRA-induced granulocytic differentiation of myeloid cells.
NB4 cells were treated for 72 hours (panel A) or the indicated amount of time (panels B and C) with vehicle, ATRA (0.1 μM), the indicated concentrations of ST1346, or the corresponding combinations of the BISIND and the retinoid. (A,B) Equivalent amounts of cell extracts were subjected to Western blot analysis with anti–STAT 1 α/β, anti–β-actin, anti–CEBPε polyclonal antibodies, or antibodies recognizing the Tyr701 phosphorylated form of STAT 1. The right graph in panel A illustrates the quantitative results obtained by densitometric analysis of the STAT 1α band in blots similar to that depicted on the left and developed with an anti–STAT 1α–specific antibody. The levels of STAT 1α protein are expressed in relative units following normalization for the intensity of the corresponding β-actin bands. Data are expressed as the means ± SD of 3 separate experiments. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Student t test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and ATRA-treated groups, following 2-way analysis of variance and measurement of the F score of the interaction (P < .01 according to Tukey test). (C) Cell extracts were subjected to gel retardation assays using double-stranded oligonucleotides corresponding to the consensus sites of binding of the transcriptional complexes Ets-1 and CREB. The competitor is a cold Ets-1 (ets) or CREB oligonucleotide present in the reaction mixture in 100-fold excess.
Influence of ST1346 and ATRA on the levels and state of transcriptional factors or complexes involved in ATRA-induced granulocytic differentiation of myeloid cells.
NB4 cells were treated for 72 hours (panel A) or the indicated amount of time (panels B and C) with vehicle, ATRA (0.1 μM), the indicated concentrations of ST1346, or the corresponding combinations of the BISIND and the retinoid. (A,B) Equivalent amounts of cell extracts were subjected to Western blot analysis with anti–STAT 1 α/β, anti–β-actin, anti–CEBPε polyclonal antibodies, or antibodies recognizing the Tyr701 phosphorylated form of STAT 1. The right graph in panel A illustrates the quantitative results obtained by densitometric analysis of the STAT 1α band in blots similar to that depicted on the left and developed with an anti–STAT 1α–specific antibody. The levels of STAT 1α protein are expressed in relative units following normalization for the intensity of the corresponding β-actin bands. Data are expressed as the means ± SD of 3 separate experiments. *Significantly higher than the corresponding group treated with vehicle (P < .01 according to the Student t test). °Significantly higher than the sum of the effects observed in the corresponding ST1346- and ATRA-treated groups, following 2-way analysis of variance and measurement of the F score of the interaction (P < .01 according to Tukey test). (C) Cell extracts were subjected to gel retardation assays using double-stranded oligonucleotides corresponding to the consensus sites of binding of the transcriptional complexes Ets-1 and CREB. The competitor is a cold Ets-1 (ets) or CREB oligonucleotide present in the reaction mixture in 100-fold excess.
Role of JNK in the differentiation of NB4 cells triggered by ATRA and enhanced by ST1346
MAP kinases, ERK, p38, and JNK are involved in the process of differentiation of myeloid cells.41-44 As illustrated in Figure 9A, undifferentiated NB4 cells show high levels of constitutive ERK phosphorylation. The predominant form of phosphorylated ERK is p42. Treatment of cells with ATRA, ST1346, and the combination of the 2 compounds for 8 or 72 hours does not affect the steady-state levels or the phosphorylation of ERK p42 and p44 proteins. As shown in Figure 9B, in conditions of complete suppression of ERK activation (data not shown), the MEK inhibitor U0126 does not affect the proportion of NBT+ NB4 cells nor the level of cell-associated NBT-R activity observed following treatment with vehicle, ATRA, ST1346, or ATRA + ST1346. This indicates that constitutive ERK phosphorylation and activation do not play any significant role in the process of granulocytic maturation activated by ATRA and enhanced by ST1346.
Influence of ST1346 and ATRA on the phosphorylation of MAP kinases and effect of MAP kinase inhibitors on the granulocytic differentiation of NB4 cells caused by ATRA and combinations of ATRA + ST1346.
(A,C,E) NB4 cells were treated for the indicated amount of time with vehicle, ATRA (0.1 μM), ST1346 (10 μM), or the combination of the 2 compounds. Total cellular extracts were subjected to Western blot analysis using polyclonal antibodies recognizing ERK, p38, or JNK and the corresponding phosphorylated forms, as indicated by the circled ‘p’. The positions of selected molecular weight markers are indicated on the right. The blots shown are representative of 5 independent experiments. (B,D,F) NB4 cells were treated for 3 days with vehicle (DMSO), ATRA (0.1μM), ST1346 (10 μM), the ERK inhibitor U0126 (10 μM), the p38 inhibitor PD169316 (10 μM), the JNK inhibitor SP600125 (10 μM), or the indicated combinations of the various compounds. Aliquots of the extracts were used for the determination of NBT-reducing activity. In the case of panel F, we also measured the level of JNK kinase activity on JNK immunoprecipitates, using c-Jun as a substrate. Equivalent amounts of immunoprecipitates were electrophoresed on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), blotted on nitrocellulose, and challenged with antibodies recognizing the phosphorylated form of c-JUN (serine/63) or JNK. The results are shown underneath the column graph. The number of NBT+ cells is also shown above each panel. Results are expressed as the means ± SD of 3 separate culture dishes and are representative of at least 3 independent experiments. **Significantly higher or lower than the corresponding group treated with ATRA + ST1346 (P < .01 according to the Studentt test).
Influence of ST1346 and ATRA on the phosphorylation of MAP kinases and effect of MAP kinase inhibitors on the granulocytic differentiation of NB4 cells caused by ATRA and combinations of ATRA + ST1346.
(A,C,E) NB4 cells were treated for the indicated amount of time with vehicle, ATRA (0.1 μM), ST1346 (10 μM), or the combination of the 2 compounds. Total cellular extracts were subjected to Western blot analysis using polyclonal antibodies recognizing ERK, p38, or JNK and the corresponding phosphorylated forms, as indicated by the circled ‘p’. The positions of selected molecular weight markers are indicated on the right. The blots shown are representative of 5 independent experiments. (B,D,F) NB4 cells were treated for 3 days with vehicle (DMSO), ATRA (0.1μM), ST1346 (10 μM), the ERK inhibitor U0126 (10 μM), the p38 inhibitor PD169316 (10 μM), the JNK inhibitor SP600125 (10 μM), or the indicated combinations of the various compounds. Aliquots of the extracts were used for the determination of NBT-reducing activity. In the case of panel F, we also measured the level of JNK kinase activity on JNK immunoprecipitates, using c-Jun as a substrate. Equivalent amounts of immunoprecipitates were electrophoresed on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), blotted on nitrocellulose, and challenged with antibodies recognizing the phosphorylated form of c-JUN (serine/63) or JNK. The results are shown underneath the column graph. The number of NBT+ cells is also shown above each panel. Results are expressed as the means ± SD of 3 separate culture dishes and are representative of at least 3 independent experiments. **Significantly higher or lower than the corresponding group treated with ATRA + ST1346 (P < .01 according to the Studentt test).
Figure 9C demonstrates that NB4 cells express detectable amounts of p38 protein, and the quantity of the polypeptide is not altered by any of the treatments considered. Following 72 hours of challenge, ATRA increases p38 phosphorylation. ST1346 has no detectable effect on the ATRA-induced level of p38 phosphorylation. Figure 9D demonstrates that the specific p38 inhibitor PD169316 enhances the induction of NBT-R activity observed in the presence of ATRA. Interestingly, addition of the inhibitor to ST1346 and ATRA results in a level of NBT-R activity that is slightly but significantly higher than that observed in the presence of the combination of the BISIND and the retinoid. Altogether, our results indicate that activation of p38 plays a negative role in the process of granulocytic maturation of APL cells. However, they suggest that enhancement of NB4 cell differentiation by ST1346 does not involve p38 inhibition.
As shown in Figure 9E, 2 forms of JNK, p46 and p54, are synthesized by undifferentiated NB4 cells. Both forms are phosphorylated in basal conditions, although the level of p54 phosphorylation is higher than that of p46. Following 8 or 72 hours of treatment with ATRA, ST1346, or the combination of the 2 compounds, no significant variation in the amounts of p46 and p54 are observed. At 8 hours, the level of p46 or p54 is similar in all the experimental groups considered; at 72 hours, ATRA inhibits JNK phosphorylation. This down-regulation is relieved in cells treated with ATRA + ST1346. As expected and as illustrated in Figure 9F, inhibition of JNK phosphorylation by ATRA is associated with a decrease in the corresponding c-JUN kinase activity. JNK activity is brought back to control levels by the combination of ST1346 + ATRA. Treatment of NB4 cells with the JNK inhibitor SP600125 blocks JNK activity in all the experimental conditions considered. This is accompanied by a decrease in the percentage of NBT-R–positive NB4 cells observed upon challenge with ATRA + ST1346 to a level similar to that observed following treatment with ATRA alone. The effect is evident not only in terms of the number of NBT-R–positive cells but also in terms of cell-associated enzymatic activity. Thus, prolonged JNK activation by ATRA + ST1346 may be crucial for the enhancement of the retinoid cytodifferentiating effect by the BISIND.
In vivo efficacy of the combination of ATRA + ST1346
To evaluate whether the potentiating effect of ST1346 on ATRA cytodifferentiating activity has an impact on the therapeutic efficacy of the retinoid, we performed experiments in SCID mice that received transplanted NB4 cells intraperitoneally.22 As shown in Table 3, treatment of animals with a suboptimal dose of ATRA (5 mg/kg) or 2 doses of ST1346 (15 and 30 mg/kg) does not cause a significant increase in the mean survival time (MST). Coadministration of ST1346 and ATRA results in an increase of MST, which is statistically significant at the higher dose of BISIND. The effect on MST and increased lifespan (ILS) obtained with ATRA + ST1346 (30 + 5 mg/kg) is similar to that observed with a maximally tolerated dose of ATRA (15 mg/kg) (data not shown). No dosage regimen is accompanied by significant toxicity as determined by the lack of body weight loss and by the absence of toxic deaths. These results suggest that enhancement of ATRA-induced cytodifferentiation by BISIND can be exploited to increase the in vivo efficacy of the retinoid.
In vivo activity of ST1346 and ATRA in the SCID mouse receiving transplanted NB4 cells
| Treatment . | Dose, mg/kg . | Lethality . | BWL, % . | MST (range) . | MST . | ILS, % . |
|---|---|---|---|---|---|---|
| Vehicle | — | 0/7 | 0 | (39-46) | 43 | — |
| ATRA | 5 | 0/7 | 0 | (42-63) | 45 | 5 |
| ST1346 | 15 | 0/7 | 0 | (34-48) | 39 | — |
| ST1346 | 30 | 0/6 | 0 | (36-64) | 40 | — |
| ATRA + ST1346 | 5 + 15 | 0/7 | 0 | (42-65) | 49 | 14 |
| ATRA + ST1346 | 5 + 30 | 0/7 | 0 | (46-70) | 603-150,3-151 | 40 |
| Treatment . | Dose, mg/kg . | Lethality . | BWL, % . | MST (range) . | MST . | ILS, % . |
|---|---|---|---|---|---|---|
| Vehicle | — | 0/7 | 0 | (39-46) | 43 | — |
| ATRA | 5 | 0/7 | 0 | (42-63) | 45 | 5 |
| ST1346 | 15 | 0/7 | 0 | (34-48) | 39 | — |
| ST1346 | 30 | 0/6 | 0 | (36-64) | 40 | — |
| ATRA + ST1346 | 5 + 15 | 0/7 | 0 | (42-65) | 49 | 14 |
| ATRA + ST1346 | 5 + 30 | 0/7 | 0 | (46-70) | 603-150,3-151 | 40 |
SCID mice were xenografted with NB4 cells intraperitoneally and treated as stated in “Materials and methods” with the indicated dosages of ATRA and/or ST1346 5 times per week for 3 weeks. BWL% max indicates maximum body weight loss during the treatment; MST, mean survival time; ILS%, increase in lifespan (MST treated/MST control ×100) – 100.
Significantly higher than the vehicle–treated experimental group (P < .01, according to the Mann–Whitney test following analysis of variance [ANOVA]).
Significantly higher than the ATRA–treated experimental group (P < .05, according to the Mann–Whitney test following ANOVA).
Discussion
Here, we report on the identification of a novel class of experimental agents, BISINDs, that potentiate the cytodifferentiating activity of ATRA in APL and other acute myeloid leukemia (AML) cells. The BISIND prototypical molecule ST1346 exerts a general stimulating effect on many aspects of the granulocytic maturation program set in motion by ATRA. The potentiating effect of ST1346 is specifically directed toward ATRA-induced cytodifferentiation, as the compound leaves unaltered the antiproliferative and apoptogenic activity of the retinoid. Administration of ST1346 to SCID mice xenografted with NB4 cells dose-dependently stimulates the therapeutic effect of an otherwise ineffective dose of ATRA and significantly increases the lifespan of leukemia-bearing animals. These results are consistent with the fact that enhancement of the cytodifferentiating potential of ATRA is a viable strategy to increase the in vivo therapeutic efficacy of the retinoid.
As ATRA acts predominantly through modulation of target gene expression, we established the number and type of ATRA-responsive genes regulated by ST1346. Profiling of NB4 gene expression with microarrays demonstrates that the BISIND enhances and/or prolongs the ATRA-dependent up-regulation of a number of gene products. Approximately 3% of the mRNAs analyzed are significantly up-regulated by ATRA either early and/or late during the granulocytic maturation process. The expression levels of about one half of these transcripts are enhanced by coincubation of the cells with ATRA + ST1346. The list includes mRNAs coding for myeloid differentiation markers (MRP-8, MRP-14, MCP-1, G-CSF–receptor, and ICAM-1), transcription factors (Id-2, IRF-7, and Net), signal transducers (c-raf, Tyk-2, protein kinase C–related 1 (PRK-1), LIM-kinase, and LD78 guanosine 5′-triphosphate [GTP]-binding protein), and a miscellaneous group of proteins among which the microtubule-associated protein EB1 and the integrin ICAM-1 stand out.
In the conditions of the microarray experiments, the effect of ST1346 on genes negatively regulated by ATRA is difficult to determine, because the expression of most of them is already turned off by the retinoid. Nevertheless, in the case of the limited number of ATRA down-regulated genes for which we have data on the respective protein products, we observed significant interactions between the retinoid and the BISIND. Decreased secretion of the proinflammatory cytokines TNF and IL-1 by ATRA and/or ST1346 may be of clinical relevance, as these types of products are causally related to the life-threatening ATRA syndrome.45
The results obtained with microarrays highlight another important point, that is, there are genes neither responsive to ATRA nor to ST1346 that are significantly induced by ATRA + ST1346. Among these, the Na+/H+ antiporter APNH1 may be of some functional relevance, as changes in the intracellular pH have been associated with the process of granulocytic maturation.46
ST1346 modulates the level and the state of activation of some transcription factors potentially involved in the granulocytic maturation of leukemic blasts. ATRA increases the steady-state levels of STAT1, and this effect is preceded by a transient activation of the protein by tyrosine phosphorylation.37 STAT1 activation and induction are responsible for the increased expression of interferon-responsive genes, such as the IRF1,37 which plays a role in myeloid differentiation. ST1346 enhances the ATRA-dependent induction of activated STAT1, demonstrating that this transcription factor is one of those retinoid-modulated genes that is responsive to the BISIND. The observed up-regulation of the Id-2 transcript by ATRA and its further stimulation by ST1346 and ATRA may also be of interest in this context. Id-2 is a helix-loop-helix (HLH) transcriptional factor with inhibitory effects on the activity of other HLH.47 Most importantly, ST1346 prevents the down-modulation of the CREB transcription complex afforded by ATRA, through an as-yet-unknown mechanism. CREB complexes (cAMP-responsive element binding complexes) can be activated by the cAMP-dependent protein kinase A. The cyclic nucleotide, the corresponding kinase, and the downstream transcriptional factors are permissive determinants of APL differentiation.18,19 48Hence, sustained CREB activation may favor ATRA-induced granulocytic maturation.
The primary molecular targets underlying the pharmacologic activity of BISINDs are still unknown. However, the data obtained in COS-7 cells are not consistent with any direct effect of ST1346 on the transcriptional activity of RARα or PML-RARα (Figure 7). In addition, contrary to 2 other retinoid sensitizers like ST157120 and sodium butyrate,36 ST1346 does not inhibit the degradation of RARα or PML-RARα and is not a general activator of histone acetylation.
Because MAP kinases play a major role in the processes of cell growth and differentiation of various cell types,41-44 we focused our attention on ERKs, p38, and JNKs. As to the first family, prolonged activation of ERKs has been associated with ATRA-induced granulocytic maturation of HL-60 cells.49 In NB4 cells, we demonstrate that ERKs are constitutively phosphorylated and activated. ATRA, ST1346, and the combination of the 2 compounds do not alter the level of ERK phosphorylation. Furthermore, inhibition of ERK phosphorylation with the selective MEK inhibitor U0126 has no impact on the cytodifferentiating effects of ATRA or its combinations with ST1346. This indicates that constitutive ERK activation is not a prerequisite for ATRA-induced granulocytic maturation of APL cells and demonstrates that the enzymatic system is not involved in the potentiating action of ST1346.
As to the second family, ATRA treatment of NB4 cells results in long-term phosphorylation of p38, which is not further enhanced by ST1346. Since p38 inhibition stimulates ATRA-dependent granulocytic maturation of HL-60 cells,50 it is not surprising that we observed a similar effect in PD169316-treated NB4 cells. Moreover, when the p38 inhibitor is present in the culture medium along with ST1346 and ATRA, the level of the myeloid differentiation marker, NBT-R activity, is slightly but significantly higher than following treatment with ATRA + ST1346 or ATRA + PD169316. Thus, although activation of p38 seems to play a negative role in the process of ATRA-induced granulocytic maturation, the enhancing effect of ST1346 is likely to be independent of p38 activity.
As to JNKs, the third family of MAP kinases, they are involved in many cellular processes.51,52 JNKs activate the AP1 transcriptional complex,53 and ATRA represses AP1 activation in part by acting on JNKs.53,54 These phenomena may have relevance in terms of APL cell growth and maturation, as the 2 processes are tightly associated.15 Our data indicate that ATRA causes a late suppression of the constitutive level of JNK phosphorylation. The phenomenon is relieved by ST1346, which reinstates levels of JNK phosphorylation similar to those observed in basal conditions. This, along with the results obtained with the JNK inhibitor SP600125, indicates that activation of the kinase is necessary for ST1346 to enhance the cytodifferentiating effect of ATRA.
In conclusion, though, the primary molecular targets of BISINDs are unknown, and the biologic action is likely to be novel and may ultimately modulate the CREB and JNK intracellular pathways. Nevertheless, the in vitro and in vivo pharmacologic activity of this novel class of agents may be of clinical benefit in the treatment of APL and, perhaps, other types of AML. BISINDs may increase the therapeutic index of ATRA by reducing the dosage of the retinoid administered. In more general terms, given the low level of toxicity, ST1346 may find application in several settings where the chronic use of ATRA is called for, such as in chemoprevention of neoplasia.
We are thankful to Prof Silvio Garattini, Dr Mario Salmona, and Dr Anne Dejean (Institut Pasteur, Paris) for critical reading of the manuscript. We also thank Prof P. Chambon and Dr Cecile Rochette-Egly (Institut de Genetique et de Biologie Moleculaire et Cellulaire [IGBMC], Strasbourg, France) for the kind gift of anti-RAR antibodies. The help of Marina Sironi in the measurement of IL-1, TNF, and MCP-1 is also acknowledged. We thank Dr Michel Lanotte (Hopital St Louis, Paris, France), Dr Carlo Gambacorti-Passerini (Istituto Nazionale dei Tumori, Milano, Italy), and Dr Pier Giuseppe Pelicci (Istituto Oncologico Europeo, Milano, Italy) for the kind gift of the NB4, NB4.306, and PR9 cell lines.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2002-03-0720.
Supported by grants from the Associazione Italiana per la Ricerca contro il Cancro (AIRC) (E.G.), from the “Istituto Superiore di Sanità,” from the Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MURST), and from Sigma-Tau S.p.A. The financial support of the Weizmann-Pasteur-Negri Foundation is also acknowledged.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Enrico Garattini, Laboratory of Molecular Biology, Centro Catullo e Daniela Borgomainerio, Istituto di Ricerche Farmacologiche “Mario Negri,” via Eritrea, 62, 20157 Milano, Italy; e-mail address: egarattini@marionegri.it.

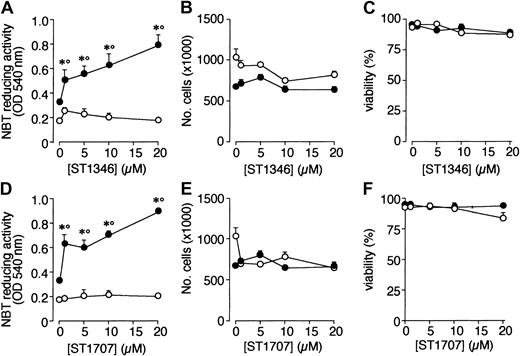
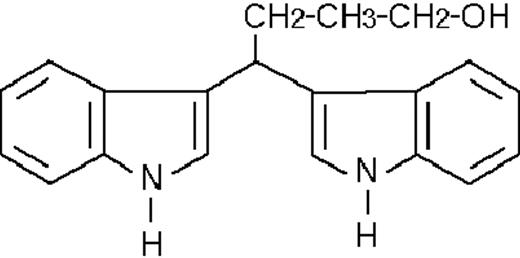
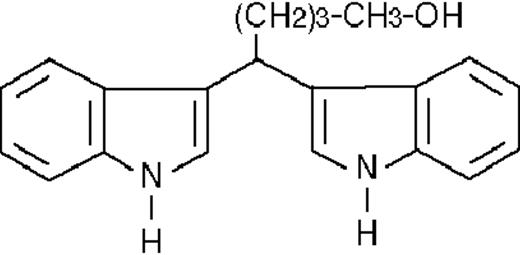
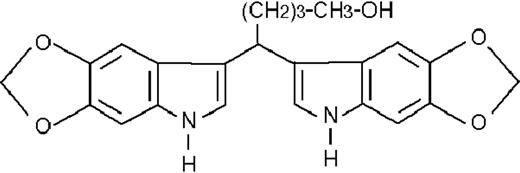
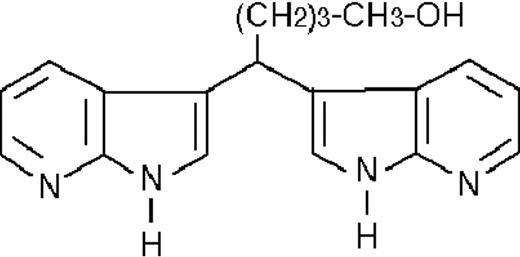
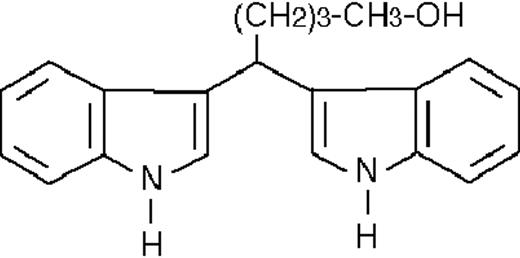
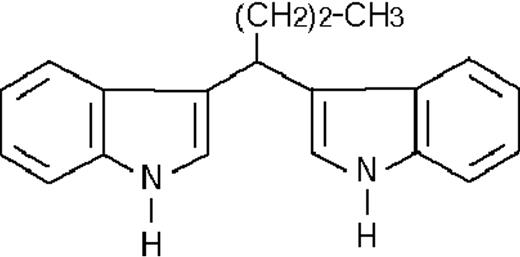
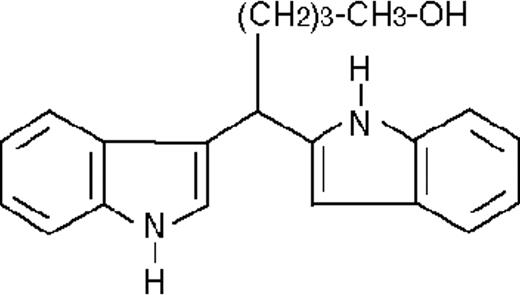
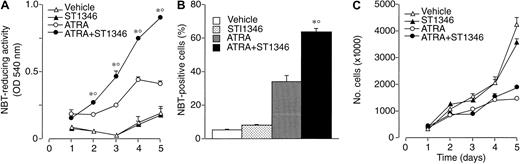
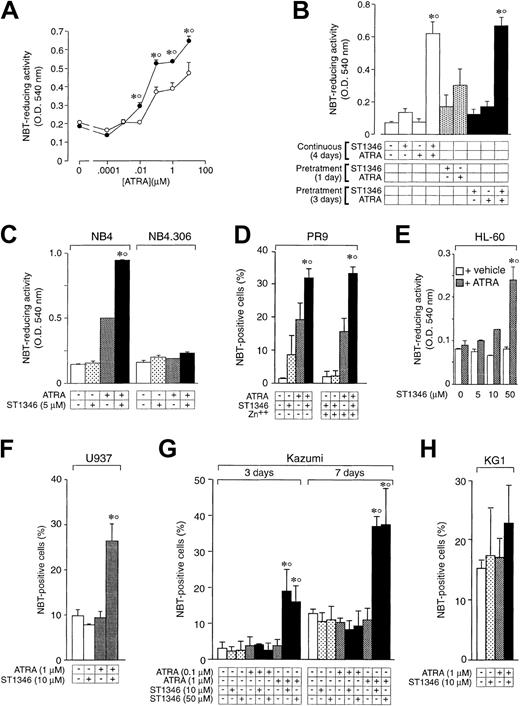
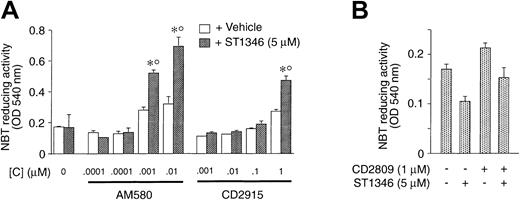
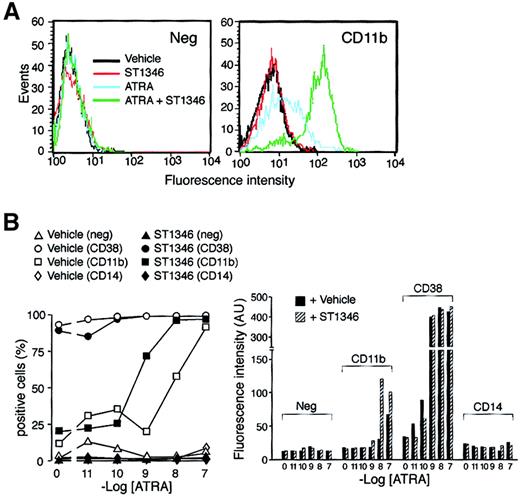
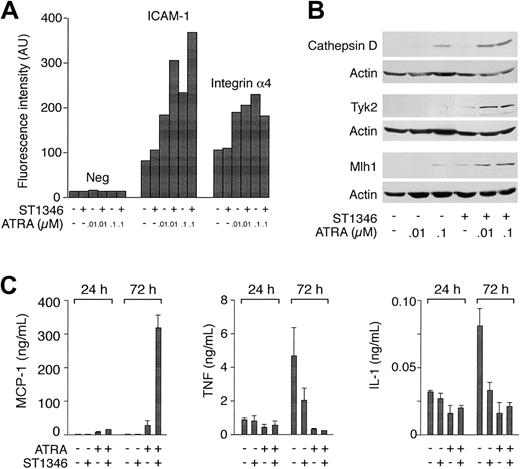
![Fig. 7. Influence of ST1346 and ATRA on the transcriptional activity of RARα in transfected COS-7 cells and on the levels of PML-RARα, RARα, and RXRα proteins as well as histone acetylation in NB4 cells. / (A) COS-7 cells were transfected with the plasmid pSG5 (void vector) and the same plasmid containing RARα or RXRα as indicated. The transfection mixture also contained the retinoid-responsive reporter construct TRE-TK-Luc (containing the luciferase reporter gene) and the β-galactosidase expression vector pCH110. Twenty-four hours following transfection, cells were treated with vehicle (dimethyl sulfoxide [DMSO]), ST1346 (5 μM), ATRA (0.1 μM), or the combination of the 2 compounds for a further 24 hours. Cell extracts were used for the determination of luciferase enzymatic activity. The results are always normalized for the transfection efficiency as assessed by determination of the β-galactosidase enzymatic activity. Results are expressed as the means ± SD of 3 separate dishes and are representative of at least 3 experiments. (B) NB4 cells were treated for 72 hours with vehicle, ATRA (0.1 μM), and ST1346 at the indicated concentrations, or combinations of ATRA + ST1346. Aliquots of cell extracts were subjected to Western blot analysis with anti-RARα, anti-RXRα, and anti–β-actin antibodies. The positions of relevant molecular weight markers are indicated on the right. (●) indicates the undegraded form; * represents a degradation band of PML-RARα. (C) NB4 cells were treated for 72 hours with vehicle, ST1346 (5 μM), sodium butyrate (1 mM), and ATRA at the indicated concentrations or combinations of ATRA + ST1346 and ATRA + sodium butyrate. Aliquots of cell extracts were subjected to Western blot analysis with antibodies recognizing histone H3 or the acetylated form of histone H3 (Ac-H3). Results are representative of 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/10/10.1182_blood-2002-03-0720/4/m_h82223379007.jpeg?Expires=1769138929&Signature=o70P1MOrnE7J9sbkUWpe6NU75d5heN1bZKQiRiyK1-CG-23~oZAslK5kA0bXjMXWUX1GXhX2k2cs~2pHJZ47yMcnEEuxy~YuItjW3fJZPBjraPiPpbGilLYb4jZ7--igQyw~DhDOCIC4Sg4nOVpfn8fdHzLUx5mUbLcierIHffIYoqXZGYgY0FKynjDoZr8icaDZ7ifM5w-armK3bb168elo0p1K5ztmqYmufFCSbkp-ZluPTcQcxNsGoEvQAn28xUCY0To7xHFD7oTlSyUNnr6dije6b28d4RHYmqAGnY0SGUnsp7wgMJV0Koz~W1nb8Cnb-B1oN6PnyZDJPxIlRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
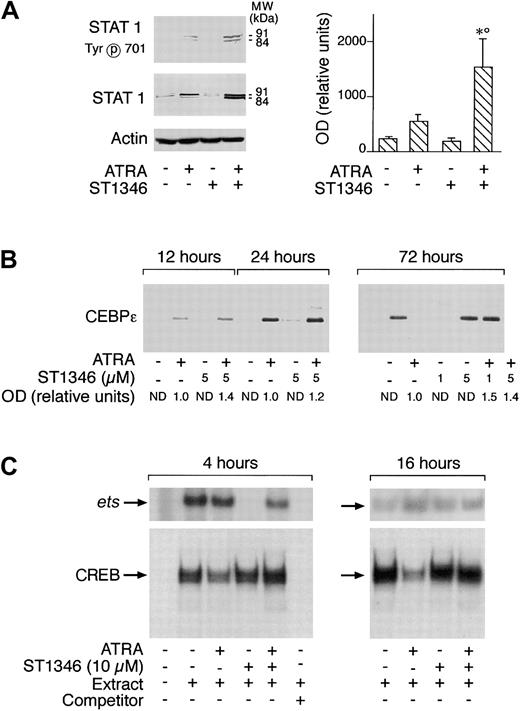
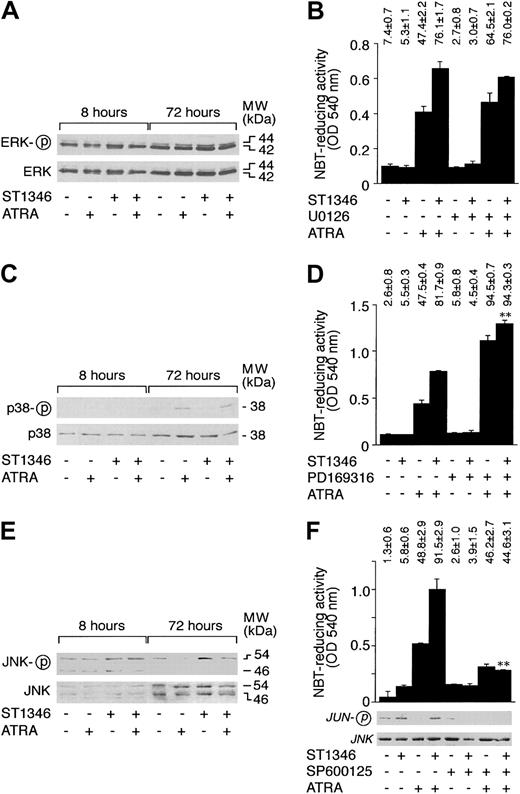
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal