We analyzed prospectively 1213 adults with de novo acute myeloid leukemia (AML) to ascertain the prognostic impact of cytogenetic abnormalities on complete remission (CR) rate, 5-year cumulative incidence of relapse (CIR), and 5-year overall survival (OS). All patients received similar induction therapy. Median follow-up for surviving patients was 8.3 years. Nonprioritized cytogenetics distinguished t(8;21) and inv(16)/t(16;16) as conferring a significantly better prognosis than normal karyotype. Prognostic impact of many abnormalities could not be determined independently because of their association with complex karyotype. Neither complex karyotype nor secondary aberrations affected outcome of patients with t(8;21), inv(16)/t(16;16), or t(9;11). Among other patients, those with complex karyotypes had significantly worse outcomes than cytogenetically normal patients. Based on outcome for specific cytogenetic abnormalities and karyotype complexity, patients were divided into 3 risk groups: favorable (CR 88%, CIR 54%, OS 55%), intermediate (CR 67%, CIR 67%, OS 24%), and adverse (CR 32%, CIR 92%, OS 5%). Multivariate analyses confirmed the major contribution of cytogenetics to the probability of attaining CR, CIR, and OS. For the adverse-risk group, the probability of achieving CR was 4.0 and 11.9 times lower, the probability of relapse 3.0 and 4.4 times higher, and the risk of death 2.1 and 4.3 times higher than those for the intermediate and favorable groups, respectively. We conclude that although the prognostic impact of many recurring abnormalities has not been ascertained independently of complex karyotype, cytogenetics is among the most useful factors predicting attainment of CR, CIR, and long-term survival in adult AML.
Introduction
Acute myeloid leukemia (AML), once recognized as a single disease, is a heterogeneous disorder with regard to morphology and chromosome aberrations detected in the leukemic cells. The concept of classifying AML according to pretreatment karyotype has recently become acceptable to most leukemia investigators. This is based on the ability of karyotype to predict response to induction therapy, relapse risk, and overall survival (OS).1-15 Treatment of certain cytogenetic subsets, such as patients with t(15;17)(q22;q21) using all-trans retinoic acid (ATRA)16-19 and patients with t(8;21)(q22;q22) or inv(16)(p13q22)/t(16;16)(p13;q22) using repetitive doses of high-dose cytarabine, has resulted in markedly improved outcome.10,12 In contrast, patients with other aberrations, such as −5/del(5q), −7, abnormalities of 3q, or a complex karyotype, have been shown to have inferior complete remission (CR) rates and OS.1-8,10,11,13-15 These observations have led to a recently published World Health Organization classification of AML20 21 that specifically separates patients with cytogenetic (or molecular) evidence of t(8;21), inv(16)/t(16;16), t(15;17), and 11q23 abnormalities into specific disease subsets.
However, despite publication of large AML cytogenetic series,11,13 15 questions still remain regarding the prognostic significance of some recurrent aberrations, for example, +8, del(9q), or translocations involving band 11q23 and specific partner chromosomes. Similarly, the importance of secondary abnormalities in patients with t(8;21), inv(16)/t(16;16), and t(9;11)(p22;q23) as their primary chromosome aberrations is debatable, and it is still unclear which definition of a complex karyotype—that is, 3 or more as opposed to 5 or more abnormalities in a leukemic clone—is more relevant clinically.
To identify patients at highest risk for induction failure, relapse, and shortened OS who might be candidates for novel treatment approaches, we analyzed data from a prospective cytogenetic study performed by Cancer and Leukemia Group B (CALGB). We describe the outcome of 1213 adults with de novo AML categorized into cytogenetic groups comprising at least 5 cases. We also propose a stratification system that categorizes AML patients into favorable, intermediate, and adverse risk groups with regard to probability of achieving CR, remaining in remission, and long-term OS according to pretreatment cytogenetic findings.
Patients and methods
Patients
All patients included in this analysis were enrolled on CALGB 8461, a prospective cytogenetics trial initiated in 1984.22 Consecutive patients who had the primary diagnosis of AML as defined by the French-American-British (FAB) classification23-25 and enrolled on 5 sequential treatment studies, 8221, 8525, 8923, 9022, and 9222, were examined for this analysis.26-30 The details of these trials are provided in Table 1. Pathological diagnoses were reviewed centrally. Patients with a prior history of myelodysplasia, other antecedent hematologic malignancies, prior nonsteroidal cytotoxic chemotherapy or radiation therapy, preexisting liver disease, or uncontrolled infection were excluded. Eligibility criteria for these studies were similar, with the exception that CALGB 9022 29 and CALGB 9222 30 excluded patients who were 60 years or older and those who had acute promyelocytic leukemia, while CALGB 8923 28 excluded patients under the age of 60. Patients with t(15;17) (n = 88) and t(9;22) (n = 10) have subsequently been excluded from the analysis because molecularly targeted therapy with ATRA or imatinib mesylate, respectively, is now considered a standard treatment option for these AML subtypes. Written informed consent was obtained from all patients according to the guidelines set forth by institutions participating in the study.
Summary of therapy administered
| Treatment details . | Treatment protocol (reference no.) . | |||||
|---|---|---|---|---|---|---|
| 8221 (Mayer et al26) . | 8525 (Mayer et al27) . | 8923 (Stone et al28) . | 9022 (Moore et al29) . | 9222 (Moore et al30) . | ||
| Accrual period for 8461 | 8/84–10/85 | 11/85–9/90 | 2/90–11/93 | 10/90–3/92 | 7/92–12/95 | |
| Induction 1 | ||||||
| Daunorubicin 45 mg/m2 ivp × 3 d | Yes | Yes | Yes* | Yes | Yes | |
| Cytarabine 200 mg/m2 by ci × 7 d | Yes | Yes | Yes | Yes | Yes | |
| Induction 2, if needed | ||||||
| Daunorubicin 45 mg/m2 ivp × 2 d | Yes | Yes | Yes | Yes | Yes | |
| Cytarabine 200 mg/m2 by ci × 5 d | Yes | Yes | Yes | Yes | Yes | |
| Intensification therapy randomized | No | Yes; arms A, B, C | Yes; arms A, B | No | Yes; arms A, B | |
| Intensification therapy 1 | HDAC or IDAC | A: SDAC | A: SDAC | HDAC | A: HDAC | |
| B: IDAC | B: M + IDAC | B: HDAC | ||||
| C: HDAC | ||||||
| Intensification therapy 2 | HDAC or IDAC | A: SDAC | A: SDAC | E + C | A: HDAC | |
| B: IDAC | B: M + IDAC | B: E + C | ||||
| C: HDAC | ||||||
| Intensification therapy 3 | HDAC or IDAC | A: SDAC | A: SDAC | M + D | A: HDAC | |
| B: IDAC | B: NA | B: M + D | ||||
| C: HDAC | ||||||
| Intensification therapy 4 | HDAC or IDAC | A: SDAC | A: SDAC | NA | NA | |
| B: IDAC | B: NA | |||||
| C: HDAC | ||||||
| Maintenance therapy | Yes | Yes | No | No | No | |
| Treatment details . | Treatment protocol (reference no.) . | |||||
|---|---|---|---|---|---|---|
| 8221 (Mayer et al26) . | 8525 (Mayer et al27) . | 8923 (Stone et al28) . | 9022 (Moore et al29) . | 9222 (Moore et al30) . | ||
| Accrual period for 8461 | 8/84–10/85 | 11/85–9/90 | 2/90–11/93 | 10/90–3/92 | 7/92–12/95 | |
| Induction 1 | ||||||
| Daunorubicin 45 mg/m2 ivp × 3 d | Yes | Yes | Yes* | Yes | Yes | |
| Cytarabine 200 mg/m2 by ci × 7 d | Yes | Yes | Yes | Yes | Yes | |
| Induction 2, if needed | ||||||
| Daunorubicin 45 mg/m2 ivp × 2 d | Yes | Yes | Yes | Yes | Yes | |
| Cytarabine 200 mg/m2 by ci × 5 d | Yes | Yes | Yes | Yes | Yes | |
| Intensification therapy randomized | No | Yes; arms A, B, C | Yes; arms A, B | No | Yes; arms A, B | |
| Intensification therapy 1 | HDAC or IDAC | A: SDAC | A: SDAC | HDAC | A: HDAC | |
| B: IDAC | B: M + IDAC | B: HDAC | ||||
| C: HDAC | ||||||
| Intensification therapy 2 | HDAC or IDAC | A: SDAC | A: SDAC | E + C | A: HDAC | |
| B: IDAC | B: M + IDAC | B: E + C | ||||
| C: HDAC | ||||||
| Intensification therapy 3 | HDAC or IDAC | A: SDAC | A: SDAC | M + D | A: HDAC | |
| B: IDAC | B: NA | B: M + D | ||||
| C: HDAC | ||||||
| Intensification therapy 4 | HDAC or IDAC | A: SDAC | A: SDAC | NA | NA | |
| B: IDAC | B: NA | |||||
| C: HDAC | ||||||
| Maintenance therapy | Yes | Yes | No | No | No | |
Maintenance therapy indicates daunorubicin 45 mg/m2by intravenous bolus (ivb) on day 1 plus cytarabine 100 mg/m2 subcutaneously (sq) twice daily on days 1 to 5 repeated monthly × 4; ivp indicates intravenous push; HDAC, cytarabine 3 g/m2 by ivb over 3 hours twice daily on days 1, 3, and 5; IDAC, cytarabine 400 mg/m2 by continuous infusion (ci) every day × 5 days; SDAC, cytarabine 100 mg/m2 by ci every day × 5 days; M + IDAC, mitoxantrone plus alternative IDAC; E + C, etoposide 1800 mg/m2 by ci on day 1 plus cyclophosphamide 50 mg/kg ivb on days 2 and 3; NA, not applicable; M + D, mitoxantrone 12 mg/m2 ivb plus diaziquone 24 mg/m2 by ci on days 1, 2, and 3 plus filgrastim 5 μg/kg sq on days 4 to 28.
Fifty percent of enrolled patients received GM-CSF during induction.
Cytogenetic studies
Cytogenetic analyses of bone marrow (BM) or blood were performed in institutional CALGB cytogenetics laboratories, and karyotypes were centrally reviewed biannually. Specimens were obtained at diagnosis from all patients and processed using unstimulated short-term (24-, 48-, and 72-hour) cultures, a direct method, or both. G-banding was usually done although Q-banding was also acceptable. The criteria used to describe a cytogenetic clone and description of karyotype followed the recommendations of the International System for Human Cytogenetic Nomenclature.31 At least 20 BM metaphase cells were analyzed in patients designated as having a normal karyotype. Pretreatment karyotype was not available in 410 patients because cytogenetic analyses were not performed (n = 67), failed, yielding no analyzable mitoses (n = 67), or were deemed inadequate on CALGB Central Karyotype Review because the quality of banded chromosomes was poor (n = 172), the normal result was obtained only from study of blood samples (no BM, n = 35) or from analyses of BM that yielded fewer than 20 metaphases from short-term cultures (n = 49), or for other reasons (n = 20).
Patients with adequate cytogenetics were grouped according to the presence of a recurrent abnormality noted in at least 5 patients, and clinical features and outcome were examined for each group. Hence, patients with 2 or more abnormalities may appear in more than one group. We observed in a number of patients that loss of material from the chromosome arms 5q, 7q, 17p, and 20q was due not only to deletions or the loss of the whole chromosome (monosomy) but also to various structural aberrations, for example, unbalanced translocations with a known partner chromosome, the presence of additional material of unknown origin (add), isochromosomes, and others. Because it is at present unknown whether clinical characteristics of patients with such aberrations are comparable to or different from those of patients with deletions of these chromosome arms, we categorized the patients with the aforementioned unbalanced aberrations other than del(5q), del(7q), and del(20q) separately and designated these groups as “loss of” material from the respective arm (eg, “loss of 5q”). To be included in a given karyotypic category, it was sufficient for the patient to have the aberration in only 1 of 2 or more clones identified in their karyotype. A complex karyotype was defined in our final risk stratification system as the presence of 3 or more chromosome abnormalities. Each reciprocal translocation was regarded as 1 abnormality. We also analyzed outcome data for patients with 3 or 4 abnormalities and for those with 5 or more abnormalities. Outcome of patients in each cytogenetic group was compared with the outcome of patients with a normal karyotype to identify groups that do better than, similar to, and worse than this well-recognized prognostic group.
Treatment
The therapies administered to patients in this study have been previously described and are summarized in Table 1.26-30Induction therapy was identical in each of the 4 trials for patients under the age of 60 years. Postremission therapy differed among the trials, although high-dose cytarabine was administered in a similar fashion in each of the trials. Following sequential cycles of cytarabine consolidation therapy, all patients enrolled on CALGB 8221 and 8525 were assigned to receive 4 cycles of standard-dose cytarabine and daunorubicin as previously described.26 27 During treatment, patients underwent a BM aspiration following completion of cytarabine consolidation and maintenance therapy. Thereafter, patients were followed with BM testing every 3 months for 1 year, every 6 months for 2 years, and then every year for 2 additional years. Patients were followed yearly after 5 years of remission, with BM examinations being performed only if the blood counts suggested relapse of AML.
Criteria for response and definition of relapse
A CR was defined as the presence of morphologically normal BM and at least 1.5 × 109/L granulocytes and 100 × 109/L platelets in the blood. Relapse was defined as at least 5% leukemic blasts in a BM aspirate or new extramedullary leukemia in patients with a previously documented CR as defined previously.32 Failure to attain CR was divided into categories of early death (death within 30 days, more likely representative of regimen-related toxicity) and late death (more than 30 days, more likely to be associated with resistant disease).
Statistical analyses
The major objectives of this study were to examine the relationship between pretreatment cytogenetics and clinical end points in a prospectively studied group of AML patients with prolonged follow-up. The main end points analyzed were CR rate, cumulative incidence of relapse (CIR), and OS. We also analyzed the outcome of patients with 3 or 4 cytogenetic abnormalities, with complex karyotype with at least 3 and at least 5 abnormalities, and those with and without secondary abnormalities accompanying t(8;21), inv(16)/t(16;16), and t(9;11). Exploratory analyses were performed on cytogenetic subgroups with at least 5 patients by comparing them with patients with a normal karyotype with respect to the main end points.
Patients were categorized into favorable, intermediate, and adverse risk groups with respect to CR rate, CIR, and OS. Patients with a normal karyotype were classified as intermediate risk. The classification process was then carried out by first identifying cytogenetic subgroups that were significantly associated with favorable outcome. Then, after removing those patients, the adverse risk group of patients with a complex karyotype with 3 or more abnormalities was identified, and these patients also were removed. Thereafter, the CR rates, CIR, and OS of remaining patients with other abnormalities were re-evaluated and compared with those of patients with a normal karyotype. The favorable risk group included patients with abnormalities conferring a significantly better CR rate, CIR, or OS (P < .05), and patients in the intermediate risk group had a similar (P ≥ .05) and those in the adverse group a significantly worse (P < .05) CR rate, CIR, or OS relative to karyotypically normal AML patients.
Comparison of the proportions of complete responders between each cytogenetic group and normal-karyotype patients was based on the Fisher 2-tailed exact test.33 Comparisons of median age and leukocyte count between different cytogenetic groups and normal-karyotype patients were based on the Wilcoxon rank sum test.33 The CIR analysis included only patients that achieved a CR with time calculated from date of CR until relapse. Patients alive without relapse were censored, whereas those who died without relapse were counted as a competing cause of failure. The CIR and its standard error (SE) was estimated by the method of Gray, and differences between groups were analyzed using a test developed by Gray.34 The estimation of OS distributions was performed using the Kaplan-Meier method,35 and the differences between groups were analyzed using the log-rank statistic.36 Ninety-five percent confidence intervals (CIs) for OS probabilities were calculated according to the method of Simon and Lee.37 For OS, an event was death from any cause with patients alive at last follow-up censored. The outcome data were current as of January 2002.
The relationship between clinical and laboratory factors and the probability of attaining CR was analyzed with the logistic regression model.38 The relationship between clinical and laboratory factors and CIR and OS was analyzed with the Cox regression model.39 The factors examined included treatment protocol, age (< 60, ≥ 60 years); sex (male, female); splenomegaly (no, yes); hepatomegaly (no, yes); infection at study entry (no, yes); cytogenetic risk group (favorable, intermediate, or adverse); and the following continuous variables: leukocyte count, percent marrow blasts, hemoglobin, and platelet count. For CIR and OS, the number of induction courses (1, 2) was also analyzed.
Results
Patients and treatment protocol outcome
Of a total of 1795 patients registered on CALGB 8461 and the concurrent treatment studies, 1385 (77%) had adequate cytogenetic results. However, 74 of these patients were ineligible due to not having AML (n = 58), for other reasons (n = 8), or were not treated on the respective treatment trial (n = 8). Additionally, patients with t(15;17) (n = 88) and t(9;22) (n = 10) were excluded as described in “Patients and methods,” leaving 1213 patients who are included in this report. The clinical features at presentation of these patients are shown in Table 2. This analysis includes patients treated over a decade, which introduces a possible confounding variable of different supportive care issues potentially altering the outcome results. Because treatment-related deaths due to infection and other acute leukemia complications are most common during induction, we compared the CR rates among the different trials. The CR proportions by treatment study for patients less than 60 years of age were similar (P = .83) as follows: CALGB 8221, 72%; CALGB 8525, 74%; CALGB 9022, 78%; and CALGB 9222, 75%. Similarly, the CR proportions by treatment study for patients aged at least 60 years were not significantly different (P = .05) as follows: CALGB 8221, 56%; CALGB 8525, 43%; and CALGB 8923, 54%. Figure 1 shows the OS for all patients included in this analysis. Finally, performance of stem cell transplantation (SCT) off-protocol in first CR or as part of salvage therapy could potentially confound analysis of OS. A total of 131 patients underwent SCT in first CR (n = 29) or following relapse (n = 102). Outcome for OS for the entire group of patients was similar irrespective of the inclusion or exclusion of patients who underwent SCT.
Clinical characteristics at presentation of 1213 patients with de novo AML
| Age, y | |
| Median | 52 |
| Range | 15-86 |
| % patients older than 60 | 36 |
| % female | 47 |
| Median WBCs, × 109/L | 18.2 |
| % patients with WBCs more than 50 × 109/L | 29 |
| % patients with WBCs more than 100 × 109/L | 12 |
| Median hemoglobin, g/dL | 9.2 |
| Median platelets, × 109/L | 57 |
| % hepatomegaly | 11 |
| % splenomegaly | 10 |
| % extramedullary leukemia | 10 |
| FAB, no. (%) | 1115 |
| M0 | 19 (2) |
| M1 | 189 (17) |
| M2 | 413 (37) |
| M3 | 18 (2) |
| M4 | 212 (19) |
| M4Eo | 47 (4) |
| M5 | 136 (12) |
| M6/M7 | 33 (3) |
| Unclassified | 48 (4) |
| % with at least grade 3 bleeding | 6 |
| % with at least grade 3 infection | 4 |
| No. of deaths in first 7 d | 25 |
| Second induction (% CR) | 349 (52) |
| Age, y | |
| Median | 52 |
| Range | 15-86 |
| % patients older than 60 | 36 |
| % female | 47 |
| Median WBCs, × 109/L | 18.2 |
| % patients with WBCs more than 50 × 109/L | 29 |
| % patients with WBCs more than 100 × 109/L | 12 |
| Median hemoglobin, g/dL | 9.2 |
| Median platelets, × 109/L | 57 |
| % hepatomegaly | 11 |
| % splenomegaly | 10 |
| % extramedullary leukemia | 10 |
| FAB, no. (%) | 1115 |
| M0 | 19 (2) |
| M1 | 189 (17) |
| M2 | 413 (37) |
| M3 | 18 (2) |
| M4 | 212 (19) |
| M4Eo | 47 (4) |
| M5 | 136 (12) |
| M6/M7 | 33 (3) |
| Unclassified | 48 (4) |
| % with at least grade 3 bleeding | 6 |
| % with at least grade 3 infection | 4 |
| No. of deaths in first 7 d | 25 |
| Second induction (% CR) | 349 (52) |
Overall survival of 1213 adult de novo AML patients enrolled on CALGB 8461 and included in this analysis.
Overall survival of 1213 adult de novo AML patients enrolled on CALGB 8461 and included in this analysis.
Frequency of karyotypic abnormalities in adults with de novo AML
Table 3 summarizes the absolute frequency of recurrent chromosome abnormalities among the 1213 patients studied. Of these patients, 582 (48%) had a normal karyotype, whereas 631 (52%) had 1 or more clonal abnormalities. The nonprioritized classification demonstrates that +8, inv(16)/t(16;16), t(8;21), −Y, −7, and del(5q) constitute the only recurring abnormalities with a frequency above 3%. In Table 3, we also provide the percentages of patients in whom each abnormality occurred as a sole chromosome change and of those in whom each abnormality was part of a complex karyotype with at least 3 or at least 5 abnormalities. These proportions vary greatly among cytogenetic groups. In general, specific reciprocal translocations—that is, t(6;9)(p23;q34), t(6;11)(q27;q23), t(8;21), t(9;11), t(11;19)(q23;p13.1), and inv(16)/t(16;16)—were seen less frequently as part of a complex karyotype, and most such rearrangements, except t(8;21) and t(9;11), occurred as the sole aberration in most patients carrying them. In contrast, unbalanced structural and numeric aberrations (except +11) were detected predominantly in conjunction with other aberrations, and several of them, including +4, −5/5q−, del(7q), loss of 7q, abn(12p), del(13q), +14, −17/17p−, −18, −20, and loss of 20q, were seen as part of a complex karyotype with 3 or more abnormalities in at least two thirds of patients in the respective groups. These data suggest that assessment of outcome measures (CR, CIR, and OS) of recurring abnormalities will have to consider the impact of complex karyotype.
Frequency and characteristics of nonprioritized cytogenetic subgroups
| Cytogenetic abnormality . | No. of patients (%) . | Median age (% younger than 60 y) . | No. of patients with a sole abnormality (%) . | No. of patients with 3 or more abnormalities (%) . | No. of patients with 5 or more abnormalities (%) . |
|---|---|---|---|---|---|
| Normal | 582 (48.0) | 54 (64) | NA | NA | NA |
| +8 | 123 (10.1) | 58 (54) | 41 (31) | 43 (35) | 20 (16) |
| inv(16) or t(16;16) | 96 (7.9) | 40 (84) | 64 (67) | 8 (8) | 1 (1) |
| −7/7q− | 95 (7.8) | 64 (38) | 17 (18) | 62 (65) | 47 (49) |
| −7 | 47 (3.9) | 65 (38) | 11 (23) | 26 (55) | 19 (40) |
| Loss of 7q | 29 (2.4) | 65 (28) | 3 (10) | 20 (69) | 17 (59) |
| del(7q) | 19 (1.6) | 59 (53) | 3 (16) | 16 (84) | 11 (58) |
| −5/5q− | 86 (7.1) | 66 (30) | 4 (5) | 77 (90) | 74 (86) |
| del(5q) | 42 (3.5) | 66 (31) | 1 (2) | 37 (88) | 34 (81) |
| −5 | 26 (2.1) | 64 (27) | 1 (4) | 25 (96) | 25 (96) |
| Loss of 5q | 18 (1.5) | 62 (33) | 2 (11) | 15 (83) | 15 (83) |
| t(8;21) | 81 (6.7) | 34 (90) | 22 (27) | 14 (17) | 2 (2) |
| −17/17p− | 62 (5.1) | 64 (34) | 0 (0) | 60 (97) | 58 (94) |
| −17 | 32 (2.6) | 66 (28) | 0 (0) | 30 (94) | 29 (91) |
| Loss of 17p | 30 (2.5) | 63 (40) | 0 (0) | 30 (100) | 29 (97) |
| −Y | 58 (4.8) | 49 (62) | 14 (29) | 17 (29) | 7 (12) |
| Balanced abn(11q23) | 54 (4.5) | 42 (83) | 27 (50) | 15 (28) | 4 (7) |
| t(9;11) | 27 (2.2) | 42 (89) | 12 (44) | 8 (30) | 3 (11) |
| t(11;v)3-150 | 13 (1.1) | 36 (77) | 5 (38) | 6 (46) | 1 (8) |
| t(6;11) | 8 (0.7) | 46 (100) | 5 (63) | 1 (13) | 0 (0) |
| t(11;19)(q23;p13.1) | 6 (0.5) | 57 (50) | 5 (83) | 0 (0) | 0 (0) |
| −20/20q− | 39 (3.2) | 60 (44) | 6 (15) | 29 (74) | 28 (72) |
| del(20q) | 16 (1.3) | 65 (44) | 5 (31) | 9 (56) | 8 (50) |
| −20 | 15 (1.2) | 60 (47) | 1 (7) | 12 (80) | 12 (80) |
| Loss of 20q | 8 (0.7) | 61 (38) | 0 (0) | 8 (100) | 8 (100) |
| del(9q) | 33 (2.7) | 37 (82) | 11 (33) | 10 (30) | 2 (6) |
| abn(12p) | 33 (2.7) | 64 (36) | 1 (3) | 25 (76) | 21 (64) |
| Unbalanced abn(12p) | 25 (2.1) | 64 (32) | 1 (4) | 20 (80) | 16 (64) |
| Balanced abn(12p) | 8 (0.7) | 61 (50) | 0 (0) | 5 (63) | 5 (63) |
| +22 | 31 (2.6) | 59 (55) | 1 (3) | 19 (61) | 14 (45) |
| −18 | 30 (2.5) | 64 (30) | 1 (3) | 29 (97) | 28 (93) |
| +13 | 29 (2.4) | 59 (55) | 13 (45) | 12 (41) | 10 (34) |
| +21 | 28 (2.3) | 59 (54) | 5 (24) | 14 (50) | 10 (36) |
| +11 | 20 (1.6) | 65 (20) | 12 (60) | 7 (35) | 6 (30) |
| −X | 18 (1.5) | 44 (67) | 0 (0) | 8 (44) | 5 (28) |
| +4 | 14 (1.2) | 55 (57) | 2 (14) | 10 (71) | 6 (43) |
| +14 | 12 (1.0) | 64 (42) | 0 (0) | 10 (83) | 9 (75) |
| del(11q) | 12 (1.0) | 42 (58) | 4 (33) | 5 (42) | 3 (25) |
| inv(3) or t(3;3) | 12 (1.0) | 43 (83) | 2 (17) | 4 (33) | 1 (8) |
| del(13q) | 9 (0.7) | 44 (56) | 1 (11) | 7 (78) | 5 (56) |
| t(6;9) | 8 (0.7) | 33 (100) | 6 (75) | 0 (0) | 0 (0) |
| Cytogenetic abnormality . | No. of patients (%) . | Median age (% younger than 60 y) . | No. of patients with a sole abnormality (%) . | No. of patients with 3 or more abnormalities (%) . | No. of patients with 5 or more abnormalities (%) . |
|---|---|---|---|---|---|
| Normal | 582 (48.0) | 54 (64) | NA | NA | NA |
| +8 | 123 (10.1) | 58 (54) | 41 (31) | 43 (35) | 20 (16) |
| inv(16) or t(16;16) | 96 (7.9) | 40 (84) | 64 (67) | 8 (8) | 1 (1) |
| −7/7q− | 95 (7.8) | 64 (38) | 17 (18) | 62 (65) | 47 (49) |
| −7 | 47 (3.9) | 65 (38) | 11 (23) | 26 (55) | 19 (40) |
| Loss of 7q | 29 (2.4) | 65 (28) | 3 (10) | 20 (69) | 17 (59) |
| del(7q) | 19 (1.6) | 59 (53) | 3 (16) | 16 (84) | 11 (58) |
| −5/5q− | 86 (7.1) | 66 (30) | 4 (5) | 77 (90) | 74 (86) |
| del(5q) | 42 (3.5) | 66 (31) | 1 (2) | 37 (88) | 34 (81) |
| −5 | 26 (2.1) | 64 (27) | 1 (4) | 25 (96) | 25 (96) |
| Loss of 5q | 18 (1.5) | 62 (33) | 2 (11) | 15 (83) | 15 (83) |
| t(8;21) | 81 (6.7) | 34 (90) | 22 (27) | 14 (17) | 2 (2) |
| −17/17p− | 62 (5.1) | 64 (34) | 0 (0) | 60 (97) | 58 (94) |
| −17 | 32 (2.6) | 66 (28) | 0 (0) | 30 (94) | 29 (91) |
| Loss of 17p | 30 (2.5) | 63 (40) | 0 (0) | 30 (100) | 29 (97) |
| −Y | 58 (4.8) | 49 (62) | 14 (29) | 17 (29) | 7 (12) |
| Balanced abn(11q23) | 54 (4.5) | 42 (83) | 27 (50) | 15 (28) | 4 (7) |
| t(9;11) | 27 (2.2) | 42 (89) | 12 (44) | 8 (30) | 3 (11) |
| t(11;v)3-150 | 13 (1.1) | 36 (77) | 5 (38) | 6 (46) | 1 (8) |
| t(6;11) | 8 (0.7) | 46 (100) | 5 (63) | 1 (13) | 0 (0) |
| t(11;19)(q23;p13.1) | 6 (0.5) | 57 (50) | 5 (83) | 0 (0) | 0 (0) |
| −20/20q− | 39 (3.2) | 60 (44) | 6 (15) | 29 (74) | 28 (72) |
| del(20q) | 16 (1.3) | 65 (44) | 5 (31) | 9 (56) | 8 (50) |
| −20 | 15 (1.2) | 60 (47) | 1 (7) | 12 (80) | 12 (80) |
| Loss of 20q | 8 (0.7) | 61 (38) | 0 (0) | 8 (100) | 8 (100) |
| del(9q) | 33 (2.7) | 37 (82) | 11 (33) | 10 (30) | 2 (6) |
| abn(12p) | 33 (2.7) | 64 (36) | 1 (3) | 25 (76) | 21 (64) |
| Unbalanced abn(12p) | 25 (2.1) | 64 (32) | 1 (4) | 20 (80) | 16 (64) |
| Balanced abn(12p) | 8 (0.7) | 61 (50) | 0 (0) | 5 (63) | 5 (63) |
| +22 | 31 (2.6) | 59 (55) | 1 (3) | 19 (61) | 14 (45) |
| −18 | 30 (2.5) | 64 (30) | 1 (3) | 29 (97) | 28 (93) |
| +13 | 29 (2.4) | 59 (55) | 13 (45) | 12 (41) | 10 (34) |
| +21 | 28 (2.3) | 59 (54) | 5 (24) | 14 (50) | 10 (36) |
| +11 | 20 (1.6) | 65 (20) | 12 (60) | 7 (35) | 6 (30) |
| −X | 18 (1.5) | 44 (67) | 0 (0) | 8 (44) | 5 (28) |
| +4 | 14 (1.2) | 55 (57) | 2 (14) | 10 (71) | 6 (43) |
| +14 | 12 (1.0) | 64 (42) | 0 (0) | 10 (83) | 9 (75) |
| del(11q) | 12 (1.0) | 42 (58) | 4 (33) | 5 (42) | 3 (25) |
| inv(3) or t(3;3) | 12 (1.0) | 43 (83) | 2 (17) | 4 (33) | 1 (8) |
| del(13q) | 9 (0.7) | 44 (56) | 1 (11) | 7 (78) | 5 (56) |
| t(6;9) | 8 (0.7) | 33 (100) | 6 (75) | 0 (0) | 0 (0) |
NA indicates not applicable.
v denotes various chromosomes (other than chromosomes 6, 9, and 19 with a break at p13.1) participating in translocations with chromosome 11 at band q23.
Treatment outcome by nonprioritized cytogenetic group
Table 4 demonstrates that CR rates for patients with nonprioritized cytogenetic abnormalities varied considerably, from 94% to 17% depending on the kind of pretreatment abnormality. Compared with the normal karyotype group, whose CR rate was 68%, patients with t(8;21), inv(16)/t(16;16), and del(9q) had significantly higher CR rates. In contrast, patients with inv(3)/t(3;3), −5/5q−, −7, loss of 7q, +8, abn(12p), −17/17p−, −18, −20, and loss of 20q had a significantly lower frequency of attaining CR than patients with a normal karyotype. In most of these cytogenetic subgroups, there was a higher frequency of resistant disease as opposed to early death (Table 4). The CR rates of patients with other chromosome abnormalities listed in Table 4 did not differ significantly from that of karyotypically normal patients.
Induction treatment outcome for nonprioritized cytogenetic groups
| Cytogenetic group . | No. of patients . | No. of CRs (%) . | P4-150 . | Early death4-151(%) . | Resistant disease‡ (%) . |
|---|---|---|---|---|---|
| Normal | 582 | 395 (68) | 66 (11) | 121 (21) | |
| del(9q) | 33 | 31 (94) | .001 | 0 (0) | 2 (6) |
| Sole del(9q) | 11 | 10 (91) | .19 | 0 (0) | 1 (9) |
| t(8;21) | 81 | 74 (91) | <.001 | 2 (2) | 5 (6) |
| inv(16) or t(16;16) | 96 | 82 (85) | <.001 | 10 (10) | 4 (4) |
| −X | 18 | 15 (83) | .20 | 1 (6) | 2 (11) |
| Balanced abn(11q23) | 54 | 40 (74) | .37 | 7 (13) | 7 (13) |
| t(9;11) | 27 | 23 (85) | .09 | 3 (11) | 1 (4) |
| t(11;v)4-153 | 13 | 9 (69) | 1.00 | 1 (8) | 3 (23) |
| t(6;11) | 8 | 5 (63) | .72 | 3 (38) | 0 (0) |
| t(11;19)(q23;p13.1) | 6 | 3 (50) | .39 | 0 (0) | 3 (5) |
| del(13q) | 9 | 6 (67) | 1.00 | 0 (0) | 3 (33) |
| −Y | 58 | 38 (66) | .77 | 8 (14) | 12 (21) |
| Sole −Y | 14 | 10 (71) | 1.00 | 4 (29) | 0 (0) |
| +22 | 31 | 20 (65) | .84 | 4 (13) | 7 (23) |
| +8 | 123 | 71 (58) | .04 | 24 (20) | 28 (23) |
| +21 | 28 | 16 (57) | .30 | 2 (7) | 10 (36) |
| +13 | 29 | 16 (55) | .16 | 6 (21) | 7 (24) |
| del(11q) | 12 | 6 (50) | .22 | 3 (25) | 3 (25) |
| +4 | 14 | 7 (50) | .25 | 5 (36) | 2 (14) |
| +11 | 20 | 9 (45) | .05 | 4 (20) | 7 (35) |
| +14 | 12 | 5 (42) | .07 | 3 (25) | 4 (33) |
| −7/7q− | 95 | 41 (43) | <.001 | 14 (15) | 40 (42) |
| −7 | 47 | 20 (43) | <.001 | 5 (11) | 22 (47) |
| Loss of 7q | 29 | 10 (34) | <.001 | 5 (17) | 14 (48) |
| del(7q) | 19 | 11 (58) | .46 | 4 (21) | 4 (21) |
| −20/20q− | 39 | 15 (38) | <.001 | 8 (21) | 16 (41) |
| del(20q) | 16 | 8 (50) | .17 | 1 (6) | 7 (44) |
| −20 | 15 | 5 (33) | .007 | 4 (27) | 6 (40) |
| Loss of 20q | 8 | 2 (25) | .02 | 3 (38) | 3 (38) |
| t(6;9) | 8 | 3 (38) | .12 | 1 (13) | 4 (50) |
| abn(12p) | 33 | 11 (33) | <.001 | 9 (27) | 13 (39) |
| Unbalanced abn(12p) | 25 | 9 (36) | .001 | 7 (28) | 9 (36) |
| Balanced abn(12p) | 8 | 2 (25) | .02 | 2 (25) | 4 (50) |
| −5/5q− | 86 | 27 (31) | <.001 | 22 (26) | 37 (43) |
| del(5q) | 42 | 11 (26) | <.001 | 9 (21) | 22 (52) |
| −5 | 26 | 12 (46) | .03 | 8 (31) | 6 (23) |
| Loss of 5q | 18 | 4 (22) | <.001 | 5 (28) | 9 (50) |
| −17/17p− | 62 | 19 (31) | <.001 | 17 (27) | 26 (42) |
| −17 | 32 | 9 (28) | <.001 | 11 (34) | 12 (38) |
| Loss of 17p | 30 | 10 (33) | .001 | 6 (20) | 14 (47) |
| −18 | 30 | 8 (27) | <.001 | 8 (27) | 14 (47) |
| inv(3) or t(3;3) | 12 | 2 (17) | <.001 | 3 (25) | 7 (58) |
| Cytogenetic group . | No. of patients . | No. of CRs (%) . | P4-150 . | Early death4-151(%) . | Resistant disease‡ (%) . |
|---|---|---|---|---|---|
| Normal | 582 | 395 (68) | 66 (11) | 121 (21) | |
| del(9q) | 33 | 31 (94) | .001 | 0 (0) | 2 (6) |
| Sole del(9q) | 11 | 10 (91) | .19 | 0 (0) | 1 (9) |
| t(8;21) | 81 | 74 (91) | <.001 | 2 (2) | 5 (6) |
| inv(16) or t(16;16) | 96 | 82 (85) | <.001 | 10 (10) | 4 (4) |
| −X | 18 | 15 (83) | .20 | 1 (6) | 2 (11) |
| Balanced abn(11q23) | 54 | 40 (74) | .37 | 7 (13) | 7 (13) |
| t(9;11) | 27 | 23 (85) | .09 | 3 (11) | 1 (4) |
| t(11;v)4-153 | 13 | 9 (69) | 1.00 | 1 (8) | 3 (23) |
| t(6;11) | 8 | 5 (63) | .72 | 3 (38) | 0 (0) |
| t(11;19)(q23;p13.1) | 6 | 3 (50) | .39 | 0 (0) | 3 (5) |
| del(13q) | 9 | 6 (67) | 1.00 | 0 (0) | 3 (33) |
| −Y | 58 | 38 (66) | .77 | 8 (14) | 12 (21) |
| Sole −Y | 14 | 10 (71) | 1.00 | 4 (29) | 0 (0) |
| +22 | 31 | 20 (65) | .84 | 4 (13) | 7 (23) |
| +8 | 123 | 71 (58) | .04 | 24 (20) | 28 (23) |
| +21 | 28 | 16 (57) | .30 | 2 (7) | 10 (36) |
| +13 | 29 | 16 (55) | .16 | 6 (21) | 7 (24) |
| del(11q) | 12 | 6 (50) | .22 | 3 (25) | 3 (25) |
| +4 | 14 | 7 (50) | .25 | 5 (36) | 2 (14) |
| +11 | 20 | 9 (45) | .05 | 4 (20) | 7 (35) |
| +14 | 12 | 5 (42) | .07 | 3 (25) | 4 (33) |
| −7/7q− | 95 | 41 (43) | <.001 | 14 (15) | 40 (42) |
| −7 | 47 | 20 (43) | <.001 | 5 (11) | 22 (47) |
| Loss of 7q | 29 | 10 (34) | <.001 | 5 (17) | 14 (48) |
| del(7q) | 19 | 11 (58) | .46 | 4 (21) | 4 (21) |
| −20/20q− | 39 | 15 (38) | <.001 | 8 (21) | 16 (41) |
| del(20q) | 16 | 8 (50) | .17 | 1 (6) | 7 (44) |
| −20 | 15 | 5 (33) | .007 | 4 (27) | 6 (40) |
| Loss of 20q | 8 | 2 (25) | .02 | 3 (38) | 3 (38) |
| t(6;9) | 8 | 3 (38) | .12 | 1 (13) | 4 (50) |
| abn(12p) | 33 | 11 (33) | <.001 | 9 (27) | 13 (39) |
| Unbalanced abn(12p) | 25 | 9 (36) | .001 | 7 (28) | 9 (36) |
| Balanced abn(12p) | 8 | 2 (25) | .02 | 2 (25) | 4 (50) |
| −5/5q− | 86 | 27 (31) | <.001 | 22 (26) | 37 (43) |
| del(5q) | 42 | 11 (26) | <.001 | 9 (21) | 22 (52) |
| −5 | 26 | 12 (46) | .03 | 8 (31) | 6 (23) |
| Loss of 5q | 18 | 4 (22) | <.001 | 5 (28) | 9 (50) |
| −17/17p− | 62 | 19 (31) | <.001 | 17 (27) | 26 (42) |
| −17 | 32 | 9 (28) | <.001 | 11 (34) | 12 (38) |
| Loss of 17p | 30 | 10 (33) | .001 | 6 (20) | 14 (47) |
| −18 | 30 | 8 (27) | <.001 | 8 (27) | 14 (47) |
| inv(3) or t(3;3) | 12 | 2 (17) | <.001 | 3 (25) | 7 (58) |
CR indicates complete remission.
For comparison of a given abnormality with normal cytogenetics.
Death within 30 days of initiating induction.
Survival beyond 30 days without attaining CR.
v denotes various chromosomes (other than chromosomes 6, 9, and 19 with a break at p13.1) participating in translocations with chromosome 11 at band q23.
The analysis of OS and CIR demonstrates that patients with t(8;21) and inv(16)/t(16;16) had a significantly improved probability of 5-year survival and remaining relapse free, and patients with del(9q) had an improved chance of 5-year survival compared with patients with a normal karyotype (Table 5). Patients with −5/5q−, t(6;11), −7, abn(12p), −17/17p−, −18, and −20/20q− had a significantly higher CIR and shorter survival, and those with inv(3)/t(3;3), t(6;9), loss of 7q, +11, t(11;19)(q23;p13.1), and +13 had a significantly lower probability of 5-year survival compared with the normal group.
Long-term treatment outcome for nonprioritized cytogenetic groups
| Cytogenetic group . | No. of patients . | No. of CRs . | 5-y CIR (SE) . | P5-150 . | Median OS, y . | 5-y OS (95% CI) . | P5-150 . |
|---|---|---|---|---|---|---|---|
| Normal | 582 | 395 | 0.66 (0.02) | NA | 1.3 | 24 (20-27) | NA |
| del(9q) | 33 | 31 | 0.58 (0.09) | .41 | >4.6 | 55 (38-71) | <.001 |
| Sole del(9q) | 11 | 10 | 0.50 (0.17) | .28 | >4.6 | 55 (28-79) | .02 |
| t(8;21) | 81 | 74 | 0.47 (0.05) | .009 | 5.1 | 52 (40-63) | <.001 |
| inv(16) or t(16;16) | 96 | 82 | 0.55 (0.05) | .02 | 7.9 | 57 (47-67) | <.001 |
| −X | 18 | 15 | 0.53 (0.14) | .41 | 1.3 | 44 (25-66) | .08 |
| Balanced abn(11q23) | 54 | 40 | 0.75 (0.07) | .008 | 0.8 | 20 (11-34) | .14 |
| t(9;11) | 27 | 23 | 0.57 (0.11) | .80 | 1.1 | 41 (24-60) | .25 |
| t(11;v)5-151 | 13 | 9 | 1.00 (0.11) | <.001 | 0.7 | 0 | .02 |
| t(6;11) | 8 | 5 | 1.00 (0.20) | .002 | 0.6 | 0 | .003 |
| t(11;19)(q23;p13.1) | 6 | 3 | ND | ND | 0.6 | 0 | .03 |
| del(13q) | 9 | 6 | 0.83 (0.20) | .12 | 0.7 | 11 (3-33) | .16 |
| −Y | 58 | 38 | 0.58 (0.08) | .40 | 1.2 | 27 (17-41) | .96 |
| Sole −Y | 14 | 10 | 0.80 (0.15) | .47 | 1.7 | 29 (13-52) | .99 |
| +22 | 31 | 20 | 0.12 (0.31) | .31 | 1.2 | 32 (19-50) | .66 |
| +8 | 123 | 71 | 0.69 (0.06) | .83 | 1.0 | 20 (14-29) | .12 |
| +21 | 28 | 16 | 0.88 (0.10) | .002 | 1.0 | 10 (3-25) | .17 |
| +13 | 29 | 16 | 0.75 (0.12) | .52 | 1.1 | 7 (2-18) | .04 |
| del(11q) | 12 | 6 | 0.67 (0.23) | .90 | 0.6 | 17 (6-39) | .27 |
| +4 | 14 | 7 | 1.00 (0.14) | .10 | 0.9 | 14 (5-35) | .09 |
| +11 | 20 | 9 | 0.89 (0.14) | .28 | 0.9 | 5 (1-17) | .03 |
| +14 | 12 | 5 | 0.40 (0.25) | .30 | 0.5 | 25 (10-49) | .72 |
| −7/7q− | 95 | 41 | 0.83 (0.06) | <.001 | 0.5 | 7 (4-14) | <.001 |
| −7 | 47 | 20 | 0.90 (0.09) | <.001 | 0.5 | 0 | <.001 |
| Loss of 7q | 29 | 10 | 0.70 (0.16) | .30 | 0.4 | 10 (4-24) | <.001 |
| del(7q) | 19 | 11 | 0.82 (0.13) | .16 | 0.9 | 21 (9-41) | .16 |
| −20/20q− | 39 | 15 | 0.93 (0.15) | .003 | 0.3 | 5 (2-14) | <.001 |
| del(20q) | 16 | 8 | 0.88 (0.28) | .49 | 0.3 | 13 (4-31) | .01 |
| −20 | 15 | 5 | 1.00 (0.20) | <.001 | 0.3 | 0 | <.001 |
| Loss of 20q | 8 | 2 | ND | ND | 0.1 | 0 | <.001 |
| t(6;9) | 8 | 3 | ND | ND | 0.9 | 0 | .03 |
| abn(12p) | 33 | 11 | 0.82 (0.13) | <.001 | 0.3 | 3 (1-10) | <.001 |
| Unbalanced abn(12p) | 25 | 9 | 0.78 (0.16) | .009 | 0.3 | 4 (1-13) | <.001 |
| Balanced abn(12p) | 8 | 2 | ND | ND | 0.2 | 0 | .01 |
| −5/5q− | 86 | 27 | 0.93 (0.06) | <.001 | 0.3 | 6 (3-12) | <.001 |
| del(5q) | 42 | 11 | 0.82 (0.13) | .002 | 0.3 | 5 (2-13) | <.001 |
| −5 | 26 | 12 | 1.00 (0.08) | .01 | 0.2 | 4 (1-13) | <.001 |
| Loss of 5q | 18 | 4 | ND | ND | 0.2 | 11 (4-28) | <.001 |
| −17/17p− | 62 | 19 | 1.00 (0.05) | <.001 | 0.2 | 2 (1-6) | <.001 |
| −17 | 32 | 9 | 1.00 (0.11) | .002 | 0.1 | 3 (1-11) | <.001 |
| Loss of 17p | 30 | 10 | 1.00 (0.10) | <.001 | 0.3 | 0 | <.001 |
| −18 | 30 | 8 | 1.00 (0.13) | <.001 | 0.2 | 0 | <.001 |
| inv(3) or t(3;3) | 12 | 2 | ND | ND | 0.2 | 0 | <.001 |
| Cytogenetic group . | No. of patients . | No. of CRs . | 5-y CIR (SE) . | P5-150 . | Median OS, y . | 5-y OS (95% CI) . | P5-150 . |
|---|---|---|---|---|---|---|---|
| Normal | 582 | 395 | 0.66 (0.02) | NA | 1.3 | 24 (20-27) | NA |
| del(9q) | 33 | 31 | 0.58 (0.09) | .41 | >4.6 | 55 (38-71) | <.001 |
| Sole del(9q) | 11 | 10 | 0.50 (0.17) | .28 | >4.6 | 55 (28-79) | .02 |
| t(8;21) | 81 | 74 | 0.47 (0.05) | .009 | 5.1 | 52 (40-63) | <.001 |
| inv(16) or t(16;16) | 96 | 82 | 0.55 (0.05) | .02 | 7.9 | 57 (47-67) | <.001 |
| −X | 18 | 15 | 0.53 (0.14) | .41 | 1.3 | 44 (25-66) | .08 |
| Balanced abn(11q23) | 54 | 40 | 0.75 (0.07) | .008 | 0.8 | 20 (11-34) | .14 |
| t(9;11) | 27 | 23 | 0.57 (0.11) | .80 | 1.1 | 41 (24-60) | .25 |
| t(11;v)5-151 | 13 | 9 | 1.00 (0.11) | <.001 | 0.7 | 0 | .02 |
| t(6;11) | 8 | 5 | 1.00 (0.20) | .002 | 0.6 | 0 | .003 |
| t(11;19)(q23;p13.1) | 6 | 3 | ND | ND | 0.6 | 0 | .03 |
| del(13q) | 9 | 6 | 0.83 (0.20) | .12 | 0.7 | 11 (3-33) | .16 |
| −Y | 58 | 38 | 0.58 (0.08) | .40 | 1.2 | 27 (17-41) | .96 |
| Sole −Y | 14 | 10 | 0.80 (0.15) | .47 | 1.7 | 29 (13-52) | .99 |
| +22 | 31 | 20 | 0.12 (0.31) | .31 | 1.2 | 32 (19-50) | .66 |
| +8 | 123 | 71 | 0.69 (0.06) | .83 | 1.0 | 20 (14-29) | .12 |
| +21 | 28 | 16 | 0.88 (0.10) | .002 | 1.0 | 10 (3-25) | .17 |
| +13 | 29 | 16 | 0.75 (0.12) | .52 | 1.1 | 7 (2-18) | .04 |
| del(11q) | 12 | 6 | 0.67 (0.23) | .90 | 0.6 | 17 (6-39) | .27 |
| +4 | 14 | 7 | 1.00 (0.14) | .10 | 0.9 | 14 (5-35) | .09 |
| +11 | 20 | 9 | 0.89 (0.14) | .28 | 0.9 | 5 (1-17) | .03 |
| +14 | 12 | 5 | 0.40 (0.25) | .30 | 0.5 | 25 (10-49) | .72 |
| −7/7q− | 95 | 41 | 0.83 (0.06) | <.001 | 0.5 | 7 (4-14) | <.001 |
| −7 | 47 | 20 | 0.90 (0.09) | <.001 | 0.5 | 0 | <.001 |
| Loss of 7q | 29 | 10 | 0.70 (0.16) | .30 | 0.4 | 10 (4-24) | <.001 |
| del(7q) | 19 | 11 | 0.82 (0.13) | .16 | 0.9 | 21 (9-41) | .16 |
| −20/20q− | 39 | 15 | 0.93 (0.15) | .003 | 0.3 | 5 (2-14) | <.001 |
| del(20q) | 16 | 8 | 0.88 (0.28) | .49 | 0.3 | 13 (4-31) | .01 |
| −20 | 15 | 5 | 1.00 (0.20) | <.001 | 0.3 | 0 | <.001 |
| Loss of 20q | 8 | 2 | ND | ND | 0.1 | 0 | <.001 |
| t(6;9) | 8 | 3 | ND | ND | 0.9 | 0 | .03 |
| abn(12p) | 33 | 11 | 0.82 (0.13) | <.001 | 0.3 | 3 (1-10) | <.001 |
| Unbalanced abn(12p) | 25 | 9 | 0.78 (0.16) | .009 | 0.3 | 4 (1-13) | <.001 |
| Balanced abn(12p) | 8 | 2 | ND | ND | 0.2 | 0 | .01 |
| −5/5q− | 86 | 27 | 0.93 (0.06) | <.001 | 0.3 | 6 (3-12) | <.001 |
| del(5q) | 42 | 11 | 0.82 (0.13) | .002 | 0.3 | 5 (2-13) | <.001 |
| −5 | 26 | 12 | 1.00 (0.08) | .01 | 0.2 | 4 (1-13) | <.001 |
| Loss of 5q | 18 | 4 | ND | ND | 0.2 | 11 (4-28) | <.001 |
| −17/17p− | 62 | 19 | 1.00 (0.05) | <.001 | 0.2 | 2 (1-6) | <.001 |
| −17 | 32 | 9 | 1.00 (0.11) | .002 | 0.1 | 3 (1-11) | <.001 |
| Loss of 17p | 30 | 10 | 1.00 (0.10) | <.001 | 0.3 | 0 | <.001 |
| −18 | 30 | 8 | 1.00 (0.13) | <.001 | 0.2 | 0 | <.001 |
| inv(3) or t(3;3) | 12 | 2 | ND | ND | 0.2 | 0 | <.001 |
CR indicates complete remission; CIR, cumulative incidence of relapse; OS, overall survival; CI, confidence interval; NA, not applicable; and ND, not done.
For comparison of a given abnormality with normal cytogenetics.
v denotes various chromosomes (other than chromosomes 6, 9, and 19 with a break at p13.1) participating in translocations with chromosome 11 at band q23.
Secondary aberrations and a complex karyotype do not adversely affect outcome of patients with common balanced rearrangements −t(8;21), inv(16)/t(16;16), or t(9;11)
We sought to determine whether the presence of secondary chromosome abnormalities or a complex karyotype with 3 or more abnormalities influences the outcome of patients harboring common (ie, seen in ≥ 25 patients) balanced rearrangements—that is, t(8;21), inv(16)/t(16;16), and t(9;11). There were too few patients with at least 5 abnormalities to examine whether a complex karyotype with 5 or more abnormalities alters outcome of these patients. Table6 demonstrates that for each of the aforementioned balanced rearrangements, neither the presence of secondary abnormalities nor a complex karyotype with 3 or more abnormalities adversely affect outcome. This includes t(8;21)-positive patients with del(9q), loss of a sex chromosome, +8, and those with other secondary aberrations, for all of whom CR rates, CIR, and OS were not significantly different from those of patients with an isolated t(8;21). Likewise, for each group outlined in Table 6, CR rates, CIR, and OS of patients who had at least 3 abnormalities were not significantly different from those of patients who had only 1 or 2 aberrations. These findings provide support for treating AML patients with t(8;21), inv(16)/t(16;16), and t(9;11) only by their primary abnormality irrespective of the presence or absence of secondary aberrations or a complex karyotype with 3 or more abnormalities.
Significance of the presence of secondary abnormalities among patients with inv(16) or t(16;16), t(8;21), or t(9;11)
| Cytogenetic abnormality . | No. of patients . | % CR . | P . | 5-y CIR (SE) . | P . | % 5-y OS (95% CI) . | P . |
|---|---|---|---|---|---|---|---|
| inv(16) or t(16;16) | |||||||
| Sole | 64 | 86 | 0.67 (0.06) | 53 (41-65) | |||
| With at least 1 secondary abnormality | 32 | 84 | 1.09-150 | 0.30 (0.09) | .0029-150 | 65 (47-80) | .309-150 |
| Sole or with 1 secondary abnormality | 88 | 85 | 0.56 (0.06) | 59 (48-69) | |||
| Complex with at least 3 abnormalities | 8 | 88 | 1.09-151 | 0.43 (0.21) | .429-151 | 38 (16-66) | .459-151 |
| t(8;21) | |||||||
| Sole | 22 | 95 | 0.43 (0.11) | 54 (34-73) | |||
| With del(9q) | 18 | 94 | 1.09-150 | 0.53 (0.13) | .529-150 | 61 (39-80) | .839-150 |
| With −X or −Y | 38 | 92 | 1.09-150 | 0.46 (0.09) | .889-150 | 50 (33-66) | .779-150 |
| With + 8 | 8 | 88 | 1.09-150 | 0.29 (0.19) | .819-150 | 50 (24-76) | .409-150 |
| With any other secondary abnormality | 6 | 67 | .119-150 | ND | ND | 67 (33-89) | .929-150 |
| Sole or with 1 secondary abnormality | 67 | 93 | 0.52 (0.06) | 49 (36-61) | |||
| Complex with at least 3 abnormalities | 14 | 86 | .609-151 | 0.25 (0.13) | .079-151 | 64 (40-83) | .289-151 |
| t(9;11) | |||||||
| Sole | 12 | 75 | 0.67 (0.18) | 33 (15-58) | |||
| With at least 1 secondary abnormality | 15 | 93 | .299-150 | 0.50 (0.14) | .339-150 | 47 (23-71) | .529-150 |
| Sole or with 1 secondary abnormality | 19 | 84 | 0.50 (0.13) | 42 (23-64) | |||
| Complex with at least 3 abnormalities | 8 | 88 | 1.09-151 | 0.71 (0.20) | .559-151 | 38 (14-69) | .569-151 |
| Cytogenetic abnormality . | No. of patients . | % CR . | P . | 5-y CIR (SE) . | P . | % 5-y OS (95% CI) . | P . |
|---|---|---|---|---|---|---|---|
| inv(16) or t(16;16) | |||||||
| Sole | 64 | 86 | 0.67 (0.06) | 53 (41-65) | |||
| With at least 1 secondary abnormality | 32 | 84 | 1.09-150 | 0.30 (0.09) | .0029-150 | 65 (47-80) | .309-150 |
| Sole or with 1 secondary abnormality | 88 | 85 | 0.56 (0.06) | 59 (48-69) | |||
| Complex with at least 3 abnormalities | 8 | 88 | 1.09-151 | 0.43 (0.21) | .429-151 | 38 (16-66) | .459-151 |
| t(8;21) | |||||||
| Sole | 22 | 95 | 0.43 (0.11) | 54 (34-73) | |||
| With del(9q) | 18 | 94 | 1.09-150 | 0.53 (0.13) | .529-150 | 61 (39-80) | .839-150 |
| With −X or −Y | 38 | 92 | 1.09-150 | 0.46 (0.09) | .889-150 | 50 (33-66) | .779-150 |
| With + 8 | 8 | 88 | 1.09-150 | 0.29 (0.19) | .819-150 | 50 (24-76) | .409-150 |
| With any other secondary abnormality | 6 | 67 | .119-150 | ND | ND | 67 (33-89) | .929-150 |
| Sole or with 1 secondary abnormality | 67 | 93 | 0.52 (0.06) | 49 (36-61) | |||
| Complex with at least 3 abnormalities | 14 | 86 | .609-151 | 0.25 (0.13) | .079-151 | 64 (40-83) | .289-151 |
| t(9;11) | |||||||
| Sole | 12 | 75 | 0.67 (0.18) | 33 (15-58) | |||
| With at least 1 secondary abnormality | 15 | 93 | .299-150 | 0.50 (0.14) | .339-150 | 47 (23-71) | .529-150 |
| Sole or with 1 secondary abnormality | 19 | 84 | 0.50 (0.13) | 42 (23-64) | |||
| Complex with at least 3 abnormalities | 8 | 88 | 1.09-151 | 0.71 (0.20) | .559-151 | 38 (14-69) | .569-151 |
Abbreviations are explained in a footnote to Table 5.
For comparison with the sole group.
For comparison with the group comprising patients with sole primary abnormality and those with 1 secondary abnormality.
Complex karyotype: clinical features and outcome
Different cytogenetic classifications have defined complex karyotype by the presence of at least 5 clonal aberrations11,15 or at least 3 abnormalities in the absence of t(8;21), inv(16)/t(16;16), and t(15;17).13 40 The classification of this category as at least 3 or at least 5 abnormalities is empiric, with little previous attempt to test if incremental increase in the number of chromosomal aberrations correlates with poorer outcome. To examine this clinically relevant issue, we analyzed the prognostic impact of the presence of 3 or 4 (n = 36) versus 5 or more (n = 99) abnormalities in patients who did not harbor t(8;21), inv(16)/t(16;16), or t(9;11). The influence of a complex karyotype with 3 or more abnormalities in patients with the aforementioned abnormalities was analyzed separately (Table 6). As shown in Table7, patients with 3 or 4 abnormalities were younger and had significantly better CIR and 5-year OS than those with 5 or more abnormalities. However, both the CR rate and OS of patients with 3 or 4 abnormalities were significantly lower than those of the cytogenetically normal group (P = .02 andP = .002, respectively), whereas the CIR was significantly higher (P = .002). Only 1 of the patients with 3 or 4 abnormalities remains in remission at 5 years. Therefore, despite the difference in outcome between the patients with 3 or 4 aberrations and those with 5 or more aberrations, the low CR rate, 5-year OS, and the high CIR for patients in the former group justify combining patients with 3 or 4 abnormalities with patients with 5 or more abnormalities into 1 complex karyotype category defined by the presence of 3 or more abnormalities. The survival results are depicted in Figure2.
Selected pretreatment clinical and hematologic characteristics and treatment outcome of AML patients with a complex karyotype defined by the presence of 3 or 4 or at least 5 abnormalities6-150
| No. of abnormalities . | No. of patients . | Median age, y . | % patients younger than 60 y . | Median WBC, × 109/L . | % CR . | P6-151 . | 5-y CIR (SE) . | P6-151 . | % (95% CI) 5-y OS . | P6-151 . |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 or 4 | 36 | 51 | 58 | 10.8 | 47 | .02 | 0.88 (0.09) | .002 | 8 (3-20) | .002 |
| 5 or more | 99 | 64 | 33 | 7.3 | 30 | <.001 | 0.97 (0.04) | <.001 | 2 (1-6) | <.001 |
| P6-152 | NA | .02 | .01 | .30 | .10 | NA | .04 | NA | .004 | NA |
| No. of abnormalities . | No. of patients . | Median age, y . | % patients younger than 60 y . | Median WBC, × 109/L . | % CR . | P6-151 . | 5-y CIR (SE) . | P6-151 . | % (95% CI) 5-y OS . | P6-151 . |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 or 4 | 36 | 51 | 58 | 10.8 | 47 | .02 | 0.88 (0.09) | .002 | 8 (3-20) | .002 |
| 5 or more | 99 | 64 | 33 | 7.3 | 30 | <.001 | 0.97 (0.04) | <.001 | 2 (1-6) | <.001 |
| P6-152 | NA | .02 | .01 | .30 | .10 | NA | .04 | NA | .004 | NA |
WBC indicates white blood cell count; NA, not applicable. Other abbreviations are explained in the footnote to Table 5.
Patients with t(8;21), inv(16)/t(16;16), and t(9;11) excluded.
For comparison with normal cytogenetics.
For comparison of 3 or 4 abnormalities with 5 or more abnormalities.
Overall survival of AML patients with a complex karyotype defined as 3 or more, 3 or 4, and 5 or more abnormalities (abn; excluding patients with t(8;21), inv(16)/t(16;16), and t(9;11)) and of patients with a normal karyotype for comparison.
Overall survival of AML patients with a complex karyotype defined as 3 or more, 3 or 4, and 5 or more abnormalities (abn; excluding patients with t(8;21), inv(16)/t(16;16), and t(9;11)) and of patients with a normal karyotype for comparison.
Outcome of patients with trisomy 8 is poor in the absence of t(8;21), inv(16)/t(16;16), and t(9;11)
Trisomy 8 is the most common trisomy in de novo AML (Table 3). However, the +8 group is heterogeneous, because +8 can be the sole abnormality detected, can be part of a complex karyotype, or can be the only secondary aberration accompanying primary rearrangements, including t(8;21), inv(16)/t(16;16), or t(9;11). Previous studies have shown that prognosis of AML patients with +8 depends on whether +8 is an isolated abnormality or is accompanied by aberrations bestowing favorable or adverse prognosis.11 41 We therefore examined the outcome of different subsets of patients with +8 in our series (Table 8). Patients with sole +8 and +8 with 1 additional abnormality other than t(8;21), inv(16)/t(16;16), and t(9;11) had significantly inferior OS, but not CR or CIR rates, while patients with +8 and a complex karyotype with 3 or more abnormalities had a significantly inferior CR rate, CIR, and OS compared with those with a normal karyotype. These data show that the impact of trisomy 8 is best predicted by the presence and nature of abnormalities that accompany it (Figure3).
Treatment outcome of patients with sole trisomy 8 and of those with trisomy 8 accompanying other aberrations
| Trisomy 8 . | No. of patients . | % CR . | P7-150 . | 5-y CIR (SE) . | P7-150 . | % 5-y OS (95% CI) . | P7-150 . |
|---|---|---|---|---|---|---|---|
| With t(9;11) | 10 | 100 | .04 | 0.50 (0.17) | .45 | 60 (31-83) | .07 |
| With inv(16) or t(16;16) | 12 | 83 | .36 | 0.52 (0.18) | .16 | 57 (31-80) | .009 |
| Sole | 41 | 61 | .39 | 0.77 (0.09) | .54 | 12 (5-27) | .049 |
| With 1 other abnormality | 26 | 50 | .09 | 0.77 (0.13) | .05 | 8 (3-20) | .002 |
| In complex karyotype with at least 3 abnormalities | 26 | 23 | <.001 | 1.00 (0.17) | .001 | 4 (1-13) | <.001 |
| Trisomy 8 . | No. of patients . | % CR . | P7-150 . | 5-y CIR (SE) . | P7-150 . | % 5-y OS (95% CI) . | P7-150 . |
|---|---|---|---|---|---|---|---|
| With t(9;11) | 10 | 100 | .04 | 0.50 (0.17) | .45 | 60 (31-83) | .07 |
| With inv(16) or t(16;16) | 12 | 83 | .36 | 0.52 (0.18) | .16 | 57 (31-80) | .009 |
| Sole | 41 | 61 | .39 | 0.77 (0.09) | .54 | 12 (5-27) | .049 |
| With 1 other abnormality | 26 | 50 | .09 | 0.77 (0.13) | .05 | 8 (3-20) | .002 |
| In complex karyotype with at least 3 abnormalities | 26 | 23 | <.001 | 1.00 (0.17) | .001 | 4 (1-13) | <.001 |
Abbreviations are explained in the footnote to Table 5.
Overall survival of AML patients with trisomy 8 as a sole abnormality, those with trisomy 8 as part of a complex karyotype with 3 or more abnormalities, and patients with trisomy 8 accompanied by, respectively, t(8;21), inv(16)/t(16;16), t(9;11), and 1 additional abnormality other than the aforementioned ones.
Overall survival of AML patients with trisomy 8 as a sole abnormality, those with trisomy 8 as part of a complex karyotype with 3 or more abnormalities, and patients with trisomy 8 accompanied by, respectively, t(8;21), inv(16)/t(16;16), t(9;11), and 1 additional abnormality other than the aforementioned ones.
Prioritization schema that facilitates risk assessment for untreated de novo AML
We developed a prioritization schema for the assignment of risk based on pretreatment karyotype according to the probability of achievement of CR, CIR, and OS for patients with AML. Patients with a normal karyotype were classified as intermediate risk. We classified all patients with t(8;21) and inv(16)/t(16;16) in the favorable and those with t(9;11) in the intermediate risk categories based on outcome data presented in Tables 4 and 5 and the absence of impact on outcome of complex karyotype with 3 or more abnormalities in these groups (Table 6). All other patients with a complex karyotype with 3 or more abnormalities were classified as having adverse risk. After excluding the patients thus classified, we analyzed the impact of specific aberrations on outcome for the remaining patients in cytogenetic groups that still comprised at least 5 patients (Table 9).
Treatment outcome for other cytogenetic groups after exclusion of patients with t(8;21), inv(16)/t(16;16), t(9;11), and complex ≥3 karyotype
| Cytogenetic group . | No. of patients . | No. of CRs (%) . | P8-150 . | 5-y CIR (SE) . | P8-150 . | Median OS, y . | % 5-y OS (95% CI) . | P8-150 . |
|---|---|---|---|---|---|---|---|---|
| Normal | 582 | 395 (68) | NA | 0.66 (0.01) | NA | 1.3 | 24 (20-27) | NA |
| del(9q)8-151 | 13 | 12 (92) | .07 | 0.58 (0.15) | .55 | >4.5 | 54 (29-77) | .01 |
| +21 | 11 | 8 (73) | .77 | 0.88 | .002 | 1.1 | 18 (6-42) | .96 |
| −Y | 22 | 14 (64) | .82 | 0.79 | .27 | 0.7 | 23 (11-42) | .40 |
| +8 sole | 41 | 25 (61) | .39 | 0.77 (0.09) | .54 | 1.0 | 12 (5-27) | .049 |
| +13 | 17 | 10 (59) | .60 | 0.70 (0.16) | .98 | 1.0 | 6 (2-19) | .22 |
| del(20q) | 7 | 4 (57) | .69 | ND | ND | 0.3 | 29 (10-58) | .41 |
| Loss of 7q | 7 | 4 (57) | .69 | ND | ND | 1.0 | 14 (4-40) | .62 |
| t(6;11) | 7 | 4 (57) | .69 | ND | ND | 0.3 | 0 | .002 |
| +8 with 1 other abnormality8-152 | 26 | 13 (50) | .09 | 0.77 (0.13) | .050 | 0.5 | 8 (3-20) | .002 |
| t(11;19)(q23;p13.1) | 6 | 3 (50) | .39 | ND | ND | 0.6 | 0 | .03 |
| −7 | 21 | 10 (48) | .06 | 0.80 (0.17) | .02 | 0.6 | 0 | <.001 |
| +11 | 13 | 6 (46) | .13 | 0.83 (0.20) | .74 | 0.7 | 8 (2-24) | .24 |
| del(11q) | 7 | 3 (43) | .22 | ND | ND | 0.7 | 14 (4-40) | .46 |
| del(5q) | 5 | 2 (40) | .34 | ND | ND | >3.3 | 40 (15-72) | .23 |
| t(6;9) | 8 | 3 (38) | .12 | ND | ND | 0.9 | 0 | .03 |
| abn(12p) | 7 | 2 (29) | .04 | ND | ND | 0.4 | 14 (4-40) | .53 |
| inv(3) or t(3;3) | 8 | 1 (13) | .002 | ND | ND | 0.2 | 0 | .007 |
| Cytogenetic group . | No. of patients . | No. of CRs (%) . | P8-150 . | 5-y CIR (SE) . | P8-150 . | Median OS, y . | % 5-y OS (95% CI) . | P8-150 . |
|---|---|---|---|---|---|---|---|---|
| Normal | 582 | 395 (68) | NA | 0.66 (0.01) | NA | 1.3 | 24 (20-27) | NA |
| del(9q)8-151 | 13 | 12 (92) | .07 | 0.58 (0.15) | .55 | >4.5 | 54 (29-77) | .01 |
| +21 | 11 | 8 (73) | .77 | 0.88 | .002 | 1.1 | 18 (6-42) | .96 |
| −Y | 22 | 14 (64) | .82 | 0.79 | .27 | 0.7 | 23 (11-42) | .40 |
| +8 sole | 41 | 25 (61) | .39 | 0.77 (0.09) | .54 | 1.0 | 12 (5-27) | .049 |
| +13 | 17 | 10 (59) | .60 | 0.70 (0.16) | .98 | 1.0 | 6 (2-19) | .22 |
| del(20q) | 7 | 4 (57) | .69 | ND | ND | 0.3 | 29 (10-58) | .41 |
| Loss of 7q | 7 | 4 (57) | .69 | ND | ND | 1.0 | 14 (4-40) | .62 |
| t(6;11) | 7 | 4 (57) | .69 | ND | ND | 0.3 | 0 | .002 |
| +8 with 1 other abnormality8-152 | 26 | 13 (50) | .09 | 0.77 (0.13) | .050 | 0.5 | 8 (3-20) | .002 |
| t(11;19)(q23;p13.1) | 6 | 3 (50) | .39 | ND | ND | 0.6 | 0 | .03 |
| −7 | 21 | 10 (48) | .06 | 0.80 (0.17) | .02 | 0.6 | 0 | <.001 |
| +11 | 13 | 6 (46) | .13 | 0.83 (0.20) | .74 | 0.7 | 8 (2-24) | .24 |
| del(11q) | 7 | 3 (43) | .22 | ND | ND | 0.7 | 14 (4-40) | .46 |
| del(5q) | 5 | 2 (40) | .34 | ND | ND | >3.3 | 40 (15-72) | .23 |
| t(6;9) | 8 | 3 (38) | .12 | ND | ND | 0.9 | 0 | .03 |
| abn(12p) | 7 | 2 (29) | .04 | ND | ND | 0.4 | 14 (4-40) | .53 |
| inv(3) or t(3;3) | 8 | 1 (13) | .002 | ND | ND | 0.2 | 0 | .007 |
Abbreviations are explained in the footnote to Table5.
For comparison of abnormality with normal cytogenetics.
Six of the 13 patients with del(9q) underwent SCT off-protocol. For the remaining 7 patients who did not receive SCT, 5-year CIR (SE) was 0.67 (0.05), P = .96, median OS 1.5 years, and probability of 5-year OS 29% (95% CI, 10%-58%), P = .39. Thus, the OS of AML patients with del(9q) not undergoing transplantation is not significantly different from that of the normal group.
An abnormality accompanying +8 is other than t(8;21), t(9;11), and inv(16) or t(16;16).
Patients with a significantly superior or inferior (P < .05) CR rate, CIR, or OS relative to karyotypically normal AML patients were classified in the favorable or adverse risk group, respectively. Patients with a similar (P ≥ .05) CR rate, CIR, or OS were classified in the intermediate risk group. Table10 gives the final risk assignments of 1124 classifiable patients for CR and OS and 716 classifiable patients for CIR.
Risk groups for success of induction and overall survival*
| Cytogenetic risk group . | Induction success . | Cumulative incidence of relapse10-151 . | Overall survival . |
|---|---|---|---|
| Favorable | t(8;21) | t(8;21) | t(8;21) |
| inv(16) or t(16;16) | inv(16) or t(16;16) | inv(16) or t(16;16) | |
| del(9q)10-152 | |||
| Intermediate | Normal karyotype | Normal karyotype | Normal karyotype |
| −Y | −Y | −Y | |
| del(5q) | t(9;11) | del(5q) | |
| t(6;9) | del(9q) | Loss of 7q | |
| t(6;11) | +8 sole | t(9;11) | |
| −7 | +8 with 1 other | +11 | |
| Loss of 7q | abnormality10-153 | del(11q) | |
| +8 sole | +11 | abn(12p) | |
| +8 with 1 other | +13 | +13 | |
| abnormality10-153 | del(20q) | ||
| del(9q) | +21 | ||
| t(9;11) | |||
| +11 | |||
| del(11q) | |||
| t(11;19)(q23;p13.1) | |||
| +13 | |||
| del(20q) | |||
| +21 | |||
| Adverse | Complex karyotype | Complex karyotype | Complex karyotype |
| (≥ 3 abnormalities) | (≥ 3 abnormalities) | (≥ 3 abnormalities) | |
| inv(3) or t(3;3) | −7 | inv(3) or t(3;3) | |
| abn(12p) | +21 | t(6;9) | |
| t(6;11) | |||
| −7 | |||
| +8 sole | |||
| +8 with 1 other | |||
| abnormality10-153 | |||
| t(11;19)(q23;p13.1) |
| Cytogenetic risk group . | Induction success . | Cumulative incidence of relapse10-151 . | Overall survival . |
|---|---|---|---|
| Favorable | t(8;21) | t(8;21) | t(8;21) |
| inv(16) or t(16;16) | inv(16) or t(16;16) | inv(16) or t(16;16) | |
| del(9q)10-152 | |||
| Intermediate | Normal karyotype | Normal karyotype | Normal karyotype |
| −Y | −Y | −Y | |
| del(5q) | t(9;11) | del(5q) | |
| t(6;9) | del(9q) | Loss of 7q | |
| t(6;11) | +8 sole | t(9;11) | |
| −7 | +8 with 1 other | +11 | |
| Loss of 7q | abnormality10-153 | del(11q) | |
| +8 sole | +11 | abn(12p) | |
| +8 with 1 other | +13 | +13 | |
| abnormality10-153 | del(20q) | ||
| del(9q) | +21 | ||
| t(9;11) | |||
| +11 | |||
| del(11q) | |||
| t(11;19)(q23;p13.1) | |||
| +13 | |||
| del(20q) | |||
| +21 | |||
| Adverse | Complex karyotype | Complex karyotype | Complex karyotype |
| (≥ 3 abnormalities) | (≥ 3 abnormalities) | (≥ 3 abnormalities) | |
| inv(3) or t(3;3) | −7 | inv(3) or t(3;3) | |
| abn(12p) | +21 | t(6;9) | |
| t(6;11) | |||
| −7 | |||
| +8 sole | |||
| +8 with 1 other | |||
| abnormality10-153 | |||
| t(11;19)(q23;p13.1) |
Patients are assigned to risk group according to probability of attaining a CR. Patients were assigned in the following order: normal karyotype→t(8;21), inv(16)/t(16;16), and t(9;11)→complex karyotype with at least 3 abnormalities→other groups as outlined in the text. Abnormalities not specified are not included in the risk assessment model.
Too few patients with inv(3) or t(3;3), del(5q), t(6;9), t(6;11), loss of 7q, del(11q), t(11;19)(q23;p13.1), abn(12p), and del(20q) achieved a CR to be classified according to their cumulative incidence of relapse.
del(9q) would be assigned to the intermediate-risk group if only patients not undergoing transplantation were analyzed (see footnote designated by “†” in Table 9).
An abnormality accompanying +8 is other than t(8;21), t(9;11), and inv(16) or t(16;16).
When we categorized the patients according to this risk classification system for success of induction treatment, the favorable risk group comprised 177 (16%) patients, the intermediate risk group 800 (71%) patients, and the adverse risk group 147 (13%) patients. Their CR rates were 88%, 67%, and 32%, respectively. For 5-year CIR, the favorable risk group comprised 156 (22%) patients, the intermediate risk group 498 (70%) patients, and the adverse group 62 (9%) patients. The estimated CIRs (with SE) at 5 years were 0.51 (0.04), 0.67 (0.02), and 0.92 (0.04), respectively. The CIR curves are shown in Figure 4. For 5-year OS, the favorable risk group comprised 190 (17%) patients, the intermediate risk group 686 (61%) patients, and the adverse risk group 248 (22%) patients. The estimated probabilities (with 95% CI) of 5-year OS were 55% (47%-62%), 24% (21%-27%), and 5% (3%-8%), respectively. The differences in OS are depicted in Figure5.
Cumulative incidence of relapse (CIR) of AML patients categorized into favorable, intermediate, and adverse cytogenetic risk groups using the CALGB criteria.
Cumulative incidence of relapse (CIR) of AML patients categorized into favorable, intermediate, and adverse cytogenetic risk groups using the CALGB criteria.
Overall survival in AML patients categorized into favorable, intermediate, and adverse cytogenetic risk groups using the CALGB criteria.
Overall survival in AML patients categorized into favorable, intermediate, and adverse cytogenetic risk groups using the CALGB criteria.
We next analyzed factors related to the probability of attaining CR using the logistic regression model. Univariate analysis identified the following factors to have prognostic significance: cytogenetic risk (P < .001), treatment protocol (P < .001), age (P < .001), leukocyte count (P < .001), splenomegaly (P = .001), infection at study entry (P = .002), and hepatomegaly (P = .004). A forward stepwise multivariate analysis of 1118 cases with complete data on these variables selected cytogenetic risk (P < .001), age (P < .001), leukocyte count (P < .001), splenomegaly (P = .002), and infection at study entry (P = .02) to the model as joint predictors of attaining CR, where the P value for each variable is adjusted for variables preceding it in the list. When all variables in the model were considered, patients in the adverse cytogenetic group were 4.0 (95% CI, 2.7-5.9) times less likely to achieve CR than those in the intermediate group and 11.9 (95% CI, 4.9-28.9) times less likely than patients in the favorable group. Patients in the intermediate group were 3.0 (95% CI, 1.8-4.9) times less likely to achieve CR than those in the favorable group.
The analysis of factors related to CIR was carried out using the Gray method for comparing the cumulative incidence of a competing risk. Univariate analysis identified the following factors to have prognostic significance: cytogenetic risk (P < .001), age (P < .001), leukocyte count (P < .001), treatment protocol (P = .002), and number of induction courses (P = .007). A forward stepwise multivariate Cox regression analysis of 711 cases with complete data on these variables selected cytogenetic risk (P < .001), age (P < .001), leukocyte count (P < .001), and number of induction courses (P = .01) to the model as joint predictors of CIR, where the P value for each variable is adjusted for variables preceding it in the list. When all variables in the model were considered, patients in the adverse cytogenetic group were 3.0 (95% CI, 2.2-4.0) times more likely to relapse than those in the intermediate group and 4.4 (95% CI, 2.6-7.8) times more likely to relapse than patients in the favorable group. Patients in the intermediate group were 1.5 (95% CI, 1.1-1.9) times more likely to relapse than those in the favorable group.
The analysis of factors related to OS was carried out using the Cox regression model. Univariate analysis identified the following factors to have prognostic significance: cytogenetic risk (P < .001), treatment protocol (P < .001), age (P < .001), leukocyte count (P < .001), infection at study entry (P < .001), number of induction courses (P = .009), and percent marrow blasts (P = .04). A forward stepwise multivariate analysis of 1099 cases with complete data on these variables selected age (P < .001), cytogenetic risk (P < .001), leukocyte count (P < .001), and infection at study entry (P = .001) to the model as joint predictors of OS, where the P value for each variable is adjusted for variables preceding it in the list. When all variables in the model were considered, patients in the adverse cytogenetic group were 2.1 (95% CI, 1.8-2.5) times more likely to die than those in the intermediate group and 4.3 (95% CI, 2.9-6.3) times more likely to die than patients in the favorable group. Patients in the intermediate group were 2.0 (95% CI, 1.6-2.5) times more likely to die than those in the favorable group.
Discussion
Our data, derived from a large group of adults with de novo AML with prolonged follow-up, show that specific cytogenetic findings at diagnosis are predictive of treatment outcome. This study is one of only a few large prospective series examining prognostic impact of cytogenetics in AML11,13,15 and includes a relatively homogeneous group of de novo AML patients who received similar induction chemotherapy, with most receiving modern intensification treatment. It is important to recognize that both our study and 3 other recent studies11,13,15 validate observations made originally by the International Workshops on Chromosomes in Leukemia in the 1980s1 2 and, it is hoped, will result in reaching a consensus with respect to the prognostic importance of specific pretreatment cytogenetic findings in AML.
What does our large series add to the literature on cytogenetics in AML? Our initial goal was to identify the clinical significance of recurring chromosome abnormalities in previously untreated adults with de novo AML. Early into the analysis, we noted that although the most common balanced rearrangements, t(8;21), inv(16)/t(16;16), and t(9;11), were infrequently associated with a complex karyotype (Table 3), considerable proportions of patients with these rearrangements harbored secondary abnormalities. Therefore, we analyzed the impact of secondary abnormalities and karyotype complexity and found that neither the presence of a single secondary abnormality nor a complex karyotype influenced the outcome of AML patients with t(8;21), inv(16)/t(16;16), and t(9;11) who received contemporary treatment regimens. Our data, similar to one study11 but in contrast to another,42 do not show that secondary del(9q) confers a poor prognosis in patients with t(8;21). However, it is still possible that other specific secondary chromosome aberrations, occurring with a frequency too low to be currently tested for outcome, might affect prognosis of patients with t(8;21), inv(16)/t(16;16), or t(9;11). Furthermore, secondary aberrations may become important when therapies targeting specific molecular rearrangements generated by primary abnormalities are used. This possibility is suggested by a recent study43 reporting that t(15;17)-positive acute promyelocytic leukemia (APL) patients with secondary aberrations were significantly less likely to benefit from treatment with ATRA than patients with t(15;17) alone. Other studies examining the impact of secondary abnormalities in APL have not confirmed this observation.44 45
We next sought to dissect the prognostic impact of the presence of 3 or 4 versus 5 or more abnormalities in patients who did not harbor t(8;21), inv(16)/t(16;16), or t(9;11). Patients with at least 5 abnormalities were older and had an inferior survival than patients with 3 or 4 abnormalities. Nonetheless, each of these patient subsets had a significantly lower probability of long-term OS than patients with a normal karyotype. Our data are in agreement with another series40 and provide justification for using at least 3 abnormalities as a definition of complex karyotype for risk stratification.
Unlike other large studies,11,13,15 we analyzed the outcome of patients with structural aberrations leading to loss of 5q, 7q, and 20q separately from outcome of patients with, respectively, −5 and del(5q), −7 and del(7q), and −20 and del(20q). We found that, similarly to deletions, unbalanced translocations or additions involving 5q, 7q, and 20q seldom occurred as isolated aberrations and that in most patients they were associated with complex karyotypes with at least 3 abnormalities. In a nonprioritized analysis, loss of 5q and 20q bestowed prognosis as poor as that of patients with −5 and del(5q) and with −20 and del(20q), respectively; the outcome of patients with loss of 7q was comparable to the poor outcome of patients with −7. However, in the final risk group assignment, patients with loss of 7q were classified in the intermediate category and those with −7 in the adverse group; the del(7q) group in our study was too small to be classified. In the Medical Research Council (MRC) study,11 patients with −7 were also considered to have adverse and those with del(7q) (in the absence of adverse karyotypic features) intermediate risk. These data are important in the context of recent studies using multicolor spectral karyotyping, which have demonstrated that many aberrations recognized by G-banding alone as deletions, including del(7q) and del(5q), are in fact cryptic unbalanced translocations resulting in loss of material from the respective chromosomes.46 47
The current study confirms and extends previous CALGB results indicating that outcome of patients with balanced 11q23 translocations depends on which partner chromosome is involved.9 While the CR rate, CIR, and OS of patients with t(9;11) were not statistically different from those of patients with a normal karyotype, the OS of patients with t(6;11) or t(11;19)(q23;p13.1) was significantly shorter, with no survivors at 5 years, despite CR rates comparable to the normal group. Thus, both our results and others3 11 support classifying patients with t(9;11) into the intermediate prognostic category, separately from patients with t(6;11) and t(11;19)(q23;p13.1) who have poor outcome. Although patients with other translocations involving 11q23 also appear to have poor outcome, the definitive assignment of risk category for patients with each of the individual 11q23 translocations will be possible only when a larger number of patients are analyzed in prospective studies.
Many other aberrations associated with poor outcome in our nonprioritized analysis either never (eg, +14, loss of 17p) or rarely (eg, +4, del(7q)) occurred as a sole abnormality and were detected mainly as part of a complex karyotype. However, other recurrent abnormalities occurred in sufficient numbers in the absence of a complex karyotype to justify risk stratification, for example, t(6;9). Our data corroborate the results of others48,49 in that AML patients with t(6;9) are younger and have a dismal prognosis, with a 38% CR rate and no patient surviving 5 years. This poor outcome is not due to secondary abnormalities, because 6 of the 8 patients had t(6;9) as their sole aberration, and the karyotype of the 2 remaining patients was not complex. These data differ from the MRC studies that included patients with t(6;9) in the intermediate prognosis group.11 15 We emphasize, however, that some of the cytogenetic groups in Tables 9 and 10, including t(6;9), comprised relatively small numbers of patients. Consequently, further large prospective studies are necessary to confirm our risk group assignment of such smaller cytogenetic categories.
Our cytogenetic risk system (Table 10) shares many common features with the MRC11 and Southwest Oncology Group/Eastern Cooperative Oncology Group13 (SWOG/ECOG) classifications but also differs in some aspects from them. For the favorable risk category, our system is like the MRC (and the German AML Study Group40) but differs from SWOG/ECOG in that we included all patients with t(8;21) in the favorable group, whereas SWOG/ECOG classified patients with t(8;21) and del(9q) or a complex karyotype in the unfavorable risk category. We also did not include patients with del(16q) in the favorable risk group, because del(16q) often differs from inv(16)/t(16;16) at the molecular level and has not been associated with a favorable outcome comparable to that of inv(16)/t(16;16).50-52 The intermediate subset in our series contrasts for several chromosome aberrations with both the MRC and SWOG/ECOG systems.11,13 This subset requires close scrutiny because therapeutic recommendations will be made relative to the appropriateness of SCT or other alternative therapy in first CR. Patients with isolated +8 or +8 with 1 abnormality other than t(8;21), inv(16)/t(16;16), or t(9;11) have been assigned to the intermediate risk group for CIR and the adverse risk group for OS. Patients with +21 have been classified as having intermediate risk for OS but adverse for CIR, whereas patients with +11 and +13 have been classified as having intermediate risk for both CIR and OS. However, each of these categories comprised all patients with a given trisomy, both those with an isolated trisomy and patients with a trisomy accompanied by other aberrations. A recent analysis from CALGB that also comprised patients enrolled on CALGB 9621, which includes SCT as postremission therapy,53 has shown that when only patients with isolated +8, +11, +13, and +21 were examined their OS was significantly worse than that of patients with a normal karyotype.54 Finally, because we desired to make definitive recommendations as to the outcome of specific karyotypes, we did not include in our risk classification system cytogenetic groups with too few patients to be analyzed. These include patients with del(7q) and +22 that were assigned intermediate risk status in the MRC classification.11
To assess the predictive value of our cytogenetic risk assessment model relative to other pretreatment prognostic factors, we performed multivariate analyses with respect to the main end points. These analyses demonstrated that cytogenetic risk, age, and presenting leukocyte count were most significantly associated with predicting attainment of CR. For CIR, cytogenetic risk, age, leukocyte count, and number of induction courses were predictive. In contrast, for survival, age entered the model before cytogenetic risk group and presenting leukocyte count, suggesting that other factors associated with elderly AML, independent of cytogenetics, contribute to outcome of patients with AML. These analyses confirm the importance of cytogenetics as a prognostic tool for predicting attainment of CR, CIR, and OS. The CALGB is currently using pretreatment cytogenetics to stratify patients for different types of postremission therapy.53 Ongoing and future studies will likely further modify prognostic categorization of AML patients by subdividing cytogenetic risk groups according to the results of molecular genetic investigations detecting such prognostically relevant mutations as an internal tandem duplication of the FLT3 gene,55,56 partial tandem duplication of the MLL gene,57 58 and other, as yet undiscovered, genetic alterations in AML.
This paper is dedicated to the memory of Richard K. Dodge.
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study and contributed at least 5 patients: Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Harold O. Goodman, and Mark J. Pettenati (grant no. CA03927); University of Maryland Cancer Center, Baltimore: David A. Van Echo, Stuart Schwartz, Joseph R. Testa, and Judith Stamberg (grant no. CA31983); Dana-Farber Cancer Institute, Boston, MA: George P. Canellos and Ramana Tantravahi (grant no. CA32291); North Shore University Hospital, Manhasset, NY: Daniel R. Budman and Prasad R. K. Koduru (grant no. CA35279); Weill Medical College of Cornell University, New York, NY: Scott Wadler and Ram S. Verma (grant no. CA07968); Duke University Medical Center, Durham, NC: Jeffrey Crawford and Sandra H. Bigner (grant no. CA47577); University of Alabama at Birmingham: Robert Diasio and Andrew J. Carroll (grant no. CA47545); University of Iowa Hospitals, Iowa City: Gerald H. Clamon and Shivanand R. Patil (grant no. CA47642); Dartmouth Medical School, Lebanon, NH: Marc S. Ernstoff and Doris H. Wurster-Hill (grant no. CA04326); University of Minnesota, Minneapolis: Bruce A. Peterson and Diane C. Arthur (grant no. CA16450); University of North Carolina, Chapel Hill: Thomas Shea and Kathleen W. Rao (grant no. CA47559); Long Island Jewish Medical Center, Lake Success, NY: Marc Citron, Alan L. Shanske, and Prasad R. K. Koduru (grant no. CA11028); University of Missouri/Ellis Fischel Cancer Center, Columbia: Michael C. Perry, Judith H. Miles, Jeffrey R. Sawyer, and Tim Huang (grant no. CA12046); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano and Constance K. Stein (grant no. CA21060); University of Tennessee Cancer Center, Memphis: Harvey B. Niell and Sugandhi A. Tharapel (grant no. CA47555); Walter Reed Army Medical Center, Washington, DC: Joseph J. Drabick and Rawatmal B. Surana (grant no. CA26806); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (grant no. CA02599); University of California, San Diego: Stephen L. Seagren and Renée Bernstein (grant no. CA11789); University of Chicago Medical Center, IL: Gini Fleming, Michelle M. LeBeau, and Diane Roulston (grant no. CA41287); Finsen Institute, Copenhagen, Denmark: Preben Philip; University of Massachusetts Medical Center, Worcester: Mary Ellen Taplin and Philip L. Townes (grant no. CA37135); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett and Michael S. Watson (grant no. CA77440); Massachusetts General Hospital, Boston: Michael L. Grossbard and Leonard L. Atkins (grant no. CA 12449); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (grant no. CA04457); Medical University of South Carolina, Charleston: Mark R. Green and Eduardo S. Cantú(grant no. CA03927); McGill Department of Oncology, Montreal, QC: Brian Leyland-Jones and Jacqueline Emond (grant no. CA31809).
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2002-03-0772.
Supported by National Cancer Institute grants CA31946, 16058, and 77658, Kimmel Cancer Research Foundation, Leukemia and Lymphoma Society of America, D. Warren Brown Foundation, and the Coleman Leukemia Research Fund.
J.C.B. and K.M. contributed equally to this work.
A complete list of the Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists who participated in this study and contributed at least 5 patients, and additional grant support for participating institutions appear in “.”
Richard K. Dodge died on August 24, 2002.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John C. Byrd, Division of Hematology and Oncology and the Comprehensive Cancer Center, The Ohio State University, B302A Starling Loving Hall, 320 West 10th Ave, Columbus, OH 43210-1240; e-mail: byrd-3@medctr.osu.edu.

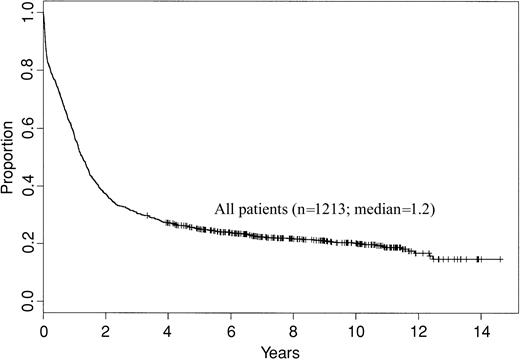
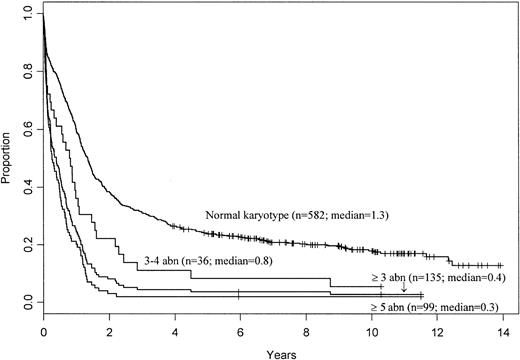
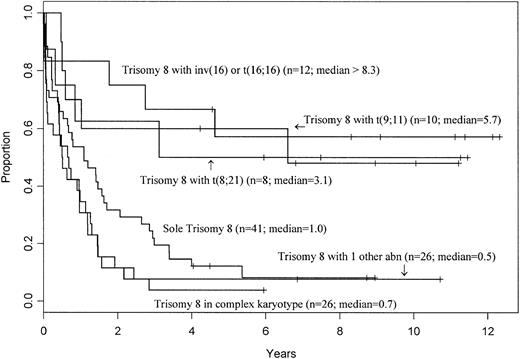
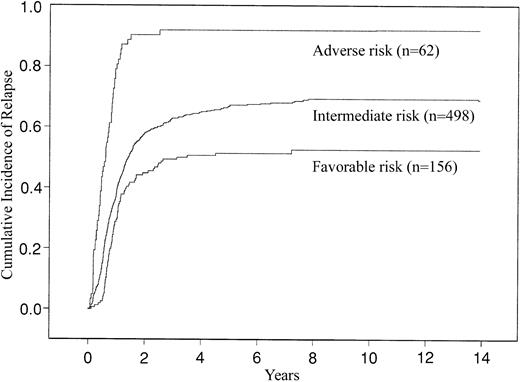

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal