Eosinophils are effector cells that play an important role in the damage induced by the allergic process by releasing inflammatory mediators and proteolytic factors after activation. Stem cell factor (SCF) is a primary cytokine involved in hematopoiesis and mast cell differentiation, proliferation, and activation. Studies have also indicated that SCF is directly involved in pathogenesis of allergic airway inflammation. In the present study, we examined the ability of SCF to activate murine eosinophils for increased mediator release and up-regulation of chemokines. Initial data demonstrated that eosinophils have significant levels of surface c-kit protein, SCF receptor. SCF-activated eosinophils degranulate and release eosinophil peroxidase and leukotriene C4 in a dose-dependent manner. In addition, SCF was further shown to induce the release of CC chemokines, RANTES, macrophagederived chemokine (MDC), macrophage inflammatory protein-1β (MIP-1β), and C10 from eosinophils. To identify the extent of SCF-induced activation of eosinophils, we also performed gene array analysis using an array containing 1153 genes related to inflammation, including cytokines and their receptors, growth factors, structural and cytoskeletal genes, signal transduction genes as well as several other classes related to immune/inflammatory responses. The gene analysis indicated that more than 150 genes were significantly up-regulated in eosinophils after SCF stimulation. The gene array results were verified using a quantitative real-time polymerase chain reaction analysis to identify the expression of several chemokine and chemokine receptor genes. Altogether, these studies indicate that SCF is a potent eosinophil degranulator and activator that may play a number of roles during an inflammatory/immune response.
Introduction
The pathophysiologic changes that occur during allergic reactions have been associated with the recruitment, activation, and degranulation of eosinophils at the site of the response. Eosinophils are granulated leukocytes predominant in a number of allergic diseases, such as allergic asthma, allergic rhinitis, and atopic dermatitis.1-3 They play an important role as effector cells in parasite infections, with their ability to release cationic proteins, lipid mediators, cytokines, and chemokines that can directly cause damage to tissue or lead to the exacerbation of the inflammatory response. Although eosinophils are hypothesized to be a primary effector cell in diseases such as asthma, the activation of these cells is still not completely understood. Thus, comprehending which mediators are involved in eosinophil activation may be important for determining the therapeutic target for treatment of allergic diseases.
Stem cell factor (SCF), a primary cytokine involved in hematopoiesis, mast cell differentiation, and mast cell activation,4,5binds to its surface receptor, c-kit, which is a member of the receptor tyrosine kinase family. Previous data have identified that structural cells such as fibroblasts, airway epithelial cells, endothelial cells, and bronchial smooth muscle cells are able to produce SCF.6-10 Endogenous SCF occurs in both transmembrane and soluble forms11 and the relative importance of either form during hematopoiesis or disease is not known. Previously, SCF has been shown to activate the adhesion of eosinophils to fibronectin and vascular cell adhesion molecule 1 in vitro,12 suggesting that SCF may activate the eosinophil to become immobilized in the tissue compartment. Furthermore, SCF appears to play a significant role in eosinophil-associated inflammation in allergic airway inflammation.13 14 In the present study, we investigated whether SCF induces activation of eosinophils to release eosinophil peroxidase (EPO), leukotriene C4 (LTC4), and CC chemokines. The additional analysis by gene array highlighted the broad class of genes that were highly up-regulated by SCF in eosinophils. Many of the gene products have been shown to be important mediators involved in allergic and chronic/fibrotic inflammation and may facilitate a number of detrimental responses.
Materials and methods
Animals
Dr Fred Lewis at the Biomedical Research Laboratory (Rockville, MD) supplied Swiss Webster mice heavily infected withSchistosoma mansoni helminth parasite. These mice display significant peripheral eosinophilia with more than 50% of circulating granulocytes as eosinophils.
Eosinophil purification
Eosinophils were elicited by injection of thioglycolate plus soluble egg antigen (SEA) into the peritoneum of mice infected withS mansoni. SEA was prepared in our laboratory by grinding isolated eggs from mice heavily infected with S mansoni as previously described.15 This injection induces a pool of circulating eosinophils recruited into the peritoneum in an antigen-specific manner. After 48 hours the mice were peritoneally lavaged and the cells collected. The initial population that was isolated from the peritoneum was about 50% eosinophils with only 2% to 5% neutrophils and about 35% to 45% mononuclear cells (lymphocytes and macrophages). Adherent cell populations were removed from the population by plastic adherence in tissue culture dishes for 1 hour. The nonadherent cells were washed and resuspended in phosphate-buffered saline/bovine serum albumin (PBS/BSA; 90 μL PBS/BSA per 107 cells) and eosinophils were purified by negative immunomagnetic bead–coupled antibodies to exclude contaminating immune cells using the magnetic-activated cell sorting (MACS) system. The antibodies used were anti-Thy1 (for T cells), anti-B220 (for B cells), and anti–class II (for antigen-presenting cells [APCs]). After the plate adherence and MACS separation, the population of cells contained more than 97% eosinophils with contaminating neutrophils (∼1%) and mononuclear cells (1%-2%). At no time did we observe contaminating mast cells as assessed by direct differential staining or by toluidine blue granular staining after the final isolation procedure.
c-kit expression by flow cytometry
The flow cytometry analysis of eosinophils was performed to determine c-kit receptor expression. The procedure was performed on ice in Dulbecco phosphate-buffered saline (D-PBS) with 1% fetal bovine serum (FBS) and 0.09% sodium azide. A total of 1 × 106cells was used to block mouse Fc receptor with Fc block (1 μg/100 μL; Pharmingen, San Diego, CA) for 20 minutes at 4°C. Pelleted cells were fixed and permeabilized for 20 minutes at 4°C. Diluted polyclonal anti–c-kit or control antibodies were incubated for 30 minutes at 4°C. After incubation, cells were stained with an antimouse c-kit fluorescein isothiocyanate (FITC)–labeled antibody or isotype control (Pharmingen) for 30 minutes at 4°C. Cells were washed and the pelleted cells resuspended in D-PBS containing 1% FBS and 0.09% sodium azide until analyzed by flow cytometry.
Stimulation of eosinophils with murine recombinant SCF
Eosinophils (3 × 106 cells/well) were incubated in complete Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum (FCS) in the presence or absence of SCF (R & D Systems, Minneapolis, MN) in different concentrations (1, 10, and 100 ng/mL) at 37°C in 5% CO2 for 6 and 18 hours. After stimulation, cells were centrifuged and the cell-free supernatant recovered to measure CC chemokines.
Measurement of EPO or LTC4
To analyze EPO or LTC4 release from the eosinophils suspended in phenol red–free DMEM, cells were allowed to incubate for 4 hours in response to SCF. The supernatant from the activated cells (2 × 106 eosinophils) was harvested and the EPO level in the cell-free supernatant was determined as previously described.16 Briefly, o-phenylenediamine (OPD; 10 mg) was dissolved into 5.5 mL distilled H2O. Then, 1.5 mL OPD solution was added to 8.5 mL of a Tris (tris(hydroxymethyl)aminomethane) buffer (pH 8.0) followed by the addition of 7.5 μL H2O2. Using a 96-well plate, 100 μL substrate solution was added to 50 μL sample. After 30 minutes the reaction was quenched with 50 μL 4 M H2SO4 and the absorbance was read at 490 nm. The relative increase in samples was then compared. As a positive control, eosinophils (2 × 106 cells) were sonicated and measured as total EPO. A negative control was used by sonicating neutrophils (2 × 106).
The LTC4 levels were analyzed by specific enzyme-linked immunosorbent assay (ELISA) LTC4 kit (Cayman Chemical, Ann Arbor, MI) according to the manufacturer's instructions. The sensitivity of the LTC4 ELISA was 2 pg/mL.
Quantification of CC chemokines
Extracellular immunoreactive murine CC chemokines (RANTES, macrophage-derived chemokine [MDC], C10, and macrophage inflammatory protein 1β [MIP-1β]) were quantified using a modified double-ligand procedure of ELISA.17 This ELISA method consistently detected CC chemokine levels over 20 pg and did not cross-react with other cytokines. The 96-well flat-bottomed microtiter plates were coated with 50 μL/well of either rabbit anti-CC chemokine antibodies (1 μg/mL in 0.6 M NaCl, 0.26 M H3BO4, and 0.08 N NaOH, pH 9.6) for 16 hours at 4°C, and then washed (PBS, pH 7.5, 0.05% Tween 20). Blocking of nonspecific binding sites was accomplished by incubating plates with PBS containing 2% BSA for 90 minutes at 37°C. Plates were rinsed thoroughly with wash buffer and aqueous samples were added. Following a 1-hour incubation at 37°C, plates were washed and biotinylated rabbit anti-CC chemokine antibody was added and incubated for 30 minutes at 37°C. Plates were then washed and chromogen substrate added, and they were subsequently read at 490 nm.
Immunoarray gene selection
cDNAs were chosen on a logical basis from IMAGE consortium clones (http://genome.wustl.edu/est/est_general/est_project_intro.html). Clones were obtained from a master set of approximately 15 000 sequence-verified, T1 phage–negative cDNA clones obtained from Research Genetics (Huntsville, AL), a commercial supplier of IMAGE consortium clones. Individual clones were selected if they were considered to play an important role in immune function or regulation, the development in the immune system, intracellular signaling, or programmed cell death. cDNA clones for a small number of genes were produced in our laboratory by polymerase chain reaction (PCR) amplification and the insert sequenced. The list of cDNA elements of the Immunoarray is publicly available at the following Web address: http://www.grc.nia.nih.gov/branches/rrb/dna/array.htm.
Insert amplification and cDNA microarray printing
Clones were grown in deep 96-well plates in Terrific Broth containing 200 mg/mL ampicillin. After overnight growth, plasmid DNA was prepared using a commercial 96-well kit (Qiagen, Valencia, CA). Clone inserts, generally between approximately 350 and 850 bp, were PCR amplified using a standard pair of forward 5′-CTGCAAGGCGATTAAGTTGGGTAAC-3′ and reverse 5′-GTGAGCG GATAACAATTTCACACAGGAAACAGC-3′ (Research Genetics, Carlsbad, CA) primers. The quality and quantity of the PCR products were accessed by running each PCR product on a 2% agarose gel. PCR products were denatured with 0.1 M NaOH and spotted onto Nytran + Supercharged nylon membranes (Schleicher and Schuell, Dassel, Germany) using a GMS417 Microarrayer (Affymetrix, Woburn, MA). Two replicates of 1152 cDNA clones were printed on each filter for a final feature number of 2304 spots per array. Each membrane had a 2.5 × 7.5-cm total dimension. The diameter of the spotting pin was 300 mM. Membranes were UV cross-linked using a Stratagene Stratalinker at 60 mJ (Stratagene, La Jolla, CA).
Hybridization
Immunoarrays were prehybridized in 4 mL Microhyb hybridization buffer (Research Genetics), containing 10 mL of 10 mg/mL human Cot I DNA (denatured at 95°C for 5 minutes prior to use; Life Technologies, Carlsbad, CA) and 10 mL of 8 mg/mL poly(dA) (denatured at 95°C for 5 minutes prior to use; Research Genetics). One to 5 mg total RNA was labeled with [33P]deoxycytidine triphosphate (dCTP) by reverse transcription (RT) for each time point or treatment. After 4 hours of prehybridization at 42°C, approximately 107cpm/mL of heat-denatured probe was added to each prehybridization mix, followed by 18 hours of further incubation at 42°C in a roller oven. Hybridized arrays were rinsed in 50 mL of 2 times standard sodium citrate (SSC) and 1% sodium dodecyl sulfate (SDS) twice at 55°C followed by 1 to 2 times of washing in 2 times SSC and 0.1% SDS at 55°C for 10 minutes. The microarrays were exposed to phosphorimager screens for 1 to 3 days. The screens were then scanned in a Molecular Dynamics STORM PhosphorImager (Sunnyvale, CA) at 50-μm resolution. Gene expression was determined from microarray experiments by capturing the pixel density (ie, volume) of each spot using ImageQuant 5.1 Software (Molecular Dynamics).
Real-time RT-PCR analysis
Five micrograms total RNA from specific samples was reversed transcribed into cDNA using a prescribed reverse transcriptase kit from PE Biosystems. Primers and probe sets for RANTES, TARC, CxCR3, CxCR5, and GADPH have been predeveloped by PE Biosystems using a patented technique for optimal and specific amplification. Briefly, during PCR, a fluorogenic probe, consisting of an oligonucleotide with both a reporter and a quencher dye attached, anneals specifically between the forward and reverse primers. When the probe is cleaved by the 5′ nuclease activity of the DNA polymerase, the reporter dye is separated from the quencher dye and a sequence-specific signal is generated. With each cycle, additional reporter dye molecules are cleaved from their respective probes, and the fluorescence intensity is monitored during the PCR. This real-time detection generates quantitative data based on the PCR at early cycles when PCR fidelity is the highest. Just as important, the real-time PCR system has a linear dynamic range of at least 5 orders of magnitude, reducing the need for serial dilutions. These computer-linked operations, using the specialized software, make the quantitation of PCR products achievable and because each well has its own internal standard (GAPDH) with a different fluorescent dye marker, the product can be instantly quantitated and compared to other samples.
Statistical and cluster analysis
Measurement of transcript abundance was compared in 2 ways. Time-course samples were analyzed by directly comparing each sample relative to a time-matched control. The raw data of each gene were log10 transformed, followed by the calculation of fold increase in the intensity of the signal generated over the results from the control-treated cells. These calculations were then compared to the intensity of the bands to ensure that the relative fold increases were biologically significant and not due to aberrant background levels. Data from cellular activation experiments for mediator release were analyzed by ANOVA and significance was determined withP < .05. Eosinophil populations from 4 different preparations were analyzed for gene array expression at the 1 hour time point and 2 different preparations at the 6-hour and 18-hour time points. For the gene to be considered to be significantly up-regulated it was required to be activated more than 2.0-fold in all repeats that were probed. Gene expression raw data and log values were averaged using mean ± SD. For all tests, statistical significance was considered to be at the P < .05 level.
Results
Expression of c-kit on eosinophils
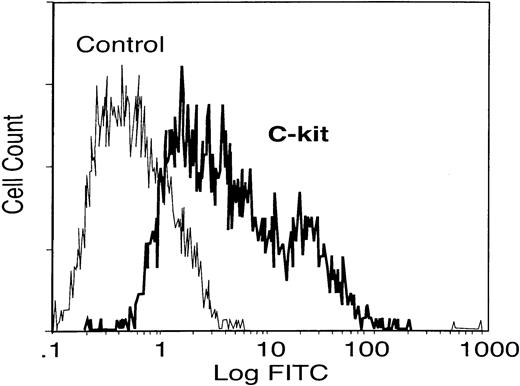
Mice infected with S mansoni were used to elicit and isolate eosinophils by intraperitoneal injection of SEAs isolated from parasite eggs. These mice have a high level of circulating eosinophils (> 50% of circulating granulocytes) as previously described.15 The peritoneal cells were harvested after 48 hours and eosinophils were isolated to about 97% purity using magnetic bead separation. To investigate whether the eosinophils can express c-kit receptor, flow cytometry analysis was used. The results in Figure1 indicate that eosinophils were strongly positive for c-kit surface protein, SCF receptor, as previously reported.12
c-kit receptor expression on elicited eosinophils.
Elicited eosinophils from peritoneum of mice infected with S mansoni were collected and the c-kit receptor expression was determined by flow cytometry analysis. Data are representative of similar results from different experiments.
c-kit receptor expression on elicited eosinophils.
Elicited eosinophils from peritoneum of mice infected with S mansoni were collected and the c-kit receptor expression was determined by flow cytometry analysis. Data are representative of similar results from different experiments.
Detection of EPO, LTC4, and chemokine release in SCF-stimulated eosinophils
Eosinophils are important effector cells that need to be activated to release their inflammatory mediators and carry out their functions. To investigate the role of SCF in eosinophil degranulation and activation, EPO and LTC4 release were assessed. EPO is an eosinophil granule protein released after cell activation and is one of several granular enzymes that are released from activated eosinophils that can lead to tissue damage. We observed that SCF induced EPO release in a dose-dependent manner in 4-hour stimulation cultures (Figure 2). These data suggest that SCF has the ability to induce eosinophil degranulation and may have a role in mediating eosinophil activation leading to tissue damage.
Degranulation of eosinophils by SCF.
Isolated eosinophils from mice heavily infected with S mansoni were subjected to activation with SCF at various concentrations (1-100 ng/mL). Culture supernatants were harvested at 4 hours after activation and assayed for EPO levels. Data represent mean ± SEM of 3 repeat activations. *P < .05.
Degranulation of eosinophils by SCF.
Isolated eosinophils from mice heavily infected with S mansoni were subjected to activation with SCF at various concentrations (1-100 ng/mL). Culture supernatants were harvested at 4 hours after activation and assayed for EPO levels. Data represent mean ± SEM of 3 repeat activations. *P < .05.
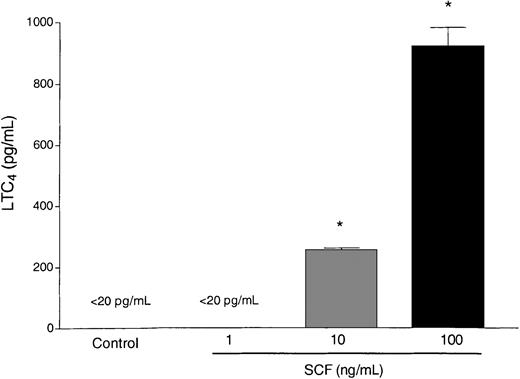
A set of mediators that has been closely linked to the pathophysiology changes in asthma are the cysteinyl leukotrienes. Eosinophils have the capacity to synthesize lipid-derived mediators, including LTC4 after activation of the lipoxygenase pathway. We observed that SCF (10 and 100 ng/mL) was able to stimulate eosinophils to release LTC4 by 4 hours after SCF stimulation (Figure3). Only minimal levels of LTC4 were found at 1 hour after activation (data not shown), indicating that LTC4 was generated de novo. Taken together these data suggest that SCF can induce eosinophil degranulation as well as initiate the activation of the lipoxygenase pathway leading to the release of LTC4. These events may have a significant impact on exacerbating airway damage and inflammation and prolong airway reactivity in lungs of individuals with asthma.
Activation of eosinophils by SCF.
Isolated eosinophils from mice heavily infected with S mansoni were subjected to activation with SCF at various concentrations (1-100 ng/mL). Culture supernatants were harvested at 4 hours after activation and assayed for LTC4 levels. Data represent mean ± SEM of 3 repeat activations. *P < .05.
Activation of eosinophils by SCF.
Isolated eosinophils from mice heavily infected with S mansoni were subjected to activation with SCF at various concentrations (1-100 ng/mL). Culture supernatants were harvested at 4 hours after activation and assayed for LTC4 levels. Data represent mean ± SEM of 3 repeat activations. *P < .05.
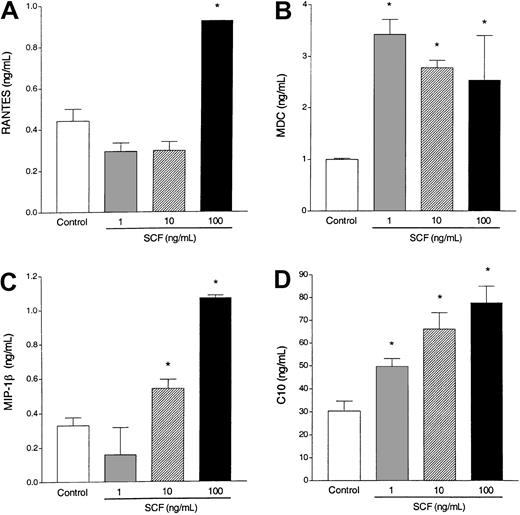
Eosinophils have previously been shown to release a number of cytokines and chemokines after specific stimulation. To investigate whether SCF is able to activate eosinophils to produce and release CC chemokines, various concentrations of SCF were used to activate the eosinophils. We observed that SCF-stimulated eosinophils differentially released significant levels of RANTES, MDC, MIP-1β, and C10 (Figure4). These chemokines have been shown to be involved in allergic eosinophilic airway responses. Thus, eosinophils stimulated by SCF may be an important source of CC chemokine production that contributes to the overall inflammation during an allergic response.
Chemokine induction.
Incubation of elicited eosinophils with SCF (1-100 ng/mL) induces RANTES (A), MDC (B), MIP-1β (C), and C10 (D) production by 6 hours (A,C) and 18 hours (B,D). Data represent the means ± SEM of triplicate cultures from one typical experiment. Similar data were obtained in 2 other experiments. * P < .001.
Chemokine induction.
Incubation of elicited eosinophils with SCF (1-100 ng/mL) induces RANTES (A), MDC (B), MIP-1β (C), and C10 (D) production by 6 hours (A,C) and 18 hours (B,D). Data represent the means ± SEM of triplicate cultures from one typical experiment. Similar data were obtained in 2 other experiments. * P < .001.
Gene array analysis of SCF activation of eosinophils
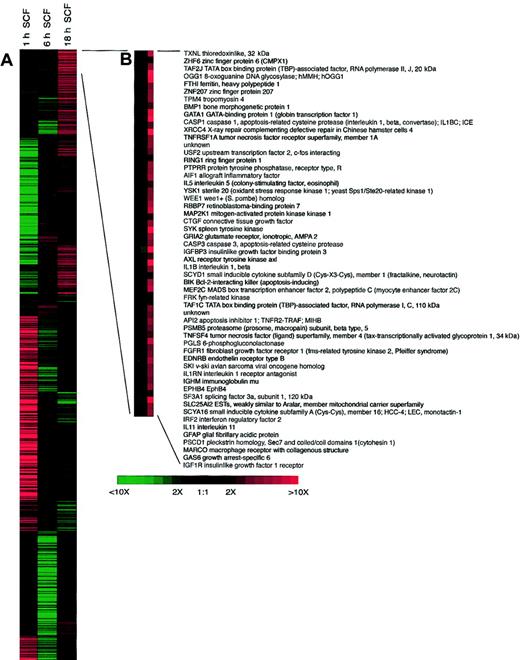
The ability to analyze the activation of a large number of genes has recently been an area of great interest for possibly identifying new targets of research and activation patterns of stimulated cells. We have begun to investigate how SCF can regulate specific families of genes previously associated with disease progression using gene array technology. In our analysis, we investigated distinct families of genes in a temporal fashion after SCF activation. Our data in Table1 indicate that SCF is a potent regulator of eosinophil gene expression, especially at 1 hour after activation. It is also very informative to examine the temporal expression of many of these genes and get a sense of downstream effects of SCF stimulation of the eosinophils. We have displayed some of the more interesting data in Table 1 that link known or hypothesized functions of eosinophils with disease progression. Interestingly, some of the genes seem to be differentially activated at the different time points, suggesting that there is likely a secondary response to the primary activation. A number of chemokines and their receptors are up-regulated by SCF in eosinophils, relating to the above data on chemokine protein release. Although there was not a significant increase in the expression of any interleukin (IL) gene, there were receptors, such as IL-11RA and IL-2RG that were significantly up-regulated, suggesting that SCF stimulation may allow the eosinophils to be more susceptible to regulation by these factors. The up-regulation of fibroblast growth factor 5 (FGF5), FGF7, and transforming growth factor β3 (TGFβ3) gene expression 1 hour after SCF stimulation demonstrates that SCF may have a role in activation of eosinophils during resolution and fibrosis. Additionally, a number of adhesion and costimulation molecules were also up-regulated, including specific α and β integrins. Altogether, we found more than 100 genes that were up-regulated by SCF by greater than 3-fold at 1 hour (many of which are not listed) indicating the potent activating role of SCF on eosinophils. The future experiments will focus on key aspects of particular genes and move to our in vivo models of allergic responses to address the impact of SCF on the expression patterns of particular genes. The fact that we observed extremely high expression of some genes compared with controls, such as RANTES, may not necessarily translate into similar increases in protein. Thus, specific data will always need to be carefully analyzed. The most highly expressed genes after 18 hours of SCF activation are expanded in Figure5 and demonstrate the differences in gene families compared with those listed in Table 1, which were highly up-regulated at 1 hour after SCF stimulation. Although we cannot completely rule out that some of the contaminating cells, such as macrophages (1%-2%), are participating in the response, our data from additional studies indicate that peritoneal macrophages are not directly activated by SCF (data not shown).
Gene array analysis of SCF-stimulated eosinophils
| Gene . | Fold expression over control . | ||
|---|---|---|---|
| 1 h* . | 6 h . | 18 h . | |
| Chemokines | |||
| CCL2/MCP-1 | 2.1-3.8 | 0.849 | 0.887 |
| CCL4/MIP-1β | 2.0-4.0 | 0.252 | 1.071 |
| CCL5/RANTES | 120-235 | 0.973 | 0.895 |
| CCL17/TARC | 2.2-25 | 0.79 | 0.84 |
| CCL21/SLC | 7.01-24.9 | 0.81 | 1.30 |
| CL22/MDC | 2.01-2.8 | 0.49 | 1.55 |
| CCR2 | 3.2-160 | 0.78 | 0.269 |
| CxCR3 | 2.69-3.5 | 0.65 | 2.48 |
| CxCR5 | 3.8-5.7 | 1.02 | 0.56 |
| IL receptors | |||
| IL-2RG | 6.1-17.9 | 0.79 | 1.99 |
| IL-11RA | 3.26-3.9 | 0.417 | 1.04 |
| Growth factors | |||
| FGF5 | 3.07-3.5 | 0.91 | 3.65 |
| FGF7 (KGF) | 2.2-4.1 | 0.634 | 1.84 |
| PDGFA | 8.5-25.6 | 1.27 | 0.41 |
| TGFβ3 | 4.7-9.2 | 0.70 | 0.69 |
| Signal transduction and promoter | |||
| MAPK4 | 3.4-13.7 | 0.58 | 0.73 |
| MAPK11 | 2.6-3.5 | 1.54 | 2.89 |
| STAT5A | 3.9-10.8 | 0.68 | 2.19 |
| STAT6 | 12.2-175.6 | 0.98 | 0.84 |
| Adhesion molecules | |||
| ITGA2 (VLA4, CD49B) | 3.7-8.1 | 0.94 | 1.07 |
| ITGAM (CD11B) | 2.02-3.5 | 0.87 | 2.935 |
| ITGB1 (CD29) | 3.5-6.8 | 0.46 | 1.76 |
| ITGB2 (CD18) | 2.4-2.6 | 0.63 | 1.67 |
| ITGB4 (β4) | 6.0-15.1 | 0.87 | 2.12 |
| Gene . | Fold expression over control . | ||
|---|---|---|---|
| 1 h* . | 6 h . | 18 h . | |
| Chemokines | |||
| CCL2/MCP-1 | 2.1-3.8 | 0.849 | 0.887 |
| CCL4/MIP-1β | 2.0-4.0 | 0.252 | 1.071 |
| CCL5/RANTES | 120-235 | 0.973 | 0.895 |
| CCL17/TARC | 2.2-25 | 0.79 | 0.84 |
| CCL21/SLC | 7.01-24.9 | 0.81 | 1.30 |
| CL22/MDC | 2.01-2.8 | 0.49 | 1.55 |
| CCR2 | 3.2-160 | 0.78 | 0.269 |
| CxCR3 | 2.69-3.5 | 0.65 | 2.48 |
| CxCR5 | 3.8-5.7 | 1.02 | 0.56 |
| IL receptors | |||
| IL-2RG | 6.1-17.9 | 0.79 | 1.99 |
| IL-11RA | 3.26-3.9 | 0.417 | 1.04 |
| Growth factors | |||
| FGF5 | 3.07-3.5 | 0.91 | 3.65 |
| FGF7 (KGF) | 2.2-4.1 | 0.634 | 1.84 |
| PDGFA | 8.5-25.6 | 1.27 | 0.41 |
| TGFβ3 | 4.7-9.2 | 0.70 | 0.69 |
| Signal transduction and promoter | |||
| MAPK4 | 3.4-13.7 | 0.58 | 0.73 |
| MAPK11 | 2.6-3.5 | 1.54 | 2.89 |
| STAT5A | 3.9-10.8 | 0.68 | 2.19 |
| STAT6 | 12.2-175.6 | 0.98 | 0.84 |
| Adhesion molecules | |||
| ITGA2 (VLA4, CD49B) | 3.7-8.1 | 0.94 | 1.07 |
| ITGAM (CD11B) | 2.02-3.5 | 0.87 | 2.935 |
| ITGB1 (CD29) | 3.5-6.8 | 0.46 | 1.76 |
| ITGB2 (CD18) | 2.4-2.6 | 0.63 | 1.67 |
| ITGB4 (β4) | 6.0-15.1 | 0.87 | 2.12 |
The fold increase over control eosinophils was calculated from repeat experiments where eosinophils were activated with SCF (100 ng/mL) for 1, 6, or 18 hours. The 1-hour time point was run a total of 4 times, whereas the 6- and 18-hour time points were analyzed only twice. The range of readings is given for only the 1-hour time point and the mean of the repeats is presented for the 6- and 18-hour time points.
CCL2 indicates C-C chemokine receptor ligand 2; MCP-1, membrane-associated cysteine protease–1; RANTES, regulated and normal T-cell expressed and presumably secreted; TARC, thymus and activation-regulated chemokine; SLC, secondary lymphoid-tissue chemokine; PDGFA, platelet-derived growth factor; MAPK4, mitogen-activated protein kinase; STAT5A, signal transducer and activator of transcription 5A; IGTA2, integrin A2; and VLA4, very-late activation protein A4.
All values at 1 hour were P < .05.
Clustergram of 1153 IMAGE Consortium ESTs differentially expressed during treatment eosinophils with 100 ng/mL SCF.
Columns correspond to the time in culture and gene expression profiles are in rows. Red indicates transcriptional activation; and green, repression compared with untreated control cells cultured for the same time periods. (B) Enlargement of clusters for the most highly up-regulated genes following SCF treatment.
Clustergram of 1153 IMAGE Consortium ESTs differentially expressed during treatment eosinophils with 100 ng/mL SCF.
Columns correspond to the time in culture and gene expression profiles are in rows. Red indicates transcriptional activation; and green, repression compared with untreated control cells cultured for the same time periods. (B) Enlargement of clusters for the most highly up-regulated genes following SCF treatment.
To further verify that our gene array was not overestimating the expression of genes analyzed, we further assessed selected genes by quantitative real-time PCR procedures. As previously described, freshly isolated eosinophils (> 97% pure) were stimulated with SCF for 1 and 18 hours and the isolated mRNA was analyzed using specific predeveloped probe/primer sets for representative chemokines and chemokine receptors. The data in Figure 6 indicate that both the chemokines (RANTES, MDC, and TARC) and chemokine receptors (CxCR3 and CxCR5) demonstrated similar regulation at 1 and 18 hours after stimulation as observed by the gene array analysis. In addition, the magnitude of the up-regulation appeared to be greater than the gene array had indicated. This was especially observed with RANTES, MDC, and TARC, which had more than 1200-, 8.8-, and 230-fold increase over unstimulated control eosinophils compared with the approximate 235-, 2.0-, and 25-fold increase, respectively, as assessed by gene array. Thus, the overall analysis by gene array may actually underestimate the level of gene expression, but can be used to indicate the most highly up-regulated genes.
Real-time PCR analysis of specific chemokines and chemokine receptors in SCF-stimulated eosinophils.
Isolated eosinophils were cultured in the presence or absence of SCF (100 ng/mL) and the mRNA isolated as described. Specific predeveloped primer/probe pairs were used to examine the expression level of RANTES, TARC, CxCR3, and CxCR5, which were used as representative genes to assess the accuracy of the gene array system. Data represent mean of 3 repeat analyses. * indicates statistical significance atP < .05.
Real-time PCR analysis of specific chemokines and chemokine receptors in SCF-stimulated eosinophils.
Isolated eosinophils were cultured in the presence or absence of SCF (100 ng/mL) and the mRNA isolated as described. Specific predeveloped primer/probe pairs were used to examine the expression level of RANTES, TARC, CxCR3, and CxCR5, which were used as representative genes to assess the accuracy of the gene array system. Data represent mean of 3 repeat analyses. * indicates statistical significance atP < .05.
Discussion
SCF is a known inducer of mast cell proliferation, mast cell degranulation, and matrix protein adhesion and is required for inhibition of mast cell apoptosis.5,18-24 Our previous studies have identified that SCF production during allergic inflammation can contribute significantly to the induction of the eosinophilic inflammatory responses and the airway hyperreactivity via direct mast cell activation.13,14,25 The results from this study suggest that SCF can play an important role in eosinophil activation, inducing degranulation, leukotriene, and CC chemokine production. The finding that eosinophils express a functional c-kit receptor for SCF reveals that activation of eosinophils via this pathway during allergic responses can be an important mechanism to perpetuate the inflammatory process. A previous seminal study has demonstrated that human peripheral blood eosinophils express a functional c-kit receptor that allows better adhesion and migration.12 The activation of eosinophils by SCF may have implications during multiple diseases, including allergic and fibrotic diseases where eosinophil infiltration is often coincident with severe end-stage disease.10,26-28 Interestingly, in vitro analysis indicates that eosinophil interaction with fibroblast, which can express high levels of surface SCF, can increase eosinophil survival and activation.29-33 Thus, SCF-induced eosinophil activation may play a complimentary role in the progression of multiple inflammatory diseases locally within inflamed tissue. In this study we used antigen-elicited eosinophils from parasite-infected animals that may be representative of an activated environment normally encountered by an infiltrating eosinophil. Although the eosinophils were removed from the infected animals, we cannot discount that the parasite infection itself altered the reactivity of the eosinophils. However, the level and number of genes that were up-regulated by SCF, especially at one hour after stimulation, indicated that SCF is a significant stimulus for eosinophil activation and mediator release. For example, the up-regulation of fibroblast growth factors, such as FGF5 and FGF7 (KGF), may lead to examination of novel interactions between SCF-producing structural cells and eosinophils. The up-regulation of integrins is consistent with an original publication demonstrating up-regulation of eosinophil adherence events after SCF stimulation.12 Over the years, our laboratory has been interested in the role of chemokines and their receptors during disease progression. These initial gene expression studies have indicated novel expression of a number of genes related to inflammatory and activation events, including CxCR3 and CxCR5 that had not previously been described. The expression of these chemokine receptors was verified by quantitative real-time PCR analysis. A previous study has also examined gene expression in eosinophils after IL-5 activation and found that a number of genes related to survival, adhesion, and migration were up-regulated, but concentrated primarily on the survival/apoptosis pathways.34 Thus, these data together will allow further investigations into a number of novel areas related to SCF activation of eosinophils and the association to disease progression.
The ability of SCF/c-kit interactions to increase eosinophil adherence12 may serve as one of many initial stimuli for eosinophil migration into inflamed tissue. Once the eosinophil has migrated into the inflamed tissue, it needs to be activated to release inflammatory mediators. In the present studies, SCF was able to induce eosinophil degranulation and activate up-regulation of additional chemokine receptors, and, therefore, may be important for eosinophil migratory and tissue stasis functions. These aspects likely have a direct impact on airways disease progression. The subsequent release of mediators, such as leukotrienes and EPO, has a role in the airway hyperreactivity response and tissue damage. LTC4 has previously been identified as an inducer of airway hyperreactivity.35-38 Recently, we have found that SCF can directly induce airway hyperreactivity when instilled into the airways of normal mice via the activation of mast cells and release of cysteinyl leukotrienes, LTC4, D4, and E4.39 The fact that SCF also appears to be an important eosinophil activator for LTC4 production suggests that SCF may be an excellent target for therapy in patients with allergic asthma.
SCF was also shown to induce CC chemokine production and suggests that activation of these eosinophils may lead to increased inflammatory cell recruitment. The data in the present study have demonstrated that SCF-stimulated eosinophils can produce chemokines,39including RANTES, MIP-1β, MDC, and C10. These CC chemokines play a role in regulating leukocyte recruitment, cellular activation, and inflammatory mediator release.40,41 Interestingly, SCF up-regulated C10 production 20-fold higher than other CC chemokines examined. Although C10 was not one of genes analyzed by our gene array, C10 is a CC chemokine that acts as a macrophage and eosinophil chemoattractant and is a constitutive component of eosinophils.42 High levels of C10 have been observed after peritoneal or pulmonary inflammation, suggesting that this CC chemokine plays an important role during inflammatory processes.43-45 In addition to C10, SCF-stimulated eosinophils produced significant levels of MDC that play an important role in inflammation and airway hyperreactivity in models of allergic asthma.46 MDC is a recruitment factor for multiple leukocyte populations, including dendritic cells, monocytes, Th2 lymphocytes, and eosinophils, all important in the allergic/asthmatic response.47-53 Thus, the activation and production of these chemokines from eosinophils may constitute a significant mechanism for maintaining and intensifying the inflammatory response.
In conclusion, the present study has demonstrated an important role for SCF mediating eosinophil release of EPO, LTC4, and CC chemokines. Furthermore, SCF up-regulated a significant and diverse array of inflammation-related gene expression suggesting a significant role of SCF in the progression of eosinophil-related disease. Thus, blocking SCF may be an important approach for targeting therapy in allergic disease.
We thank Pam Lincoln and Holly Evanoff for technical assistance. The schistosome-infected mice were graciously supplied by Dr Fred Lewis at the Laboratory of Biomedical Research Institute.
Supported by National Institutes of Health grants, HL59178, AI36302, and HL31963. S.H.P.O. is a postdoctoral fellow of the Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP), Brazil.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Nicholas W. Lukacs, University of Michigan Medical School, Department of Pathology, 1301 Catherine St, Ann Arbor, MI 48109; e-mail: nlukacs@umich.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal