Ataxia telangiectasia (A-T), a genetic disorder caused by the homozygous mutation of the ATM gene, frequently associates with variable degrees of cellular and humoral immunodeficiency. However, the immune defects occurring in patients with A-T are still poorly characterized. Here we show that the T-cell receptor (TCR) variable β (BV)–chain repertoire of 9 A-T patients was restricted by diffuse expansions of some variable genes prevalently occurring within the CD4 subset and clustering to certain TCRBV genes (eg, 5.1, 11, 14, and 23). In addition, the study of the third complementarity-determining region (CDR3) showed, in all patients, significantly altered profiles in most BV genes examined suggesting diffuse oligoclonal expansions. The sequencing of TCR CDR3 regions revealed completely normal V(D)J coding joints and confirmed a reduced diversity of the antigen-receptor repertoire. The B-cell repertoire was similarly restricted and skewed by diffuse oligoclonal expansions with normal V(D)J joints. Thymic output, evaluated by measuring TCR rearrangement excision circles, was extremely low. The majority of peripheral T cells had the phenotype and the function of effector memory cells, indicating that in vivo they are able to respond normally by terminal differentiation to antigenic stimulation. These results indicate that ATM mutation limits the generation of a wide repertoire of normally functioning T and B cells.
Introduction
Ataxia telangiectasia (A-T) is an autosomal recessive multisystem disorder characterized by cerebellar ataxia, conjunctival and cutaneous telangiectasias, immune deficiency, chromosome aberrations, radiation hypersensitivity, and a high incidence of tumors, mainly of lymphoid origin.1 Immune deficiency of A-T patients is heterogeneous and involves both cellular and humoral responses.2,3 Typically, humoral immunodeficiency consists of the reduction of IgG2, IgA, and IgE with normal to increased IgM. The thymus is hypoplastic, and circulating T cells are usually decreased with a predominance of those expressing the γδ T-cell receptor (TCR) relative to those expressing the αβ TCR.4 These immune defects lead to frequent sinopulmonary infections whose incidence and severity varies widely between patients.
The gene responsible for A-T, which has been identified by positional cloning and denoted ATM (ataxia-telangiectasia mutated), encodes a protein with phosphatidylinositol 3-kinase (PI3K) signature at the carboxyl terminus.5 The PI3K family includes several proteins involved in cell cycle control, telomere maintenance, and repair of DNA double-strand breaks (DSBs).6 ATM is localized mostly to the nucleus and associates with DNA, with particular affinity for DNA ends. In the presence of DNA DSB damage, ATM phosphorylates a variety of protein targets, including p53, cAbl, replication protein A (RPA), Chk1, and Chk2, and activates multiple signal transduction pathways.7
Mice with disrupted ATM gene (ATM−/−) display immune defects and extremely high incidence of T-cell lymphomas.8-10
The mechanism(s) of the immunodeficiency consequent to inactivation of the ATM gene have not yet been fully elucidated.11 Chromosomal translocations involving immunoglobulin and TCR genes at 7p14, 7q35, 14q11.2, and 14q32 are commonly observed in neoplastic and nonneoplastic lymphocytes of patients with A-T. This suggests that illegitimate joining during V(D)J recombination might underlie both impaired lymphocyte development and enhanced lymphomagenesis.12-14 The finding that T-cell number and function can be rescued in ATM−/− mice by introducing a functional TCRαβ transgene15 indicates that a defect in TCR recombination is responsible for impaired T-cell development. Some evidence suggests that ATMmutation induces subtle defects in the efficiency and, especially, in the fidelity of DSB rejoining,16 although cells from A-T patients are proficient in supporting a V(D)J recombination reaction.17 Impaired responsiveness of peripheral T and B cells due to defective signal transduction in response to mitogenic stimulation18-20 is a possible additional mechanism for immunodeficiency in A-T.
To gain further insight into the mechanism(s) causing immunodeficiency in A-T, we investigated the repertoires of TCR β-chain variable region (TCRBV) and immunoglobulin heavy-chain (IgH) variable genes, the structure of V(D)J coding joints, the rate of thymic output by measurement of T-cell receptor rearrangement excision circles (TRECs), and the phenotype and function of peripheral T cells in a group of patients with A-T.
Patients and methods
Patients
All patients had classical A-T phenotype with early-onset ataxia, raised α1-fetoprotein levels, reduced IgA levels, and recurrent sinopulmonary infections. Two patients (nos. 2 and 4) are siblings. All patients were receiving intravenous γ-globulin replacement therapy at the time of the study. Because different ATM mutations may result in remarkably different disease phenotypes,21 we molecularly characterized the ATM mutations of all patients, except no. 1, by sequencing both gDNA and cDNA. Four patients were identified as compound heterozygotes and 4 as homozygotes. The detected mutations originate a truncation of the transcript in 7 of the 8 patients studied. Patient no. 9 is a compound heterozygote with one missense mutation detected; due to the absence of ATM protein at Western blot analysis (not shown), we argue that the second mutation is a truncating one.
The patients with partial DiGeorge syndrome and typical 22q11.2 deletion have been described in a previous publication.22
All experiments were done using anticoagulant-treated peripheral blood obtained by venipuncture. This study was conducted according to the good clinical practice guidelines of the Italian Ministry of Health. Informed consent was obtained from all patients or parents.
Flow cytometric analysis of CD4+ and CD8+T-cell subsets and TCRBV repertoire
Whole blood (500 μL) was lysed using 10 mL Ortho lysing reagent (Ortho-Clinical Diagnostics, Raritan, NJ), washed, labeled with a cocktail of 4 monoclonal antibodies (mAbs) for 30 minutes at 4°C, and fixed within 1 hour from blood collection. Anti-CD4 allophycocyanin (APC), anti-CD8 peridinin chlorophyll protein (PerCP), anti-CD45RA and anti-CD45R0 fluorescein isothiocyanate (FITC), anti-CD62L phycoerythrin (PE), and anti-HLA–DR FITC were purchased from Becton Dickinson Immunocytometry Systems (San Jose, CA). FITC-labeled anti-Fas was obtained from MBL (Medical and Biological Laboratories, Nagoya, Japan). Direct staining with anti-TCRBV antibodies (IOTest Beta Mark; Immunotech, Marseille, France) was performed according to the manufacturer's instruction. After staining, cells were washed once in phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) and analyzed on a FACSCalibur cytofluorometer (Becton Dickinson Immunocytometry Systems) using the Cell Quest software. To determine marker expression on CD4+ and CD8+ cells, total lymphocytes were first identified and gated by forward and side scatter. The cells were then additionally gated for CD4 or CD8 expression; 20 000 gated events were collected for each sample. Appropriate isotypic negative controls were run in parallel.
Single-cell analysis of cytokine production
Analysis of cytokine production at the single-cell level was performed as previously described.23 Briefly, peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by Ficoll-Isopaque (Lymphoprep-Nycomed, Oslo, Norway) gradient centrifugation, counted, and resuspended at 1 × 106 cells/mL in RPMI 1640 medium (Gibco Laboratories, Grand Island, NY) supplemented with 10% FBS (Sigma, St Louis, MO), 2 mM glutamine (Sigma), and 50 μg/mL gentamycin (Life Technologies, Gaithersburg, MD). Cells were then stimulated for 16 hours with 1 μg/mL ionomycin (Sigma) and 25 ng/mL phorbol myristate acetate (Sigma) in the presence of 10 μg/mL brefeldin A to inhibit cytokine secretion. After a wash in PBS, cells were fixed with 4% paraformaldehyde by incubation for 5 minutes at room temperature, permeabilized with FACS permeabilizing solution (Becton Dickinson, Immunocytometry Systems) for 10 minutes, washed, and stained. The following cytokine-specific mAbs were used: FITC-labeled anti–human interferon γ (anti–hIFN-γ; IgG2b), FITC-labeled anti–human interleukin 2 (anti–hIL-2; IgG1), and PE-labeled anti–hIL-4 (IgG1). Surface phenotyping was performed with anti-CD4 APC and anti-CD8 PerCP. All the mAbs were purchased from Becton Dickinson Immunocytometry Systems. After staining cells were washed once in PBS containing 10% FBS and analyzed on a FACSCalibur cytofluorometer (Becton Dickinson, Immunocytometry Systems) using the Cell Quest software. To determine the frequency of cytokine-producing T cells, total lymphocytes were first gated by forward and side scatter and then additionally gated for CD4 or CD8 expression; 20 000 gated events were collected for each sample. Appropriate isotypic negative controls were run in parallel.
Molecular studies
CD4+ and CD8+ T cells were separated by using CD4 and CD8 MicroBeads and MACS columns according to the manufacturer's protocols (Miltenyi Biotec, Bergisch Gladbach, Germany). Total mRNA was extracted directly from 106 to 107 bead-coated cells using Trizol-LS Reagent (Gibco-BRL, Grand Island, NY) and Micro-carrier (Molecular Research Center, Cincinnati, OH) and precipitated with isopropyl alcohol. The pelleted RNA was resuspended in diethyl-pyrocarbonate–treated water and the poly-(A)+ portion of total RNA was converted into cDNA using 2.5 μM oligo-deoxythymidine (dT) as primer for reverse transcription (RT), 50 mM KCl, 10 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, 5 mM MgCl2, 1 mM of each deoxyribonucleoside triphosphates (dNTPs), 1 U/μL RNase inhibitor, and 2.5 U/μL murine leukemia virus (MULV) reverse transcriptase (Applied Biosystems, Foster City, CA).
To analyze the TCRBV transcript size patterns cDNA samples were amplified by using a TCRB C1/C2-specific primer (CGG GCT GCT CCT TGA GGG GCT GCG) and a set of 24 TCRBV-specific primers (BV 1, 2, 3, 4, 5.1, 5.3, 6.1, 6.2, 7, 8, 9, 11, 12, 13, 14, 15, 16, 17, 18, 20, 21, 22, 24).24 Briefly, 2 μL of the RT product was brought to a final reaction volume of 50 μL containing 50 mM KCl, 10 mM Tris-HCl 1,5 mM MgCl2, 0,2 mM of each dNTPs, 25 pmol of each oligonucleotide, and 2 U Taq DNA polymerase (AmpliTaq; Applied Biosystems). After an initial denaturation step of 3 minutes at 95°C the reactions were subjected to 35 cycles of polymerase chain reaction (PCR; 30 seconds at 94°C, 30 seconds at 60°C, 30 seconds at 72°C) followed by a final elongation step for 10 minutes at 72°C. Aliquots of the unlabeled PCR products were then labeled by 10 cycles of elongation in a 10 μL “run-off” reaction with the FAM TCRBC primer (CTG CAC CTC CTT CCC ATT) mixed with deionized formamide and TAMRA 500 size standard (Applied Biosystems). Finally, run-off products were electrophoresed for 24 minutes on a 310 ABI PRISM automated sequencer by using a 47-cm capillary and POP-4 polymer. The third complementarity-determining region (CDR3) profile was then analyzed with the Genescan software (Applied Biosystems). For IgH CDR3 spectratyping, amplifications were done with oligonucleotide primers specific for constant μ chain (Cμ) and for a conserved sequence in the human VH framework region 3.25
Analysis of the level of perturbation of TCRBV repertoire of A-T patients was performed according to Gorochov et al26 with minor modifications. Briefly, the CDR3 length profiles (spectratypes) were first translated into probability distributions as function of the area under the profile for each CDR3 length. A control profile, representing the nonperturbed repertoire, was determined for each BV by calculating the average distribution of the corresponding CD4 and CD8 profiles from 5 healthy blood donors. The extent of perturbation for each CDR3 fragment was then calculated by the difference between the sample's distribution and the control's distribution. Finally, the TCR repertoire perturbation per BV family was defined as the sum of the absolute values of the differences between each sample's CDR3 length and the corresponding control distribution. Values of BV perturbation greater than the sum of the SDs calculated in healthy blood donors for each CDR3 profile were considered abnormal.
For CDR3 sequencing, amplification products were inserted into a plasmid vector (pCRII-TOPO; Invitrogen, Carlsbad, CA), cloned inEscherichia coli and sequenced onto an Applied Biosystems Sequencer Model 377-96. Sequence similarities were identified using the multiple sequence alignment application, Align X, of the Vector NTI Suite 6.0 (Informax, North Bethesda, MD) based on the CLUSTAL W algorithm.27 The identification of the V(D)J junctions was performed comparing the sequences with those reported in IMGT, the International ImMunoGeneTics database http://imgt.cines.fr:8104(Initiator and coordinator: Marie-Paule Lefranc, Montpellier, France).28
For measuring TRECs, α1 circles29 were amplified from patients' DNA by real-time PCR using a Model 7700 Sequence Detector and the GeneAmp software (Applied Biosystems). Amplification of the GAPDH gene was used to normalize for DNA content. The amount of TREC/100 ng DNA was calculated on the basis of a standard curve (from 104 to 100 copies) of α1 circles cloned into a plasmid and diluted into DNA from a human cell line devoid of TRECs. The lower limit of detection of the method was 2 copies/100 ng gDNA.
Results
T-cell phenotypes
In most of the patients investigated the peripheral distribution of CD4+ T cells was lower than observed in age-matched controls, whereas that of CD8+ T cells generally fell within normal range (Table1). The large majority of T cells expressed CD95 (for CD4+: 86% ± 18% versus 39% ± 11% in healthy controls, P < .001; for CD8+: 87% ± 18% versus 36% ± 12% in controls), denoting an activated phenotype. The distribution of naive and memory T cells was investigated by the differential expression of CD45 isoforms and of CD62L. All A-T patients, with the exception of patient no. 7, showed a marked predominance of T cells with a memory phenotype (CD45RA−CD62L−, CD45RA+CD62L−, CD45RA−CD62L+; Table 1). This was seen in association with a corresponding reduction in the frequency of naive T cells, particularly pronounced within the CD4 subset.
Demographic data and major immunologic findings in A-T patients
| Patient . | Age, y/sex . | Lymphs/μL . | CD19 . | CD4 . | CD8 . | T-cell phenotype . | Cytokine production . | TRECs . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Naive . | Memory . | “Effector” memory . | CD4 . | CD8 . | |||||||||||||||
| CD4 . | CD8 . | CD4 . | CD8 . | CD4 . | CD8 . | IL-2 . | IFN-γ . | IL-4 . | IL-2 . | IFN-γ . | IL-4 . | CD4 . | CD8 . | ||||||
| 1 | 7/M | 961 | 34 | 22 | 24 | 2 | 16 | 98 | 84 | 22 | 39 | 55 | 41 | 9 | 18 | 76 | 9 | 4.3 | 5.3 |
| 2 | 10/F | 710 | ND | 38 | 17 | 6 | 15 | 93 | 86 | 27 | 54 | ND | ND | ND | ND | ND | ND | 4 | 7.9 |
| 3 | 11/F | 1260 | 1 | 23 | 30 | 3 | 20 | 95 | 79 | 26 | 53 | 56 | 55 | 3 | 18 | 81 | 1 | 0 | 18.5 |
| 4 | 12/M | 660 | ND | 37 | 21 | 10 | 24 | 89 | 76 | 27 | 43 | ND | ND | ND | ND | ND | ND | 6 | <2 |
| 5 | 13/F | 976 | 5 | 21 | 10 | 2 | 10 | 97 | 91 | 27 | 51 | 51 | 51 | 13 | 9 | 85 | 3 | <2 | <2 |
| 6 | 17/F | 860 | 3 | 29 | 14 | 5 | 25 | 94 | 75 | 15 | 29 | 67 | 35 | 15 | 50 | 63 | 19 | ND | ND |
| 7 | 21/F | 1140 | 7 | 48 | 9 | 54 | 23 | 46 | 77 | 13 | 63 | 57 | 10 | 0.7 | 14 | 61 | 2 | 5.9 | <2 |
| 8 | 21/M | 872 | 5 | 38 | 27 | 25 | 26 | 75 | 74 | 18 | 37 | 56 | 13 | 6 | 37 | 39 | 14 | 4 | 2.8 |
| 9 | 32/M | 1320 | 6 | 23 | 27 | 16 | 40 | 83 | 60 | 17 | 40 | 50 | 30 | 1.2 | 11 | 24 | 0.1 | ND | ND |
| Normal* | |||||||||||||||||||
| 6-12 | >1200 | 9-20 (12) | 32-51 (41) | 10-24 (21) | 53-87 (65) | 60-91 (74) | 13-47 (30) | 9-39 (23) | 1-10 (7) | 7-28 (19) | 41-55 (49) | 3-12 (7) | 1-3 (2) | 11-19 (16) | 9-48 (17) | 0-1 (1) | |||
| 13-35 | >1200 | 5-21 (11) | 39-54 (45) | 13-31 (19) | 37-69 (49) | 49-79 (69) | 31-63 (51) | 21-51 (30) | 5-16 (10) | 12-38 (20) | 61-75 (63) | 12-19 (14) | 3-6 (5) | 27-36 (28) | 32-40 (34) | 1-2.3 (1.5) | 120-340 (170) | 90-240 (145) | |
| Patient . | Age, y/sex . | Lymphs/μL . | CD19 . | CD4 . | CD8 . | T-cell phenotype . | Cytokine production . | TRECs . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Naive . | Memory . | “Effector” memory . | CD4 . | CD8 . | |||||||||||||||
| CD4 . | CD8 . | CD4 . | CD8 . | CD4 . | CD8 . | IL-2 . | IFN-γ . | IL-4 . | IL-2 . | IFN-γ . | IL-4 . | CD4 . | CD8 . | ||||||
| 1 | 7/M | 961 | 34 | 22 | 24 | 2 | 16 | 98 | 84 | 22 | 39 | 55 | 41 | 9 | 18 | 76 | 9 | 4.3 | 5.3 |
| 2 | 10/F | 710 | ND | 38 | 17 | 6 | 15 | 93 | 86 | 27 | 54 | ND | ND | ND | ND | ND | ND | 4 | 7.9 |
| 3 | 11/F | 1260 | 1 | 23 | 30 | 3 | 20 | 95 | 79 | 26 | 53 | 56 | 55 | 3 | 18 | 81 | 1 | 0 | 18.5 |
| 4 | 12/M | 660 | ND | 37 | 21 | 10 | 24 | 89 | 76 | 27 | 43 | ND | ND | ND | ND | ND | ND | 6 | <2 |
| 5 | 13/F | 976 | 5 | 21 | 10 | 2 | 10 | 97 | 91 | 27 | 51 | 51 | 51 | 13 | 9 | 85 | 3 | <2 | <2 |
| 6 | 17/F | 860 | 3 | 29 | 14 | 5 | 25 | 94 | 75 | 15 | 29 | 67 | 35 | 15 | 50 | 63 | 19 | ND | ND |
| 7 | 21/F | 1140 | 7 | 48 | 9 | 54 | 23 | 46 | 77 | 13 | 63 | 57 | 10 | 0.7 | 14 | 61 | 2 | 5.9 | <2 |
| 8 | 21/M | 872 | 5 | 38 | 27 | 25 | 26 | 75 | 74 | 18 | 37 | 56 | 13 | 6 | 37 | 39 | 14 | 4 | 2.8 |
| 9 | 32/M | 1320 | 6 | 23 | 27 | 16 | 40 | 83 | 60 | 17 | 40 | 50 | 30 | 1.2 | 11 | 24 | 0.1 | ND | ND |
| Normal* | |||||||||||||||||||
| 6-12 | >1200 | 9-20 (12) | 32-51 (41) | 10-24 (21) | 53-87 (65) | 60-91 (74) | 13-47 (30) | 9-39 (23) | 1-10 (7) | 7-28 (19) | 41-55 (49) | 3-12 (7) | 1-3 (2) | 11-19 (16) | 9-48 (17) | 0-1 (1) | |||
| 13-35 | >1200 | 5-21 (11) | 39-54 (45) | 13-31 (19) | 37-69 (49) | 49-79 (69) | 31-63 (51) | 21-51 (30) | 5-16 (10) | 12-38 (20) | 61-75 (63) | 12-19 (14) | 3-6 (5) | 27-36 (28) | 32-40 (34) | 1-2.3 (1.5) | 120-340 (170) | 90-240 (145) | |
All data are expressed as percentages except for absolute lymphocyte counts and TRECs (number of copies/100 ng DNA).
M indicates male; F, female; and ND, not determined.
Range (median) for the indicated age groups. The age range of the control group for TRECs is 10 to 20 years.
The expression of CD62L, a lymph node homing receptor, may distinguish “central memory” T cells, which are CD62L+ and produce mainly IL-2, from terminally differentiated “effector memory” T cells, which are CD62L− and have acquired the capacity to produce IFN-γ or IL-4.30 In A-T patients, the proportions of effector memory T cells were significantly higher than those observed in healthy controls (P < .002 both for CD4 and for CD8 cells; Table 1).
The pattern of activation-induced cytokine production by patient's T cells (Table 1) was consistent with their predominant memory phenotype. In fact, compared to healthy controls, there was a significant increase of cells polarized toward the production of IFN-γ and IL-4, a characteristic of effector memory T cells.30 As opposite the frequency of IL-2+ cells was in general normal, both in CD4+ and in CD8+ T cells.
Taken together, our findings indicate that most circulating T cells of A-T patients are memory cells, predominantly with the features of terminally differentiated effectors, whereas naive T cells are very scarce.
Thymic function
During V(D)J T-cell receptor rearrangement, DNA extrachromosomal excision products (also known as TRECs) are generated. These products are not replicated during mitosis so that their number decreases with each round of cell division. For this reason TRECs are usually used as a marker of recent thymic emigrants (RTEs).29 Based on these notions, we investigated whether the defective numbers of naive T cells we found in A-T patients could be attributed to low thymic output by evaluating the levels of TRECs in separated CD4+ and CD8+ T cells.
All A-T patients displayed extremely low levels of TRECs in comparison with age-matched healthy individuals (Table 1) and even in comparison with 4 patients with DiGeorge syndrome, an immunodeficiency caused by thymic hypoplasia, who had, respectively, 18.5, 21.7, 78, and 68 copies/100 ng DNA in CD4 cells, and 49.8, 104, 46, and 27 copies/100 ng DNA in CD8 cells.
Analysis of the TCRBV repertoire by mAbs
Anomalies in the relative TCRBV usage were investigated using a panel of BV subfamily-specific mAbs covering approximately 60% to 70% of T cells expressing TCRαβ in healthy individuals (Table 2).
TCRBV repertoire in 9 patients with A-T
| CD4 . | CD8 . | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vβ . | Normal range (median) . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | Normal range (median) . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . |
| 1 | 2.1-4.0 (3.4) | 2.4 | 2.6 | 1.7 | 3.0 | 2.0 | 2.5 | 2.3 | 2.6 | 3.4 | 2.5-7.9 (3.8) | 0.9 | 4.9 | 0.5 | 5.6 | 0.9 | 1.4 | 4.0 | 1.3 | 1.9 |
| 2 | 3.8-13.1 (9.6) | 0.5 | 2.6 | 5.5 | 7.8 | 7.5 | 3.0 | 4.0 | 5.7 | 8.0 | 0.7-20.5 (5.6) | 0.1 | 0.5 | 2.8 | 3.3 | 0.8 | 0.6 | 17.0 | 1.3 | 0.9 |
| 3 | 0.6-9.8 (4.1) | 5.2 | 4.3 | 2.3 | 6.5 | 4.1 | 3.1 | 1.4 | 0.5 | 2.0 | 0.1-19.3 (3.7) | 29.3 | 5.0 | 7.7 | 8.2 | 7.8 | 3.7 | 4.0 | 10.6 | 3.7 |
| 4 | 1.5-2.5 (1.8) | ND | 0.8 | ND | 1.4 | ND | ND | 0 | ND | 0 | 0.3-2.0 (1.4) | ND | 0.5 | ND | 0.4 | ND | ND | 0 | ND | 0 |
| 5.1 | 3.8-8.6 (6.1) | 6.5 | 9.2 | 4.7 | 10.8 | 5.9 | 15.6 | 5.0 | 7.2 | 8.9 | 1.2-6.1 (2.7) | 0.8 | 3.0 | 0.3 | 2.0 | 1.2 | 2.8 | 3.7 | 1.4 | 1.2 |
| 5.2 | 0.1-2.7 (0.8) | 0.7 | 0.8 | 1.1 | 1.3 | 0.4 | 0.7 | 0.4 | 1.0 | 1.5 | 0.2-4.0 (0.8) | 0.6 | 0.3 | 0.2 | 3.8 | 0.2 | 0.6 | 0.9 | 0.2 | 0.8 |
| 5.3 | 0.1-3.4 (1.8) | 1.0 | 1.0 | 0.7 | 0.8 | 1.2 | 0.9 | 0.3 | 1.0 | 1.2 | 0.4-21.6 (1.5) | 1.1 | 0.6 | 0.1 | 0 | <0.1 | 0.5 | 0.2 | 0.6 | 0.1 |
| 7.1 | 1.8-2.8 (2.0) | ND | 1.1 | ND | 0.8 | ND | ND | 0.8 | 1.0 | 5.0 | 1.9-4.6 (3.7) | ND | 14.3 | ND | 1.5 | ND | ND | 6.5 | 3.4 | 1.3 |
| 7.2 | 0.1-1.7 (0.9) | 2.6 | 1.5 | 0.6 | 1.2 | 1.6 | 0.6 | <0.1 | ND | 1.5 | 0.1-5.7 (0.9) | 7.6 | 1.0 | 8.9 | 0.4 | 0.1 | 2.6 | <0.1 | ND | 47 |
| 8 | 3.2-10.0 (4.9) | 7.5 | 4.4 | 5.8 | 5.6 | 3.2 | 2.0 | 1.9 | 3.6 | 3.3 | 1.0-12.4 (3.2) | 1.8 | 5.5 | <0.1 | 2.3 | 0.5 | 2.8 | 2.0 | 1.4 | 0.6 |
| 9 | 2.5-14.5 (3.8) | 7.0 | 3.9 | 3.7 | 2.4 | 3.7 | 3.0 | 1.7 | 2.6 | 3.3 | 1.2-5.2 (2.3) | 2.6 | 1.0 | 1.0 | 0.4 | 0.8 | 1.8 | 2.2 | 1.0 | 1.5 |
| 11 | 0.5-1.1 (0.8) | 2.3 | 0.9 | 1.6 | 0.5 | 0.4 | 0.5 | 0.7 | 3.2 | 0.8 | 0.4-2.0 (0.6) | 0.8 | 0.4 | 0.2 | 0.4 | 2.8 | 0.5 | 1.3 | 2.2 | 0.1 |
| 12 | 0.7-3.3 (1.8) | 1.2 | 4 | 1.6 | 4 | 2.6 | 1.8 | 2.3 | 2.4 | 2.7 | 0.3-4.5 (1.3) | 1.5 | 3.1 | 0.3 | 5.3 | <0.1 | 1.4 | 1.3 | 1.4 | 0.7 |
| 13.1 | 1.0-4.5 (3.6) | 4.6 | 4.5 | 3.9 | 3.3 | 3.1 | 3.3 | 1.8 | 3.1 | 4.4 | 1.1-7.1 (3.4) | 0.9 | 13.8 | 2.4 | 1.4 | 1.1 | 2.3 | 3.0 | 2.2 | 5.6 |
| 13.2 | 1.4-4.0 (2.8) | ND | 2.3 | ND | 2.6 | ND | ND | 0.2 | ND | 1.3 | 1.1-6.3 (2.8) | ND | 1.6 | ND | 2 | ND | ND | 0.4 | ND | 0.4 |
| 13.6 | 0.8-4.3 (2.0) | 5.0 | 1.6 | 4.8 | 1.3 | 2.4 | 2.7 | 1.2 | 1.9 | 2.8 | 0.1-3.6 (1.5) | 2.9 | 0.3 | 1.6 | 6.0 | 1.1 | 0.4 | 0.5 | 0.8 | 0.6 |
| 14 | 1.0-3.5 (2.6) | 8.0 | 2.2 | 5.0 | 2.4 | 2.0 | 1.1 | 1.9 | 2.5 | 4.8 | 2.1-10.6 (4.2) | 10 | 2.1 | 5.3 | 5.7 | 1.0 | 5.2 | 3.0 | 4.5 | 2.7 |
| 16 | 0.7-3.8 (1.2) | 2.7 | 3.0 | 0.7 | 4.2 | 0.9 | 0.4 | 1.0 | 0.8 | 2.3 | 0.1-3.8 (0.9) | 0.4 | 3.2 | <0.1 | 2.8 | 0.1 | 0.8 | 1.1 | 6.2 | 3.8 |
| 17 | 2.6-8.2 (5.5) | 2.2 | 4.5 | 3.2 | 6.0 | 11.5 | 2.6 | 2.4 | 4.1 | 4.5 | 1.7-23.2 (4.7) | 5.4 | 13 | 6.4 | 2.0 | 0.2 | 16.5 | 2.0 | 10.6 | 1.7 |
| 18 | 0.4-4.3 (1.7) | 0.9 | 1.2 | 0.4 | 1.1 | 1.4 | 1.5 | 0.5 | 1.9 | 1.1 | 0.1-4.1 (0.6) | 0.3 | <0.1 | <0.1 | <0.1 | 0.6 | <0.1 | 0.2 | 1.2 | 0.4 |
| 20 | 1.9-3.9 (2.7) | 4.1 | 0 | 3.9 | 0 | 1.4 | 2.0 | 1.6 | 3.1 | 1.7 | 0.6-3.1 (1.8) | 2.0 | 0 | 1.1 | 0 | 0.5 | 1.7 | 1.2 | 1.3 | 0.9 |
| 21.3 | 1.6-3.0 (2.3) | 2.6 | 1.8 | 1.8 | 4.4 | 1.1 | 0.7 | 1.2 | 1.6 | 2.0 | 1.0-5.6 (1.7) | 1.0 | 2.3 | 0.2 | 1.0 | <0.1 | 0.9 | 3.2 | 1.1 | 0.6 |
| 22 | 2.4-9.8 (4.1) | 4.9 | 2.4 | 3.1 | 3.0 | 2.6 | 5.6 | 1.8 | 0.8 | 3.8 | 0.7-6.7 (2.9) | 0.4 | 3.9 | 4.3 | 0.4 | 0.4 | 1.5 | 1.6 | 0.1 | 1.2 |
| 23 | 0.1-1.2 (0.4) | 1.7 | 1.4 | 1.4 | 0.4 | <0.1 | 0.9 | 0.2 | 0.5 | 0.7 | 0.1-18.9 (1.1) | 0.6 | 1.2 | 0.3 | 1.6 | 0.7 | 1.2 | 1.2 | 0.7 | 0.9 |
| Total | 70.7 | 73.7 | 61.3 | 56.5 | 74.6 | 58.2 | 54.4 | 34.6 | 51 | 70.6 | 57.1 | 70.3 | 81.3 | 43.5 | 56.4 | 20.4 | 49.2 | 60.5 | 53.4 | 78.6 |
| CD4 . | CD8 . | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vβ . | Normal range (median) . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | Normal range (median) . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . |
| 1 | 2.1-4.0 (3.4) | 2.4 | 2.6 | 1.7 | 3.0 | 2.0 | 2.5 | 2.3 | 2.6 | 3.4 | 2.5-7.9 (3.8) | 0.9 | 4.9 | 0.5 | 5.6 | 0.9 | 1.4 | 4.0 | 1.3 | 1.9 |
| 2 | 3.8-13.1 (9.6) | 0.5 | 2.6 | 5.5 | 7.8 | 7.5 | 3.0 | 4.0 | 5.7 | 8.0 | 0.7-20.5 (5.6) | 0.1 | 0.5 | 2.8 | 3.3 | 0.8 | 0.6 | 17.0 | 1.3 | 0.9 |
| 3 | 0.6-9.8 (4.1) | 5.2 | 4.3 | 2.3 | 6.5 | 4.1 | 3.1 | 1.4 | 0.5 | 2.0 | 0.1-19.3 (3.7) | 29.3 | 5.0 | 7.7 | 8.2 | 7.8 | 3.7 | 4.0 | 10.6 | 3.7 |
| 4 | 1.5-2.5 (1.8) | ND | 0.8 | ND | 1.4 | ND | ND | 0 | ND | 0 | 0.3-2.0 (1.4) | ND | 0.5 | ND | 0.4 | ND | ND | 0 | ND | 0 |
| 5.1 | 3.8-8.6 (6.1) | 6.5 | 9.2 | 4.7 | 10.8 | 5.9 | 15.6 | 5.0 | 7.2 | 8.9 | 1.2-6.1 (2.7) | 0.8 | 3.0 | 0.3 | 2.0 | 1.2 | 2.8 | 3.7 | 1.4 | 1.2 |
| 5.2 | 0.1-2.7 (0.8) | 0.7 | 0.8 | 1.1 | 1.3 | 0.4 | 0.7 | 0.4 | 1.0 | 1.5 | 0.2-4.0 (0.8) | 0.6 | 0.3 | 0.2 | 3.8 | 0.2 | 0.6 | 0.9 | 0.2 | 0.8 |
| 5.3 | 0.1-3.4 (1.8) | 1.0 | 1.0 | 0.7 | 0.8 | 1.2 | 0.9 | 0.3 | 1.0 | 1.2 | 0.4-21.6 (1.5) | 1.1 | 0.6 | 0.1 | 0 | <0.1 | 0.5 | 0.2 | 0.6 | 0.1 |
| 7.1 | 1.8-2.8 (2.0) | ND | 1.1 | ND | 0.8 | ND | ND | 0.8 | 1.0 | 5.0 | 1.9-4.6 (3.7) | ND | 14.3 | ND | 1.5 | ND | ND | 6.5 | 3.4 | 1.3 |
| 7.2 | 0.1-1.7 (0.9) | 2.6 | 1.5 | 0.6 | 1.2 | 1.6 | 0.6 | <0.1 | ND | 1.5 | 0.1-5.7 (0.9) | 7.6 | 1.0 | 8.9 | 0.4 | 0.1 | 2.6 | <0.1 | ND | 47 |
| 8 | 3.2-10.0 (4.9) | 7.5 | 4.4 | 5.8 | 5.6 | 3.2 | 2.0 | 1.9 | 3.6 | 3.3 | 1.0-12.4 (3.2) | 1.8 | 5.5 | <0.1 | 2.3 | 0.5 | 2.8 | 2.0 | 1.4 | 0.6 |
| 9 | 2.5-14.5 (3.8) | 7.0 | 3.9 | 3.7 | 2.4 | 3.7 | 3.0 | 1.7 | 2.6 | 3.3 | 1.2-5.2 (2.3) | 2.6 | 1.0 | 1.0 | 0.4 | 0.8 | 1.8 | 2.2 | 1.0 | 1.5 |
| 11 | 0.5-1.1 (0.8) | 2.3 | 0.9 | 1.6 | 0.5 | 0.4 | 0.5 | 0.7 | 3.2 | 0.8 | 0.4-2.0 (0.6) | 0.8 | 0.4 | 0.2 | 0.4 | 2.8 | 0.5 | 1.3 | 2.2 | 0.1 |
| 12 | 0.7-3.3 (1.8) | 1.2 | 4 | 1.6 | 4 | 2.6 | 1.8 | 2.3 | 2.4 | 2.7 | 0.3-4.5 (1.3) | 1.5 | 3.1 | 0.3 | 5.3 | <0.1 | 1.4 | 1.3 | 1.4 | 0.7 |
| 13.1 | 1.0-4.5 (3.6) | 4.6 | 4.5 | 3.9 | 3.3 | 3.1 | 3.3 | 1.8 | 3.1 | 4.4 | 1.1-7.1 (3.4) | 0.9 | 13.8 | 2.4 | 1.4 | 1.1 | 2.3 | 3.0 | 2.2 | 5.6 |
| 13.2 | 1.4-4.0 (2.8) | ND | 2.3 | ND | 2.6 | ND | ND | 0.2 | ND | 1.3 | 1.1-6.3 (2.8) | ND | 1.6 | ND | 2 | ND | ND | 0.4 | ND | 0.4 |
| 13.6 | 0.8-4.3 (2.0) | 5.0 | 1.6 | 4.8 | 1.3 | 2.4 | 2.7 | 1.2 | 1.9 | 2.8 | 0.1-3.6 (1.5) | 2.9 | 0.3 | 1.6 | 6.0 | 1.1 | 0.4 | 0.5 | 0.8 | 0.6 |
| 14 | 1.0-3.5 (2.6) | 8.0 | 2.2 | 5.0 | 2.4 | 2.0 | 1.1 | 1.9 | 2.5 | 4.8 | 2.1-10.6 (4.2) | 10 | 2.1 | 5.3 | 5.7 | 1.0 | 5.2 | 3.0 | 4.5 | 2.7 |
| 16 | 0.7-3.8 (1.2) | 2.7 | 3.0 | 0.7 | 4.2 | 0.9 | 0.4 | 1.0 | 0.8 | 2.3 | 0.1-3.8 (0.9) | 0.4 | 3.2 | <0.1 | 2.8 | 0.1 | 0.8 | 1.1 | 6.2 | 3.8 |
| 17 | 2.6-8.2 (5.5) | 2.2 | 4.5 | 3.2 | 6.0 | 11.5 | 2.6 | 2.4 | 4.1 | 4.5 | 1.7-23.2 (4.7) | 5.4 | 13 | 6.4 | 2.0 | 0.2 | 16.5 | 2.0 | 10.6 | 1.7 |
| 18 | 0.4-4.3 (1.7) | 0.9 | 1.2 | 0.4 | 1.1 | 1.4 | 1.5 | 0.5 | 1.9 | 1.1 | 0.1-4.1 (0.6) | 0.3 | <0.1 | <0.1 | <0.1 | 0.6 | <0.1 | 0.2 | 1.2 | 0.4 |
| 20 | 1.9-3.9 (2.7) | 4.1 | 0 | 3.9 | 0 | 1.4 | 2.0 | 1.6 | 3.1 | 1.7 | 0.6-3.1 (1.8) | 2.0 | 0 | 1.1 | 0 | 0.5 | 1.7 | 1.2 | 1.3 | 0.9 |
| 21.3 | 1.6-3.0 (2.3) | 2.6 | 1.8 | 1.8 | 4.4 | 1.1 | 0.7 | 1.2 | 1.6 | 2.0 | 1.0-5.6 (1.7) | 1.0 | 2.3 | 0.2 | 1.0 | <0.1 | 0.9 | 3.2 | 1.1 | 0.6 |
| 22 | 2.4-9.8 (4.1) | 4.9 | 2.4 | 3.1 | 3.0 | 2.6 | 5.6 | 1.8 | 0.8 | 3.8 | 0.7-6.7 (2.9) | 0.4 | 3.9 | 4.3 | 0.4 | 0.4 | 1.5 | 1.6 | 0.1 | 1.2 |
| 23 | 0.1-1.2 (0.4) | 1.7 | 1.4 | 1.4 | 0.4 | <0.1 | 0.9 | 0.2 | 0.5 | 0.7 | 0.1-18.9 (1.1) | 0.6 | 1.2 | 0.3 | 1.6 | 0.7 | 1.2 | 1.2 | 0.7 | 0.9 |
| Total | 70.7 | 73.7 | 61.3 | 56.5 | 74.6 | 58.2 | 54.4 | 34.6 | 51 | 70.6 | 57.1 | 70.3 | 81.3 | 43.5 | 56.4 | 20.4 | 49.2 | 60.5 | 53.4 | 78.6 |
Analyses were done with Vβ-specific mAbs; the nomenclature adopted is from Wei et al.52 Results are the percentages of TCRαβ T cells expressing the indicated TCRBV gene.
ND indicates determinations not done; shading, underexpressed TCRBV genes; and italics, overexpressed TCRBVs.
Among the 24 TCRBV genes investigated, significant expansions (ie, above the highest values observed in healthy subjects) were observed in 9.5% of Vβs expressed by CD4+ cells and in 4.2% of Vβs expressed by CD8+ cells (Table 2). The predominance of expansions in CD4+ rather than in CD8+ T cells contrasts with findings in healthy individuals showing occasional antigen-driven expansions exclusively within the CD8 subset.31,32 We observed clustering of expansions to certain TCRBV genes, particularly evident in the CD4 subset. In fact, Vβ5.1 was expanded in 4 patients, and Vβ11, Vβ14, and Vβ23 in 3 patients. Some patients had evidence for expansions that passed undetected to our analysis with mAbs. For example, in patient no. 5 our typing panel covered only 20% of the repertoire in CD8+ cells, suggesting the presence of one or more undetected clones. Another patient (no. 7) is known to carry a large CD4 clone33 not typed by our panel of Vβ-specific antibodies that, in fact, covered only 34.6% of the repertoire.
To investigate the capacity of A-T patients to generate a normally wide TCRBV repertoire, we looked for “holes” in the expression of Vβ genes. Although all patients had several underexpressed Vβs (Table2), we did not detect clear-cut repertoire holes. In addition, in some patients underexpression of several Vβs was clearly caused by the predominance of expanded clones (eg, within CD4+ cells of patient no. 7 and within CD8+ cells of patient no. 9). Nevertheless, it is worth noting that most of the Vβs commonly underrepresented in our A-T patients are located in the portion of the TCRBV locus more distal from the diversity-joining-constant genes cluster (Vβ1, Vβ5.3, Vβ7.1, Vβ9, Vβ13.2, Vβ20, Vβ21.3, and Vβ22) or 3′ to it (Vβ20). (The physical map of the TCRB locus used for this analysis is from IMGT, http://imgt.cines.fr:8104.) This finding is in keeping with the previous suggestion4 that complex V(D)J rearrangements, such as those involving large intervening sequences or an inversional mechanism,34 may be preferentially impaired in A-T.
Molecular analysis of the TCRB and IgH repertoires
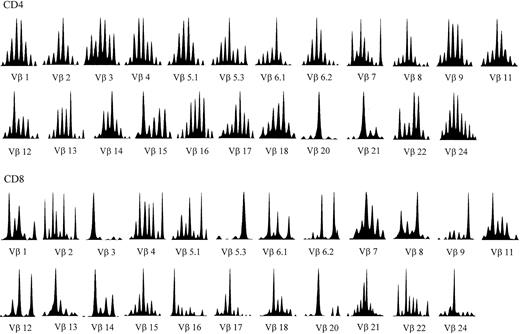
Heterogeneity of the TCRB repertoire was further investigated by CDR3 “spectratyping,” that is, by the quantitative analysis of CDR3s with different sizes generated by the random insertion/deletion of nucleotides during V(D)J rearrangement.24 A normal, polyclonal repertoire results in a histogram with a gaussianlike distribution of CDR3 lengths, whereas abnormal patterns display one or more predominant fragments outside the peak of median length (Figure1). Mathematical analysis of the deviation of patients' histograms from the normal distribution revealed that all A-T patients had significantly altered patterns in most Vβ genes examined (Figure 2A-I). These alterations are indicative of Vβ repertoires restricted by diffuse clonal expansions. As expected, the profiles of CDR3 size distribution were more perturbed within the CD8+ subset, with oligoclonal peaks found in each BV family investigated (Figure 1). In fact, disruption of the gaussianlike CDR3 profile of some BV families is often observed in CD8+ lymphocytes of healthy individuals. In healthy subjects these idiosyncratic peaks are constant over time and, therefore, do not necessarily represent ongoing immune responses. However, in A-T patients perturbations of the TCRBV repertoire of CD8+ cells were much more prominent and systematic than in healthy controls, indicating that they were most likely related to the underlying immunologic disorder.
TCRBV CDR3 spectratyping histograms in a representative A-T patient (no. 2).
The majority of the TCRBVs have altered CDR3 profiles compared to the quasi-gaussian distribution of peaks observed in normal spectratypes (eg, Vβ1 and Vβ24 in this patient). The Vβ nomenclature adopted is from Wei et al.52
TCRBV CDR3 spectratyping histograms in a representative A-T patient (no. 2).
The majority of the TCRBVs have altered CDR3 profiles compared to the quasi-gaussian distribution of peaks observed in normal spectratypes (eg, Vβ1 and Vβ24 in this patient). The Vβ nomenclature adopted is from Wei et al.52
The CDR3 spectratypes obtained by amplification of IgH-specific sequences were also altered in most patients with A-T (Figure 2J), suggesting the presence of B-cell oligoclones. B-cell oligoclonality in A-T could be due either to restricted central generation or to dysregulated peripheral expansion caused by defects in regulatory T cells. To discriminate between these 2 possibilities, we examined patients with DiGeorge syndrome, whose immunodeficiency is solely attributable to a thymic defect, and observed a normal distribution of IgH CDR3 spectratypes (not shown). This suggests that decreased diversity of the B-cell repertoire in A-T patients is due to an intrinsic developmental defect, in agreement with the observation that ATM−/− mice have reduced pre-B cells.10
Diversity of the TCRBV and IgH repertoires analyzed by CDR3 spectratyping.
The extent of TCRBV repertoire perturbation is represented as the percent difference between the patient's CDR3 distribution and the corresponding control distribution. A value of perturbation greater than the sum of the SDs relative to each CDR3 length found in normal blood donors is considered abnormal. Panels A to I show TCRBV data in patients nos. 1 to 9, respectively. The bars on the left of the doublets represent CD4 cells and the bars on the right, CD8 cells. ░ indicates, abnormal pattern in CD4 cells; ▪, abnormal pattern in CD8 cells; ■ plus *, normal pattern. Panel J represents IgH data in patients nos. 1 to 9; ░ indicates abnormal patterns; and ■ plus *, a normal pattern.
Diversity of the TCRBV and IgH repertoires analyzed by CDR3 spectratyping.
The extent of TCRBV repertoire perturbation is represented as the percent difference between the patient's CDR3 distribution and the corresponding control distribution. A value of perturbation greater than the sum of the SDs relative to each CDR3 length found in normal blood donors is considered abnormal. Panels A to I show TCRBV data in patients nos. 1 to 9, respectively. The bars on the left of the doublets represent CD4 cells and the bars on the right, CD8 cells. ░ indicates, abnormal pattern in CD4 cells; ▪, abnormal pattern in CD8 cells; ■ plus *, normal pattern. Panel J represents IgH data in patients nos. 1 to 9; ░ indicates abnormal patterns; and ■ plus *, a normal pattern.
To gain further information on the TCRB and IgH repertoires as well as on the V(D)J joining process in A-T, we sequenced over 250 CDR3 regions from selected TCRBV and IgH amplification products from 3 patients (nos. 5, 8, and 9), and an equivalent number from a healthy donor.
The A-T patients displayed an oligoclonal repertoire of productively rearranged TCRBV genes (Figure 3). The most restricted repertoire was observed within CD4 Vβ16 from patient no. 5, where the repertoire diversity, expressed as the percentage ratio between the number of different sequences obtained and the total number of sequences, was 39%, compared to 98% observed in the healthy control. A somewhat lower variability was observed within CD4 Vβ14 from patient no. 5 (78% versus 96% in the control). The repertoire variability within CD8 Vβ14 of patient no.8 was about 41%; however, similarly to previous reports,31 32 also the healthy control showed a predominant clonotype resulting in a variability of about 75% (not shown).
Sequence analysis of TCRB and IgH V(D)J coding joints.
CDR3 sequences detected more than once in each patient's sample are shown. The corresponding spectratype profiles are shown in the left panels. The sequence data are available from EMBL/GenBank/DDBJ under accession numbers AJ437349-AJ437359, AJ437669-AJ437678, andAJ437623-AJ437634.
Sequence analysis of TCRB and IgH V(D)J coding joints.
CDR3 sequences detected more than once in each patient's sample are shown. The corresponding spectratype profiles are shown in the left panels. The sequence data are available from EMBL/GenBank/DDBJ under accession numbers AJ437349-AJ437359, AJ437669-AJ437678, andAJ437623-AJ437634.
The IgH coding joints were analyzed in 2 A-T patients (nos. 8 and 9) and in 1 healthy subject. Patient no. 8 had a relatively normal repertoire, with 91% variability compared to 98% in a healthy subject (data not shown). By contrast, patient no. 9 had a more restricted pattern displaying 62% variability (Figure 3).
Nearly all patients' TCRBV and IgH V(D)J coding joints sequenced were productively rearranged. We observed only occasional abnormally assembled coding joints (data not shown) that are being further investigated. The TCRB and IgH CDR3 average lengths were similar in A-T patients (6-13 amino acids) and in healthy subjects (6-15 amino acids), and corresponded to previously reported estimates.35-37Also, we did not notice any preferential usage of TCRB joining genes38 in A-T patients.
Discussion
Our present findings highlight several aspects of the immunodeficiency associated with A-T. We found that the TCRBV repertoire of A-T patients is skewed by either underusage or oligoclonal expansion of most Vβ families. TCR repertoire restriction is seen, in a much more severe form, in human V(D)J recombination disorders such as Omenn syndrome39,40 and severe combined immunodeficiency.41 Thus, our finding is compatible with an as yet unidentified, subtle recombination defect in A-T.
The putative recombination defect of A-T does not appear to involve coding joint formation, because we found perfectly normal CDR3 sequences derived from endogenous TCRβ and IgH rearrangements. Our findings confirm previous observations on the proficiency of A-T cells in supporting normal rearrangements of exogenous recombination substrates17 and on the normality of endogenously rearranged alleles of TCR translocations.42 Nevertheless, the observation that T-cell development can be rescued in ATM−/− mice by introducing a transgenic functional TCRαβ13 points to a defect in TCR recombination. The emerging picture of ATM function implies a role in sensing DNA breaks, including those related to V(D)J recombination, and triggering responses ranging from cell cycle arrest to apoptosis.43In this context, a failure of the ATM-dependent pathway might allow entering in the S phase before V(D)J recombination is completed. This would lead to the carryover of unresolved recombination intermediates and, consequently, to reduced frequency of productive V(D)J rearrangements11 and increased risk of oncogenic44 or nononcogenic33 translocations.
A striking finding in our study was that the TCRBV repertoire of A-T patients is skewed by diffuse oligoclonal expansions. Severe restriction of the TCR repertoire has been observed in patients with recombination defects due to RAG mutations,39,40 in patients with complete DiGeorge syndrome who have extremely low levels of T cells because of the absence of thymic epithelium,45and in patients infected with HIV with low CD4 cell counts.26 However, patients with partial DiGeorge syndrome (Pierdominici et al46; A.G., M.P., unpublished data, 2002), or HIV-infected patients at early stages of the disease who have a moderate reduction of CD4 T cells,26,47,48 have substantially normal CD4 TCR repertoires. Thus, it is likely that restriction of the TCR repertoire in A-T depends more on constraints of TCR generation rather than on a generalized decrease of thymopoiesis. Peripheral T-cell oligoclones probably originate from the peripheral expansion of the relatively few cells that achieve functional TCR rearrangements.39 The possibility that these expansions were due to chronic infections is unlikely, because antigenic stimulation predominantly drives the expansion of CD8 cells,31,32 whereas oligoclones predominated in CD4 cells of our A-T patients. Furthermore, oligoclonal expansions are not observed in chronically infected patients with other immune defects such as major histocompatibility complex (MHC) class II deficiency.49 The phenotypic and functional data discussed below argue in favor of this hypothesis.
We found that patients' circulating T cells are predominantly represented by terminally differentiated effector memory cells. Several studies have suggested that T and B lymphocytes from A-T patients have subtle defects in intracellular signal transduction,18-20suggesting that abnormal responsiveness to antigenic stimulation might account, at least in part, for immunodeficiency. Our finding that patients' T cells have the phenotype and the polarized pattern of cytokine production typical for effector memory cells strongly argues against this interpretation, because it implies that these cells must have responded properly to in vivo antigenic stimulations to become terminally differentiated effectors. Therefore, T-cell immunodeficiency in A-T is more likely to depend on a developmental defect rather than on defects of T-cell activation. This hypothesis is consistent with the low thymic output suggested by the dramatic reduction of TRECs in our patients. Although a decrease of TRECs might be the consequence of peripheral T-cell activation rather than of low thymic output,50 our finding is more likely to reflect the thymic hypoplasia typical for A-T patients and for ATM−/− mutant mice.
B-cell development has been investigated less thoroughly in ATM−/− mice. Peripheral B cells are numerically normal8-10 although pre-B cells are reduced.10 The fact that T-independent antibody responses are normal11 and T-dependent responses are rescued by an exogenous TCR15 suggests that B cells are functionally normal in ATM−/− mice. It must be noted that ATM−/− mice might have better preserved B-cell functions than A-T patients, because the former have nearly normal immunoglobulin levels,11 whereas the latter usually lack certain immunoglobin isotypes. To our knowledge, no information on the diversification of the B-cell repertoire in A-T patients or in ATM−/− mice is available. Our findings indicate that most A-T patients display a restricted IgH repertoire. Comparison with patients with DiGeorge syndrome, whose immunodeficiency is solely attributable to a thymic defect, suggests that oligoclonality of the B-cell repertoire in A-T is caused by a developmental defect. These B-cell oligoclonal expansions may underlie the high prevalence of oligo/monoclonal gammopathies observed in A-T patients.51
We are extremely grateful to A. M. R. Taylor for critical reading of the manuscript and to Grazia Andolfi for excellent technical assistance.
Prepublished online as Blood First Edition Paper, August 8, 2002; DOI 10.1182/blood-2002-03-0976.
Supported in part by the Fondazione Istituto Pasteur-Fondazione Cenci Bolognetti at the University of Rome “La Sapienza,” by the Italian Ministry of Health Progetto Finalizzato 2000, by the European Community Grant QLG2-CT-1999-00786, by Telethon grants D.102 and and E.0764, and by the Associazione Italiana Ricerca sul Cancro.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Antonello Giovannetti, Department of Clinical Medicine, University of Rome “La Sapienza,” Viale dell'Università 37, 00185 Rome, Italy; e-mail:antonello.giovannetti@uniroma1.it.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal