To define the phosphatidylinositol glycan-class A (PIG-A) gene abnormality in precursor cells and the changes of expression of glycosylphosphatidylinositol-anchored protein and contribution of paroxysmal nocturnal hemoglobinuria (PNH) clones with PIG-A gene abnormalities among various cell lineages during differentiation and maturation, we investigated CD59 expression on bone marrow CD34+ cells and peripheral granulocytes from 3 patients with PNH and the PIG-A gene abnormalities in the CD59−, CD59+/−, and CD59+ populations by nucleotide sequence analyses. We also performed clonogeneic assays of CD34+CD59+ and CD34+CD59− cells from 2 of the patients and examined the PIG-A gene abnormalities in the cultured cells. In case 1, the CD34+ cells and granulocytes consisted of CD59− and CD59+ populations and CD59−, CD59+/−, and CD59+populations, respectively. Sequence analyses indicated that mutation 1-2 was in the CD59+/− granulocyte population (20 of 20) and the CD34+CD59− population (2 of 38). In cases 2 and 3, the CD34+ cells and granulocytes consisted of CD59+ and CD59− cells. Sequence analyses in case 3 showed that mutation 3-2 was not in CD34+CD59− cells and was present in the CD59− granulocyte population. However, PIG-A gene analysis of cultured CD34+CD59− cells showed that they had the mutation. This analysis also revealed that there were some other mutations, which were not found in CD34+CD59− cells and CD59− or CD59+/− granulocytes in vivo, and that sometimes they were distributed specifically among different cell lineages. In conclusion, our findings suggest that PNH clones might contribute qualitatively and quantitatively differentially to specific blood cell lineages during differentiation and maturation of hematopoietic stem cells.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired clonal hematologic disorder that is manifested by complement-mediated hemolysis, venous thrombosis, and bone marrow failure.1,2 It is well known that PNH erythrocytes consist of complement-sensitive (PNH II and III) and complement-insensitive (PNH I) populations as judged by the complement lysis sensitivity (CLS) test.3 Affected cells in PNH lack glycosylphosphatidylinositol-anchored proteins (GPI-APs), such as decay-accelerating factor (DAF)4 and CD59,5 on their surface membranes due to phosphatidylinositol glycan- class A (PIG-A) gene abnormalities.6 The absence of these complement regulatory proteins on PNH erythrocytes causes their hypersensitivity to complement.7,8 The phenotype of PNH erythrocytes determined with the CLS test is related to the extent of the deficiencies of DAF and CD59 on their surfaces.8-10Thus, single-color or 2-color flow cytometric analysis of erythrocytes has been used both to diagnose PNH and to determine the phenotype of PNH cells.8-11 Subsequently, GPI-AP–deficient cells were detected in all hematopoietic lineages (granulocytes, monocytes, T and B lymphocytes, natural killer cells, platelets, and erythrocytes) and CD34+ cells in patients with PNH.12-15 In addition, it has been reported that there is a decrease of progenitors and hypersensitivity of precursors to complement, and a decrease of CD34+CD59+ and CD34+CD59− cells due to their proliferative defects in PNH with or without hemolysis.16-22 These findings suggest that the abnormality in PNH cells occurs at the level of the pluripotent hematopoietic stem cell and that PNH is one of the bone marrow failure syndromes, which include aplastic anemia and myelodysplastic syndrome.2
It is unclear how a PNH clone with a PIG-A gene mutation expands in bone marrow, although some hypotheses, especially negative selection of PNH clones by cytotoxic T lymphocytes, have been proposed.23 All patients with PNH reported in the past have been shown to have somatic mutations of the PIG-A gene,24 which were examined mainly in granulocytes. However, abnormality of the PIG-A gene in precursor cells from PNH patients has not been shown directly, although the expression of GPI-APs in precursors was reported.14,15 Earlier studies on the PIG-A gene indicated that the blood cells from most PNH patients have only one PIG-A mutation, suggesting that PNH is monoclonal.25,26 On the other hand, recent studies indicated that the blood cells from some PNH patients bear 2 or more independent PIG-A mutants,27-31 suggesting that PNH is oligoclonal and that the phenotype of PNH cells is also determined by the genotype of the PIG-A gene.28,32,33 Recently, we reported that the distribution of PIG-A gene abnormalities changes independently among various cell lineages during differentiation and maturation or during selection, and that PIG-A gene abnormalities determine the phenotype of mature granulocytes, but not of erythroblasts.34 This may explain part of the heterogeneous expression of GPI-APs among the various cell lineages from peripheral blood of PNH patients. However, it is unclear whether a specific mutation of a PIG-A gene contributes to a particular cell lineage or whether it is present in a specific stage of maturation, that is, CD34+ cells capable of self-renewal.
In the present study, to investigate the PIG-A gene abnormality at the level of precursor cells and the changes in expression of GPI-AP and contribution of PNH clones with PIG-A gene abnormalities among various cell lineages during differentiation and maturation of precursor cells to mature cells in PNH patients, we examined CD59 expression and PIG-A gene mutations in bone marrow CD34+ cells and peripheral blood granulocytes from 3 patients with PNH, and compared the PIG-A gene abnormalities in each population of CD34+ cells with those of peripheral blood granulocytes, which were sorted into CD59−, CD59+, and CD59+/−populations. Also, we performed clonogeneic assays of CD34+CD59+ and CD34+CD59− cells using semisolid cultures in vitro and examined PIG-A gene mutations in the cultured cells derived from each CD34+ cell population from 2 of the 3 patients with PNH.
Patients, materials, and methods
Patients and controls
After informed consent was obtained, peripheral blood or bone marrow samples were taken from 3 male patients with PNH and 1 or 2 healthy individuals for assessing CD59 expression and PIG-A gene analyses in CD34+ cells and granulocytes in vivo or for PIG-A gene analyses in cultured cells derived from CD34+cells, respectively. A diagnosis of PNH was made on the basis of the history, clinical and laboratory findings, and a positive Ham test. At the time of the study, all patients were clinically stable with no recent severe hemolytic attacks, and they had no packed red blood cell transfusions for 12 months prior to our study. The clinical and hematologic data from the 3 patients are summarized in Table1. The 2-color analyses of DAF and CD59 expression on erythrocytes from all the patients were performed as described previously.8 9 The erythrocytes in case 1 consisted of positive and negative populations and those in cases 2 and 3 of positive, intermediate, and negative populations. The proportion of each population was 24.8% positive and 75.2% negative in case 1; 26.2% positive, 26.0% intermediate, and 47.8% negative in case 2; and 39.9% positive, 24.5% intermediate, and 35.6% negative in case 3.
Clinical and laboratory data and CD59 expression by peripheral blood granulocytes and bone marrow CD34+cells from 3 PNH patients and a healthy volunteer
| Case no. . | Age/sex . | Hb, g/dL . | Reticulocytes, % . | WBC, × 109/L . | Granulocyte count, × 109/L . | BM cellularity, × 109/L . | Granulocytes* . | CD34+cells† . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg, %‡ . | Int, %‡ . | Pos, %‡ . | Neg, %‡ . | Int, %‡ . | Pos, %‡ . | |||||||
| 1 | 69/M | 9.6 | 2.6 | 4.4 | 2.64 | 50 | 82.1 | 11.8 | 6.1 | 92.8 | ND | 7.2 |
| 2 | 56/M | 11.9 | 2.3 | 6.9 | 6.14 | 160 | 73.3 | ND | 26.7 | 82.6 | ND | 17.4 |
| 3 | 51/M | 6.1 | 3.2 | 3.7 | 1.62 | 80 | 36.1 | ND | 63.9 | 75.1 | ND | 24.9 |
| HV | 45/M | 14.7 | 1.4 | 5.5 | 2.56 | 100 | ND | ND | 98.7 | ND | ND | 98.7 |
| Case no. . | Age/sex . | Hb, g/dL . | Reticulocytes, % . | WBC, × 109/L . | Granulocyte count, × 109/L . | BM cellularity, × 109/L . | Granulocytes* . | CD34+cells† . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg, %‡ . | Int, %‡ . | Pos, %‡ . | Neg, %‡ . | Int, %‡ . | Pos, %‡ . | |||||||
| 1 | 69/M | 9.6 | 2.6 | 4.4 | 2.64 | 50 | 82.1 | 11.8 | 6.1 | 92.8 | ND | 7.2 |
| 2 | 56/M | 11.9 | 2.3 | 6.9 | 6.14 | 160 | 73.3 | ND | 26.7 | 82.6 | ND | 17.4 |
| 3 | 51/M | 6.1 | 3.2 | 3.7 | 1.62 | 80 | 36.1 | ND | 63.9 | 75.1 | ND | 24.9 |
| HV | 45/M | 14.7 | 1.4 | 5.5 | 2.56 | 100 | ND | ND | 98.7 | ND | ND | 98.7 |
Hb indicates concentration of hemoglobin; WBC, white blood cell count; BM, bone marrow; HV, healthy volunteer; and ND, not detectable.
The granulocytes were examined by flow cytometric single-color analysis using a monoclonal antibody to CD59 as described in “Patients, materials, and methods.” The results are from the profiles shown in Figure 3.
Bone marrow MNCs were stained with monoclonal antibodies to CD59, CD34, and CD45 as described in “Patients, materials, and methods,” following separation of CD34+ cells by flow cytometry as shown in Figure 1. The results are from the profiles shown in Figure 3.
Neg, Int, and Pos refer to negative, intermediate, and positive populations by single-color flow cytometric analysis of peripheral granulocytes and by 3-color analysis of bone marrow MNCs, respectively, as defined in “Patients, materials, and methods.”
Cell preparation
Bone marrow and peripheral blood samples were obtained by posterior iliac crest aspiration and by venipuncture, respectively. Peripheral blood granulocytes and bone marrow mononuclear cells (MNCs) were separated as described previously.34,35 The cells were suspended at 2 × 107 cells/mL in phosphate-buffered saline containing 1% bovine serum albumin (PBS-BSA) and used for immunofluorescence staining. The purity of the separated granulocytes and MNCs was over 95% as judged by morphologic examination after May-Giemsa staining. The viability of granulocytes and MNCs, evaluated by the trypan blue dye exclusion method,21 was over 93%.
Monoclonal antibodies
A mouse monoclonal antibody to CD59 (3E1, IgG1)5, a mouse monoclonal antibody to CD34 labeled with fluorescein isothiocyanate (HPCA-2, IgG1; Becton Dickinson, San Jose, CA), and a mouse monoclonal antibody to CD45 labeled with peridinin chlorophyll protein (HLe-1, IgG1; Becton Dickinson) were used. A goat antimouse immunoglobulin F(ab′)2 fragment labeled with R-phycoerythrin (Dako, Glostrup, Denmark) was used as the second antibody for staining CD59. Irrelevant monoclonal antibodies of the same subclass were used as negative controls.34 35
Immunofluorescent staining of bone marrow MNCs and peripheral blood granulocytes, flow cytometry, and cell sorting
Immunofluorescent staining of bone marrow MNCs with monoclonal antibodies to CD34, CD45, and CD59 was performed according to the method described by Sutherland et al36 and the manufacturer's recommendation with some modification.34,35 CD34+ cells in bone marrow MNCs were isolated with a FACSVantage (Becton Dickinson) by the gating method that uses both light-scattering parameters and CD34/CD45 fluorescence, as described by Sutherland et al.36 Then, CD59 expression was analyzed with CD34+ cells from the 3 PNH patients and 1 healthy volunteer. The cells were sorted into CD34+CD59− and CD34+CD59+ fractions using a FACSVantage equipped with a 488-nm laser (Figure 1). More than 5 × 103 or 1.2 × 104 cells in each fraction were sorted into 100 μL PBS-BSA in Eppendorf vials and used for genetic analysis or hematopoietic cell culture, respectively.
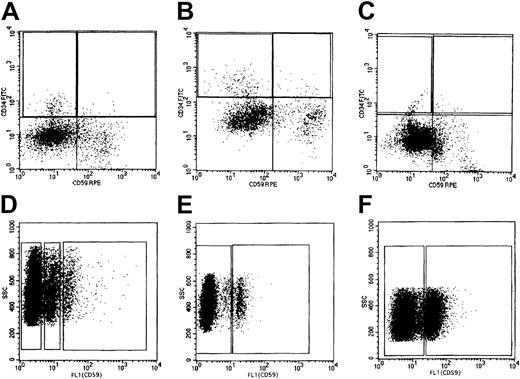
Isolation with a fluorescence-activated cell sorter of each population expressing CD59 among CD34+ cells and granulocytes.
Bone marrow MNCs were stained with monoclonal antibodies to CD59, CD34, and CD45, divided into CD34+CD59− and CD34+CD59+ populations according to the gates shown by rectangles in panels A to C (A, case 1; B, case 2; C, case 3), and sorted. Vertical and horizontal axes are the fluorescence intensities of CD34 and CD59, respectively. Peripheral blood granulocytes were stained with a monoclonal antibody to CD59, divided into CD59− CD59+/−, and CD59+populations according to the gates shown by rectangles in panels D to F (D, case 1; E, case 2; F, case 3), and sorted. Vertical and horizontal axes are the side scatter and fluorescence intensity of CD59, respectively.
Isolation with a fluorescence-activated cell sorter of each population expressing CD59 among CD34+ cells and granulocytes.
Bone marrow MNCs were stained with monoclonal antibodies to CD59, CD34, and CD45, divided into CD34+CD59− and CD34+CD59+ populations according to the gates shown by rectangles in panels A to C (A, case 1; B, case 2; C, case 3), and sorted. Vertical and horizontal axes are the fluorescence intensities of CD34 and CD59, respectively. Peripheral blood granulocytes were stained with a monoclonal antibody to CD59, divided into CD59− CD59+/−, and CD59+populations according to the gates shown by rectangles in panels D to F (D, case 1; E, case 2; F, case 3), and sorted. Vertical and horizontal axes are the side scatter and fluorescence intensity of CD59, respectively.
Peripheral blood granulocytes were stained with a monoclonal antibody to CD59 as described previously.34 The stained granulocytes were gated by forward light scattering and side light scattering to omit degraded granulocytes, analyzed for CD59 expression, and then sorted into CD59−, CD59+, or CD59+/− fractions using a FACSVantage (Figure 1). The CD59− populations of granulocytes and CD34+cells were defined as cells with a weak fluorescence intensity, similar to that of negative control cells. The CD59+ population was defined as cells with a strong fluorescence intensity, similar to that of positive normal control cells. The CD59+/− population was defined as cells with fluorescence intensity between those of CD59− and CD59+ populations. The percentage of each population expressing CD59 was calculated as follows: 100 × (number of cells in each fraction)/(total number of gated and examined granulocytes or CD34+ cells). The total numbers of granulocytes and CD34+ cells gated and examined for CD59 expression were 16 003 and 16 403 from case 1, 7593 and 4580 from case 2, and 12 635 and 20 814 from case 3, respectively.
Hematopoietic cell culture
CD34+CD59+ and CD34+CD59− cells from 2 PNH patients (cases 2 and 3) and 2 healthy volunteers were sorted as described above and cultured (3 × 103 cells/mL) in methylcellulose according to the method described by Maciejewski et al22 with a modification. Colonies of early and late erythroid precursors (erythroid burst-forming units [BFU-Es] and erythroid colony-forming units [CFU-Es,], respectively) and granulocyte-macrophage precursors (granulocyte-macrophage colony-forming units [CFU-GMs]) in 2 or 3 dishes each were counted and picked at random using an inverted microscope after 7 and 14 days of culture and washed in RPMI 1640 and PBS 3 times. More than 5 × 103 cells were used for PIG-A gene analysis. Before genetic analysis, parts of the cell suspensions were fixed with a cytocentrifuge (Shandon Elliott, Runcorn Cheshire, United Kingdom) and stained with Giemsa solution. All samples contained more than 95% erythroblasts or granulocytes and monocytes consisting of more than 90% granulocytes.
gDNA preparation
Genomic DNA (gDNA) was prepared with Instagene (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's recommendation from each sorted CD59−, CD59+, or CD59+/− cell fraction of bone marrow CD34+cells and peripheral blood granulocytes and from cultured CD34+ cells.
PCR, cloning, sequence analysis of the PIG-A gene, and direct sequencing of PCR-amplified PIG-A DNA
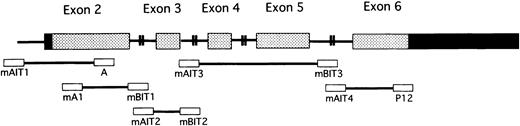
The oligonucleotide primers for polymerase chain reaction (PCR), shown in Table 2 and Figure2, were designed according to the PIG-A cDNA and gDNA sequences.6,37,38 With PIG-A gDNA obtained from each sorted cell fraction or cultured cells, the coding region of the exon and the intron region flanking the exon were amplified by PCR and sequenced. PCR of genomic PIG-A DNA and cloning were done with the methods34 described previously with some modification. At least 20 genomic PIG-A DNA bacterial clones were sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Division of Perkin Elmer, Foster City, CA) and Model 310 Genetic Analyzer (Applied Biosystems). To exclude PCR errors, only PIG-A gene mutations identified in at least 2 independent bacterial clones with 2 or more independent PCR reactions are listed. The frequencies of mutant PIG-A gene clones detected only once were 102 (14.6%) of 700 clones from granulocytes and 80 (12.9%) of 621 clones from CD34+ cells. Also, we sequenced PCR-amplified DNA39 40 from CD34+CD59− cells and CD59− and CD59+/− granulocytes from case 1, CD34+CD59− cells and CD59−granulocytes from case 2, and CD34+CD59− cells and CD59− granulocytes from case 3. CD59+granulocytes from the PNH patients and a healthy volunteer were used as controls.
Oligonucleotide primers used for PCR of the PIG-A gene
| Amplified region . | Sense primer . | Antisense primer . | ||
|---|---|---|---|---|
| Name . | Primer sequence (5′-3′) . | Name . | Primer sequence (5′-3′) . | |
| Exon 2 | mAIT1* | GAGCTGAATTCTTGTTTTAC | A* | GTATCACAAAGAGACACGG |
| Exon 2 | mA1* | CATTGCTCAGGTACCTATTTGTT | mBIT1* | AAAGTCTAGAATGCAATTATAGC |
| Exon 3 | mAIT2* | AAGTGAATTCTCAGTCGTTCTGGT | mBIT2* | CTTCTCCCTCGAGACAACATGAA |
| Exon 4, 5 | mAIT3* | ATGAAGCTTCACTCCTTTCTTCCCCTC | mBIT3* | GCACTCGAGTACGTGAAACATCAAGTA |
| Exon 6 | mAIT4* | GGTCATTGTTTCTCGAGGGACAG | P12* | ACCAGGTACCTCTTACAATCTAGGCTTCCTTC |
| Amplified region . | Sense primer . | Antisense primer . | ||
|---|---|---|---|---|
| Name . | Primer sequence (5′-3′) . | Name . | Primer sequence (5′-3′) . | |
| Exon 2 | mAIT1* | GAGCTGAATTCTTGTTTTAC | A* | GTATCACAAAGAGACACGG |
| Exon 2 | mA1* | CATTGCTCAGGTACCTATTTGTT | mBIT1* | AAAGTCTAGAATGCAATTATAGC |
| Exon 3 | mAIT2* | AAGTGAATTCTCAGTCGTTCTGGT | mBIT2* | CTTCTCCCTCGAGACAACATGAA |
| Exon 4, 5 | mAIT3* | ATGAAGCTTCACTCCTTTCTTCCCCTC | mBIT3* | GCACTCGAGTACGTGAAACATCAAGTA |
| Exon 6 | mAIT4* | GGTCATTGTTTCTCGAGGGACAG | P12* | ACCAGGTACCTCTTACAATCTAGGCTTCCTTC |
The primer sets amplify part or all of each exon and part of the intron flanking the exon.
Schematic representation of the PIG-A gene and PCR primers.
Gray and black rectangles represent coding regions and noncoding regions, respectively, in 5 exons of the PIG-A gene. Lines connecting exons represent introns. Primers (open rectangles) used for PCR are at the corresponding areas in the gene. The base sequences of these primers are shown in Table 2.
Schematic representation of the PIG-A gene and PCR primers.
Gray and black rectangles represent coding regions and noncoding regions, respectively, in 5 exons of the PIG-A gene. Lines connecting exons represent introns. Primers (open rectangles) used for PCR are at the corresponding areas in the gene. The base sequences of these primers are shown in Table 2.
Statistical analysis
All statistical analyses were performed with the Fisher exact probability test. P < .05 was considered significant.
Results
CD59 expression on peripheral blood granulocytes and bone marrow CD34+ cells
The granulocytes from case 1 consisted of CD59−, CD59+/−, and CD59+ populations and those in cases 2 and 3 of CD59− and CD59+ populations (Figure 3B-D). The granulocytes from a healthy volunteer consisted of a single positive population (Figure3A). The proportions of each population in the PNH patients and a healthy volunteer are shown in Table 1.
Flow cytometric profiles of CD59 expression on peripheral blood granulocytes and bone marrow CD34+ cells from 3 PNH patients and a healthy volunteer.
In each histogram, thick and thin lines in panels A-D or solid and dashed lines in panels E-H show reacted samples and negative controls, respectively. Horizontal and vertical axes are fluorescence intensity of CD59 (FL1) and cell counts, respectively. (A) Granulocytes from a healthy volunteer. (B) Granulocytes from case 1. (C) Granulocytes from case 2. (D) Granulocytes from case 3. (E) CD34+ cells from a healthy volunteer. (F) CD34+cells from case 1. (G) CD34+ cells from case 2. (H) CD34+ cells from case 3.
Flow cytometric profiles of CD59 expression on peripheral blood granulocytes and bone marrow CD34+ cells from 3 PNH patients and a healthy volunteer.
In each histogram, thick and thin lines in panels A-D or solid and dashed lines in panels E-H show reacted samples and negative controls, respectively. Horizontal and vertical axes are fluorescence intensity of CD59 (FL1) and cell counts, respectively. (A) Granulocytes from a healthy volunteer. (B) Granulocytes from case 1. (C) Granulocytes from case 2. (D) Granulocytes from case 3. (E) CD34+ cells from a healthy volunteer. (F) CD34+cells from case 1. (G) CD34+ cells from case 2. (H) CD34+ cells from case 3.
Nucleotide sequence analysis of the PIG-A gene and direct sequencing of PCR-amplified PIG-A DNA from sorted peripheral blood granulocytes and bone marrow CD34+ cells
The results of nucleotide sequence analyses of the PIG-A gene from each sorted cell fraction of CD34+ cells and granulocytes are shown in Table 3. Differences between the frequencies of the abnormal PIG-A gene in each population sorted from CD34+ cells and from granulocytes of 3 PNH patients were analyzed statistically. There were significant differences between the frequencies of clones with mutation 1-2 from CD34+CD59− cells and CD59+/−granulocytes and mutation 3-2 from CD34+CD59−cells and CD59−granulocytes (P < .0001 andP < .001, respectively). Subsequently, mutations 1-1 in CD34+CD59− cells (P < .0001) and CD59− granulocytes (P < .0001), 1-2 in CD59+/− granulocytes (P < .0001), 2-1 in CD34+CD59− cells (P < .0001) and CD59− granulocytes (P < .0001), 3-1 in CD34+CD59− cells (P < .05) and CD59−granulocytes (P < .05), and 3-2 in CD59− granulocytes (P < .01) were predominant compared with the other mutations in each cell population sorted from each patient. However, there were no significant differences in the frequencies of mutations 3-1 and 3-2 in CD59− granulocytes from case 3.
Mutations of the PIG-A gene in 3 PNH patients
| No. . | . | Abnormality of protein . | Frequency of mutations3-151 . | |||||
|---|---|---|---|---|---|---|---|---|
| Mutation of PIG-A gene . | BM CD34+ cells . | PB granulocytes . | ||||||
| Position3-150 . | Type . | CD59− . | CD59+ . | CD59− . | CD59+/− . | CD59+ . | ||
| 1-1 | 5′ splicing site | Deletion, GT > G | Exon 5 deletion (415)3-152 | 20:21 | 0:20 | 20:20 | 0:20 | 0:20 |
| 1-2 | 214 | Substitution, CAT > TAT | Missense (His→Tyr)3-152 | 2:38 | 0:20 | 0:20 | 20:20 | 0:20 |
| 2-1 | 548 | Substitution, TGT > TAT | Missense (Cys→Tyr)3-152 | 18:20 | 0:20 | 20:20 | — | 0:20 |
| 2-23-153 | 982 | Substitution, GTT > TTT | Missense (Val→Phe)3-152 | 0:20 | 0:20 | 0:20 | — | 0:20 |
| 2-33-153 | 1392 | Deletion, TGG > TG | Frameshift | 0:20 | 0:20 | 0:20 | — | 0:20 |
| 3-1 | 108 | Substitution, TGC > TGA | Nonsense (Cys→Stop)3-152 | 5:22 | 0:20 | 6:20 | — | 0:20 |
| 3-2 | 491 | Substitution, TCG > TTG | Missense (Ser→Leu)3-152 | 0:20 | 0:20 | 9:20 | — | 0:20 |
| 3-33-153 | 844 | Substitution, GAC > AAC | Missense (Asp→Asn)3-152 | 0:20 | 0:20 | 0:20 | — | 0:20 |
| 3-43-153 | 5′ splicing site | Deletion, GT > G | Exon 5 deletion (415)3-152 | 0:20 | 0:20 | 0:20 | — | 0:20 |
| No. . | . | Abnormality of protein . | Frequency of mutations3-151 . | |||||
|---|---|---|---|---|---|---|---|---|
| Mutation of PIG-A gene . | BM CD34+ cells . | PB granulocytes . | ||||||
| Position3-150 . | Type . | CD59− . | CD59+ . | CD59− . | CD59+/− . | CD59+ . | ||
| 1-1 | 5′ splicing site | Deletion, GT > G | Exon 5 deletion (415)3-152 | 20:21 | 0:20 | 20:20 | 0:20 | 0:20 |
| 1-2 | 214 | Substitution, CAT > TAT | Missense (His→Tyr)3-152 | 2:38 | 0:20 | 0:20 | 20:20 | 0:20 |
| 2-1 | 548 | Substitution, TGT > TAT | Missense (Cys→Tyr)3-152 | 18:20 | 0:20 | 20:20 | — | 0:20 |
| 2-23-153 | 982 | Substitution, GTT > TTT | Missense (Val→Phe)3-152 | 0:20 | 0:20 | 0:20 | — | 0:20 |
| 2-33-153 | 1392 | Deletion, TGG > TG | Frameshift | 0:20 | 0:20 | 0:20 | — | 0:20 |
| 3-1 | 108 | Substitution, TGC > TGA | Nonsense (Cys→Stop)3-152 | 5:22 | 0:20 | 6:20 | — | 0:20 |
| 3-2 | 491 | Substitution, TCG > TTG | Missense (Ser→Leu)3-152 | 0:20 | 0:20 | 9:20 | — | 0:20 |
| 3-33-153 | 844 | Substitution, GAC > AAC | Missense (Asp→Asn)3-152 | 0:20 | 0:20 | 0:20 | — | 0:20 |
| 3-43-153 | 5′ splicing site | Deletion, GT > G | Exon 5 deletion (415)3-152 | 0:20 | 0:20 | 0:20 | — | 0:20 |
BM indicates bone marrow; PB, peripheral blood; His, histidine; Tyr, tyrosine; Cys, cysteine; Val, Valine; Phe, phenylalanine; Ser, serine; Leu, leucine; Asp, aspartic acid; Asn, asparagines; and Stop, stop codon.
The nucleotides are numbered from the first nucleotide of the coding region of the PIG-A gene.
The numbers indicate the ratio of mutated clones to the total examined in each cell population.
The size of the resultant protein (amino acid residues) is indicated in parenthesis. The amino acids without and with the mutation are also indicated in parentheses when the abnormality of the PIG-A gene was a missense or nonsense mutation.
Mutations 2-2, 2-3, 3-3, and 3-4 were found only in semisolid cultures of CD34+ cells.
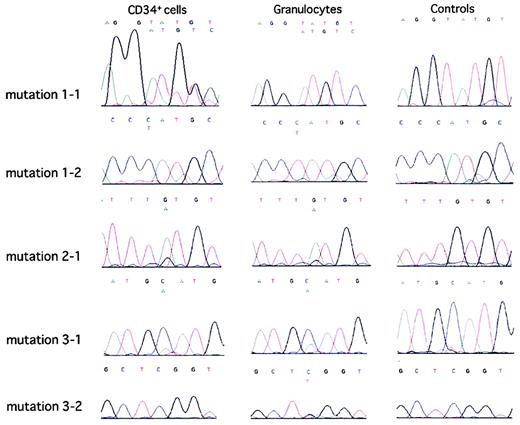
Direct sequence analyses of PCR-amplified DNA showed that mutations 1-1, 1-2, 2-1, 3-1, and 3-2 were clonal genomic abnormalities in CD34+CD59− cells and CD59− and CD59+/− granulocytes from case 1, CD34+CD59− cells and CD59−granulocytes from case 2, CD34+CD59− cells and CD59− granulocytes from case 3, and CD59−granulocytes, but not CD34+CD59− cells, from case 3 (Figure 4). In contrast, no PIG-A mutations were found in CD59+ granulocytes from the PNH patients and a healthy volunteer (Figure 4).
Direct sequence analyses of PCR-amplified PIG-A DNA in CD59-deficient CD34+ cells and granulocytes and in CD59+granulocytes from PNH patients.
Direct sequence analyses of PCR-amplified PIG-A DNA from bone marrow CD34+ cells or peripheral blood granulocytes from 3 PNH patients and a healthy volunteer were performed as described in “Patients, materials, and methods.” The results, with controls for each mutation shown in the right column, were obtained by analyses of PIG-A DNA in CD59+ populations of peripheral blood granulocytes from the PNH patients. Normal and mutated alleles are presented above each graph in the upper and lower rows, respectively.
Direct sequence analyses of PCR-amplified PIG-A DNA in CD59-deficient CD34+ cells and granulocytes and in CD59+granulocytes from PNH patients.
Direct sequence analyses of PCR-amplified PIG-A DNA from bone marrow CD34+ cells or peripheral blood granulocytes from 3 PNH patients and a healthy volunteer were performed as described in “Patients, materials, and methods.” The results, with controls for each mutation shown in the right column, were obtained by analyses of PIG-A DNA in CD59+ populations of peripheral blood granulocytes from the PNH patients. Normal and mutated alleles are presented above each graph in the upper and lower rows, respectively.
PIG-A gene mutations in cultured CD34+ cells from PNH patients
The numbers of CFU-E, BFU-E, and CFU-GM colonies from CD34+CD59+ or CD34+CD59− cells from 2 PNH patients (cases 2 and 3) were less or almost the same as those from CD34+CD59+ cells from 2 healthy individuals (Table 4). The numbers of colonies of each precursor cell from CD34+CD59+ cells from the PNH patients were less than those from CD34+CD59− cells (Table 4).
PIG-A mutations in cultured CD34+CD59+ and CD34+CD59− cells from 2 PNH patients and healthy volunteers
| Case no. . | No. of colonies4-150 . | Frequency of mutations4-151 . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD34+CD59+cells . | CD34+CD59−cells . | CD34+CD59+cells . | CD34+CD59− cells . | |||||||||||||
| CFU-E BFU-E . | CFU-GM . | CFU-E BFU-E . | CFU-GM . | CFU-E BFU-E . | CFU-GM . | CFU-E BFU-E . | CFU-GM . | |||||||||
| Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | |
| PNH patients | ||||||||||||||||
| 2 | 28 | 45 | 32 | 50 | 102 | 105 | 76 | 91 | ||||||||
| Mutation 2-1‡ | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 18:20 | 14:20 | 17:20 | 12:20 |
| Mutation 2-2 | — | — | — | — | — | — | — | — | 1:20 | 1:20 | 0:20 | 0:20 | 2:20 | 3:20 | 0:20 | 0:20 |
| Mutation 2-3 | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 4:20 |
| 3 | 24 | 37 | 59 | 34 | 131 | 116 | 80 | 116 | ||||||||
| Mutation 3-14-153 | — | — | — | — | — | — | — | — | 1:20 | 0:20 | 0:20 | 1:20 | 18:20 | 15:20 | 15:20 | 12:20 |
| Mutation 3-2 | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 4:20 | 0:20 |
| Mutation 3-34-155 | — | — | — | — | — | — | — | — | 0:20 | 1:20 | 0:20 | 0:20 | 0:20 | 3:20 | 0:20 | 0:20 |
| Mutation 3-4 | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 4:20 | 0:20 |
| Healthy volunteers | ||||||||||||||||
| 1 | 92 | 98 | 95 | 120 | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 |
| 2 | 132 | 140 | 72 | 144 | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 |
| Case no. . | No. of colonies4-150 . | Frequency of mutations4-151 . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD34+CD59+cells . | CD34+CD59−cells . | CD34+CD59+cells . | CD34+CD59− cells . | |||||||||||||
| CFU-E BFU-E . | CFU-GM . | CFU-E BFU-E . | CFU-GM . | CFU-E BFU-E . | CFU-GM . | CFU-E BFU-E . | CFU-GM . | |||||||||
| Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | Day 7 . | Day 14 . | |
| PNH patients | ||||||||||||||||
| 2 | 28 | 45 | 32 | 50 | 102 | 105 | 76 | 91 | ||||||||
| Mutation 2-1‡ | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 18:20 | 14:20 | 17:20 | 12:20 |
| Mutation 2-2 | — | — | — | — | — | — | — | — | 1:20 | 1:20 | 0:20 | 0:20 | 2:20 | 3:20 | 0:20 | 0:20 |
| Mutation 2-3 | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 4:20 |
| 3 | 24 | 37 | 59 | 34 | 131 | 116 | 80 | 116 | ||||||||
| Mutation 3-14-153 | — | — | — | — | — | — | — | — | 1:20 | 0:20 | 0:20 | 1:20 | 18:20 | 15:20 | 15:20 | 12:20 |
| Mutation 3-2 | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 4:20 | 0:20 |
| Mutation 3-34-155 | — | — | — | — | — | — | — | — | 0:20 | 1:20 | 0:20 | 0:20 | 0:20 | 3:20 | 0:20 | 0:20 |
| Mutation 3-4 | — | — | — | — | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 4:20 | 0:20 |
| Healthy volunteers | ||||||||||||||||
| 1 | 92 | 98 | 95 | 120 | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 |
| 2 | 132 | 140 | 72 | 144 | — | — | — | — | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 | 0:20 |
CFU-E, BFU-E, and CFU-GM colonies were cultured for 7 or 14 days as described in “Patients, materials, and methods.” The numbers of colonies were counted using an inverted microscope in 2 or 3 dishes for each cultured cell population. The mean values are presented.
The numbers indicate the ratio of mutated clones to the total examined in each cultured cell population.
Mutation 2-1 in CFU-E, BFU-E, 7-day CFU-GM, and 14-day CFU-GM colonies from CD34+CD59− cells was predominant compared with mutations 2-2 and 2-3 (P < .0001 versus mutations 2-2 and 2-3; P < .001 versus mutation 2-2 and P < .0001 versus mutation 2-3; P< .0001 versus mutations 2-2 and 2-3; P < .0001 versus mutation 2-2 and P < .05 versus mutation 2-3, respectively).
Mutation 3-1 in CFU-E, BFU-E, 7-day CFU-GM, and 14-day CFU-GM colonies from CD34+CD59− cells was predominant compared with the other mutations (P < .0001 versus mutations 3-2, 3-3 and 3-4; P < .0001 versus mutations 3-2 and 3-4 and P < .001 versus mutation 3-3;P < .002 versus mutations 3-2 and 3-4 andP < .0001 versus mutation 3-3; P < .0001 versus mutations 3-2, 3-3 and 3-4, respectively).
Mutation 3-3 in BFU-E colonies from CD34+CD59− cells was predominant compared with mutations 3-2 and 3-4 (P < .05).
As shown in Tables 3 and 4, we found 4 other mutations (mutations 2-2, 2-3, 3-3, and 3-4) in addition to those described above by nucleotide sequence analyses of the PIG-A gene in cells cultured from CD34+ cells from the PNH patients. In case 2, there were significant differences in the frequencies of only the 2-1 mutation in corresponding colonies from CD34+CD59+ and CD34+CD59− cells (eachP < .0001). In case 3, the frequencies of only the 3-1 mutation in CFU-E, BFU-E, 7-day CFU-GM, and 14-day CFU-GM colonies from CD34+CD59− cells were significantly higher than in those from CD34+CD59+ cells (P < .0001, P < .0001,P < .0001, P < .0005, respectively).
Discussion
In this study, we compared both CD59 expression and PIG-A gene mutations in bone marrow CD34+ cells with those in peripheral blood granulocytes from 3 patients with PNH. We found differences in the phenotypes of CD34+ cells and granulocytes in only one of the PNH patients (case 1). Previously, we reported differences in the phenotypes in erythroblasts and erythrocytes from PNH patients,35 suggesting that the phenotypes determined by GPI-AP expression can change during differentiation and maturation and the study by Rabessandratana et al41 supported this finding. In the present study, we found the PIG-A gene abnormality (mutation 1-2), which is a missense mutation believed to determine the occurrence of an intermediate population of PNH cells,29,32 in CD34+CD59−cells and CD59+/− granulocytes from case 1. This strongly suggests that the appearance of the CD59+/− population of granulocytes from case 1 could be due to a minor CD59+/−clone of CD34+ cells bearing mutation 1-2, which could not be detected clearly by flow cytometry, and that the minor clone proliferated or survived during differentiation and maturation.42 Thus, our findings with 3 PNH patients indicate that the phenotypes of granulocytes from PNH patients are consistent with those of CD34+ cells from PNH patients and that the phenotypes of both cells can be anticipated from PIG-A gene abnormalities. In fact, we reported previously that PIG-A gene abnormalities in PNH determine the phenotypes of mature granulocytes, but not of erythroblasts.34
We found a PIG-A gene abnormality (mutation 3-2) only in CD59− granulocytes, but not in CD34+CD59− cells from case 3. This was confirmed by both sequence analysis of the PIG-A gene using bacterial clones and direct sequencing of PCR-amplified PIG-A DNA. This may suggest that PIG-A gene abnormalities occur in CD34− cells in addition to CD34+ cells during the differentiation and maturation of hematopoietic stem cells from PNH patients. To confirm this hypothesis, we performed clonogeneic assays of CD34+cells using semisolid cultures and analyzed the PIG-A gene in the cultured cells because it has been reported that the GPI-AP–deficient characteristic of PNH is sometimes observed only in colonies but not in peripheral blood or bone marrow cells from patients with aplastic anemia.43 The results indicated that mutation 3-2 was in CFU-GMs cultured for 7 days, but not for 14 days, suggesting that mutation 3-2 occurred or was selected during differentiation and maturation of early to late CFU-GMs, including CD34+ cells with self-renewal capacity, but not in CD34− cells. This finding indicates that PIG-A gene abnormalities in PNH patients occur at the level of precursor cells expressing CD34 with self-renewal capacity. In addition, clonogeneic assays also revealed that there were some other mutations only in cultured cells derived from CD34+CD59− cells, but not in CD34+CD59− cells and CD59−granulocytes in vivo, and that there were some mutations even in cultured cells derived from CD34+CD59+ cells from 2 PNH patients. The latter finding may indicate that some CD34+CD59+ cells, probably border populations between CD34+CD59+ and CD34+CD59− cells in flow cytometry, contaminated the population of CD34+CD59−cells, judging from the low frequency of the mutations. The former findings lead to some interesting concepts. First, although it was reported that the frequency of monoclonality of PNH clones with a PIG-A gene mutation is much higher than that of oligoclonality in PNH patients,24-26 our results suggest that hematopoietic cells at the level of CD34+ cells from most PNH patients are oligoclonal. Nishimura et al30 also found 4 independent mutations of the PIG-A gene in cultured hematopoietic precursor cells, which were obtained from bone marrow MNCs of a patient with PNH. Second, our results showed that the mutations found only in cultured cells (mutations 2-2, 2-3, 3-3, and 3-4) occurred heterogeneously or were selected heterogeneously among erythroid and granulocyte-macrophage precursor cell lineages. On the other hand, we found the predominant PNH clones with mutations 2-1 and 3-1 in cultured cells derived from both erythroid and granulocyte-macrophage precursor cells from the PNH patients. We cannot explain how a predominant PNH clone escapes clonal selection or how a minor PNH clone is suppressed through clonal selection despite their having almost the same characteristics with respect to expression of GPI-APs. PIG-A gene analysis in semisolid cultured CD34+ cells in addition to that in fresh hematopoietic cells may be useful for answering these questions.
We found that the formation of CD34+CD59−colonies from 2 PNH patients was higher than that of their CD34+CD59+ colonies and almost the same as that of CD34+CD59+ colonies from 2 healthy volunteers, and that the formation of CD34+CD59+ colonies from the patients was less than that of CD34+CD59+ colonies from the healthy individuals. This is consistent with results reported previously with cultures of bone marrow precursor cells sorted into DAF−, DAF+, or DAF+/−fractions.44 Okuda et al45 reported that colony formation with the DAF− or DAF+fractions from bone marrow of PNH patients was greater or less compared with that from healthy volunteers, respectively. Chen et al46 also reported that CD34+CD59−cells from PNH patients proliferated to levels approaching those of normal cells, but that CD34+CD59+ cells from the patients gave rise to fewer cells in liquid culture. These findings suggest that the low numbers of colonies in cultures of unfractionated bone marrow and peripheral blood cells may be related to factors other than an intrinsic proliferative defect of hematopoietic stem cells, for example, hypersensitivity of CD34+CD59+ cells to apoptosis.46 In contrast, Maciejewski et al22 reported that formation of CD34+CD59− colonies with cells from PNH patients was less than that of CD34+CD59+colonies with cells from healthy individuals and was almost the same as that of CD34+CD59+ colonies with cells from the patients due to a proliferative defect in hematopoietic stem cells. Thus, the findings described above suggest that the proliferative capacity of CD34+ cells, including CD34+CD59+ and CD34+CD59− cells, from PNH patients is variable. How an increase or decrease in colony formation from CD34+CD59+ and CD34+CD59− cells from PNH patients occurs should be understood because this is relevant to the mechanism of expansion of a PNH clone.
In conclusion, a clonogeneic assay of CD34+ cells using semisolid cultures indicated that the PNH phenotypes determined by expression of GPI-APs on granulocytes are determined at the level of CD34+ cells, that PNH clones with PIG-A gene mutations are oligoclonal in most PNH patients, and that a minor PNH clone is selected or occurs heterogeneously among hematopoietic precursor cell lineages. These results suggest that PNH clones with PIG-A gene abnormalities might contribute qualitatively and quantitatively differentially to specific blood cell lineages during differentiation and maturation of hematopoietic stem cells.
We are grateful to Dr Teizo Fujita and Dr Minoru Takahashi (Department of Biochemistry II, Fukushima Medical University, Japan), Dr Mitsuru Munakata (Department of Pulmonary Medicine, Fukushima Medical University, Japan), Mr Akira Kuwada (Fukushima Technology Center, Japan), and Miss Makoto Yasukawa (Fukushima Technology Center, Japan) for their valuable assistance. Also, we wish to thank Dr Yuuji Sugita (Showa University, Japan) who provided the monoclonal antibody to CD59/membrane attack complex-inhibitory factor.
T.K. and T.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yukio Maruyama, First Department of Internal Medicine, Fukushima Medical University, 1 Hikariga-oka, Fukushima, Fukushima 960-1295, Japan; e-mail:t-shichi@fmu.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal