Murine leukocytes are thought to express α2-3-sialylated and α1-3-fucosylated selectin ligands such as sialyl Lewis x (sLex), although monoclonal antibodies (mAbs) to sLex or Lex reportedly do not bind to murine leukocytes. We observed that P- and E-selectin bound to pronase-sensitive ligands on murine monocytic WEHI-3 cells and murine neutrophils, indicating that the ligands for both selectins are glycoproteins. CSLEX-1, HECA-452, and other widely used mAbs to sLex and Lex did not bind to WEHI-3 cells and bound at very low levels to murine neutrophils. Only the anti-sLex mAbs 2H5 and KM93, which also recognize nonfucosylated glycans, bound to WEHI-3 cells. 2H5 and KM93 bound to pronase-resistant structures, indicating that the mAbs did not identify selectin ligands. Treatment of WEHI-3 cells with glycosidases or chlorate demonstrated that sialic acid modifications, α1-3-galactosylation, or sulfation did not mask epitopes for mAbs to sLex or Lex. Compared to human promyelocytic HL-60 cells, WEHI-3 cells and murine neutrophils expressed low α1-3-fucosyltransferase activities. Consistent with very low endogenous fucosylation, forced fucosylation of intact WEHI-3 cells or murine neutrophils by exogenous α1-3-fucosyltransferase FTVI and GDP-fucose created many new epitopes for anti-sLexmAbs such as HECA-452 and CSLEX-1. Nevertheless, forced fucosylation of intact cells did not significantly augment their ability to bind to fluid-phase P- or E-selectin or to roll on immobilized P- or E-selectin under flow. These data suggest that murine myeloid leukocytes fucosylate only a few specific glycans, which interact preferentially with P- and E-selectin.
Introduction
During inflammation, binding of selectins to cell-surface glycoconjugates mediates rolling adhesion of leukocytes on vascular surfaces.1,2 P- and E-selectin, expressed on activated platelets and/or endothelial cells, bind to ligands on leukocytes. L-selectin, expressed on leukocytes, binds to ligands on some endothelial cells and on other leukocytes. All 3 selectins recognize α2-3-sialylated and α1-3-fucosylated glycans such as sialyl Lewis x (sLex; NeuAcα2-3Galβ1-4[Fucα1-3]GlcNAcβ1-R), a terminal component of oligosaccharides that can be attached to proteins or lipids.3 On human leukocytes, the predominant ligand for P-selectin is P-selectin glycoprotein ligand-1 (PSGL-1), a sialomucin that binds to P-selectin through an N-terminal peptide bearing both tyrosine sulfates and a core-2 O-glycan capped with sLex.4 Less well-characterized are the physiologically relevant ligands for E-selectin on human leukocytes. Enzymatic desialylation of human leukocytes eliminates binding to E-selectin,5 and the selectins do not bind to leukocytes from patients with leukocyte adhesion deficiency-2, who lack fucosylated glycans.6 Human leukocytes that bind selectins express epitopes for many mAbs to sLex and related glycans. One such mAb, HECA-452, binds to an sLex-related antigen termed cutaneous lymphocyte antigen (CLA).7,8 CLA is reportedly expressed primarily on PSGL-1, and expression of CLA correlates with the ability of memory/effector T cells to bind E-selectin.9 This has led to the proposal that CLA on PSGL-1 is the physiologically relevant ligand for E-selectin.9 This hypothesis, based solely on correlative data, has been challenged by the observation that some human leukocyte cell lines bind to P- and E-selectin, yet do not express epitopes for anti-sLex mAbs such as HECA-452.10 11
Even less understood is the nature of the glycoconjugates on murine leukocytes that interact with selectins. As on human leukocytes, PSGL-1 on murine leukocytes binds preferentially to P-selectin through an N-terminal region that probably requires both tyrosine sulfation and specific O-glycosylation.1,4 Enzymatic desialylation of murine leukocytes eliminates binding to E- and P-selectin,5 and selectins do not bind to leukocytes from mice that are deficient in FTVII and FTIV, the α1-3-fucosyltransferases normally expressed in these cells.12-14 In vitro activation of a murine T-cell line leads to expression of CLA on PSGL-1, which correlates with binding to E-selectin.15 These combined data suggest that selectins recognize sLex-related glycans on murine leukocytes. However, many mAbs to sLex and Lex fail to bind to murine leukocytes.16 17 It has been widely assumed that unknown glycan modifications, perhaps unique to murine tissues, mask the epitopes for these mAbs, but no direct structural characterization of glycans on murine leukocytes has been performed. An alternative possibility is that murine leukocytes express very low levels of fucosylated glycans that bind well to selectins but not to anti-sLex mAbs.
In this study we compared the expression of fucosylated glycan epitopes and selectin ligands on murine monocytic WEHI-3 cells and murine neutrophils. Our data suggest that very limited fucosylation of specific glycans is sufficient to confer binding to P- and E-selectin. Anti-sLex mAbs such as HECA-452 cannot be used to identify selectin ligands, and enhanced expression of such epitopes does not necessarily augment selectin binding.
Materials and methods
Proteins
IgM anti–carbohydrate mAbs (all murine except rat-derived HECA-452) were as follows: anti-sLex: CSLEX-1 and HECA-452 (hybridomas from American Type Culture Collection [ATCC]), 2F3 and 2H5 (Pharmingen, Franklin Lakes, NJ), KM93 (Kamiya Biomedical Company, Seattle, WA), anti–VIM-2 (Immunotech, Fullerton, CA), and CHO-131, which binds to sLex on a core-2 O-glycan18 (a gift from Dr Bruce Walcheck, University of Minnesota); and anti-Lex: MMA (BD Bioscience), M-G1120 (Serotec, Raleigh, NC), P12 and V1MC6 (Caltag, Burlingame, CA), HI98 (Pharmingen), and SMLEX-M3 (a gift from Dr Kwame Nyame, University of Oklahoma Health Sciences Center).
Rat anti–mouse PSGL-1 mAb 4RA10 (IgG1)19 was a gift from Dr Dietmar Vestweber (University of Muenster). Anti–human PSGL-1 mAbs PL1 and PL2 (both IgG1) were prepared as described.20MOPC21, a control murine IgG1 mAb, and MOPC104E, a control murine IgM mAb, were from Pharmingen. Fluorescein isothiocyanate (FITC)–conjugated GSI-B4, a plant lectin that recognizes α1-3–linked galactose residues, was from Vector (Burlingame, CA).
The expression vector pCDM8 encoding the extracellular domain of CD45 or the lectin and epidermal growth factor (EGF) domains plus the first 2 repeats of murine P-selectin or E-selectin, each fused to the CH2, CH3, and CH4 domains of human IgM,12 were a gift from Dr John Lowe (University of Michigan). COS-7 cells grown in Dulbecco modified Eagle medium (DMEM) with 2% Nuserum (Collaborative Research, Bedford, MA) were transiently transfected with each construct, and IgM chimeras were recovered in conditioned medium.
Cells
Murine monocytic WEHI-3 cells were cultured in Iscove modified Dulbecco medium (GibcoBRL) with 10% fetal bovine serum (FBS), and human promyelocytic HL-60 cells were cultured in DMEM (GibcoBRL) with 10% FBS at 37°C.
Flow cytometry
Neutrophils were pretreated with 20 μg/mL mouse Fc block (Pharmingen) for 20 minutes at 4°C or with 20 μg/mL Fc Receptor Blocker (Accurate Chemical & Scientific Corporation). Cells (0.5-1 × 106) in 50 μL Hanks balanced salt solution containing 1% fetal bovine serum (HBSS/FBS) were incubated for 30 minutes at 4°C with 1-2 μg anti–carbohydrate mAb. Alternatively, the cells were incubated with diluted conditioned medium containing P- or E-selectin IgM chimera or an equivalent dilution of control CD45 IgM chimera. In the selectin-binding experiments, some cells were pretreated with anti–PSGL-1 mAb 4RA10 or PL1. The cells were washed with HBSS/FBS and incubated for 20 minutes at 4°C with 15 μg/mL FITC-conjugated goat F(ab)′2 fragments to mouse IgM (Caltag) or FITC-conjugated goat anti–human IgM (Chemicon). After washing, the cells were resuspended in 0.4 mL HBSS/FBS and analyzed by flow cytometry on a FACScan (Becton Dickinson). Data were collected using the CellQuest program. Light scatter–gated events (4500 to 10 000 per sample) were plotted on a log scale of fluorescence intensity. Nonviable cells, measured by uptake of propidium iodide, were excluded from analysis.
Immunoblots
Protein samples of 100 μg were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and transferred to nitrocellulose (Schleicher and Schuell). All incubations were at room temperature. The filter was blocked with 10% nonfat dry milk in 10 mM Tris (tris(hydroxymethyl)aminomethane), 150 mM NaCl, pH 7.5 for 1 hour, washed with 10 mM Tris, pH 7.5, 300 mM NaCl, 0.05% Tween 20, and incubated with 10 μg/mL mAb to sLex (CSLEX-1, HECA-452, KM93, or 2H5) or to Lex (P12 or HI98) in 10 mM Tris, pH 7.5, 150 mM NaCl, 1% nonfat dry milk, 0.05% Tween 20 for 1 hour. The filter was washed and incubated with a 1:15 000 dilution of goat anti–mouse IgM conjugated to peroxidase (Pierce) for 1 hour. After washing, bound antibodies were detected by enhanced chemiluminescence (Amersham).
Sodium chlorate treatment
WEHI-3 cells were cultured in sulfate-deficient RPMI medium (GibcoBRL) containing 20% dialyzed FBS and 10 mM sodium chlorate (Sigma) for 72 hours.23 Control experiments confirmed that this incubation blocked uptake of [35S]sulfate into newly synthesized proteins.
Sialidase, α-galactosidase, and pronase treatment
Cells (5-10 × 106/0.5 mL) were treated with 0.3 U sialidase from Arthrobacter ureafaciens (Boehringer) plus 2.5 μg sialidase from Vibrio cholera (Serva) or with 500 μg α-galactosidase from green coffee beans (Calbiochem) in HBSS containing 1% human serum albumin (HBSS/HSA) for 1 hour at 37°C. Alternatively, cells (5-10 × 106/0.5 mL) were treated with 500 μg pronase (Calbiochem) in HBSS containing 0.1% HSA for 45 minutes at 37°C. The cells were then washed with HBSS/HSA and analyzed by flow cytometry.
α1-3-fucosyltransferase treatment of intact cells
To introduce sLex determinants on the cell surface, 5-10 × 106 WEHI-3 cells or murine neutrophils were treated with 1 mM guanosine diphosphate (GDP)–fucose, 20 mU/mL α1-3-fucosyltransferase VI (FTVI) (Calbiochem), and 10 mM MnCl2 in 0.5 mL HBSS/HSA for 45 minutes at 37°C.24
Enzymatic synthesis of glycopeptides
The peptide backbone of glycopeptides GP-4 and GP-5 corresponds to residues 45-62 of human PSGL-1, EYEYLDYDFLPET*EPPEM, where the asterisk indicates Thr57 modified with an O-glycan. Sialylated and nonsialylated core-2 O-glycans at Thr57 on GP-4 and GP-5, respectively, were synthesized enzymatically25 (for O-glycan structures, see Figure 7). In matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra, the observed m/z for the [M-H]− molecular ion of GP-4 was 3009.8 (calculated m/z 3010.1), and the observed m/z for the [M-H]−molecular ion of GP-5 was 3301.1 (calculated m/z 3301.4).
α1-3-fucosyltransferase assay
Cell pellets were suspended in 50 mM sodium cacodylate, pH 7.0, containing Complete protease inhibitor cocktail (Roche Diagnostics) and 2 mM phenylmethylsulfonyl fluoride (PMSF). Cells were sonicated for 3 10-second bursts followed by addition of Triton X-100 (final concentration of 1%). The cell extracts were incubated on ice for 45 minutes and then assayed immediately. α1-3-fucosyltransferase assays were performed using 0.5 mM glycopeptide acceptor (GP-4 or GP-5), 100 pmol GDP-[3H]Fuc (2700 cpm/pmol; American Radiolabeled Chemicals), and 100 μg cell protein in 20 μL 0.1 M sodium cacodylate, pH 7.0, containing 20 mM MnCl2, 5 mM adenosine triphosphate (ATP), 15 mM Fuc, 0.5% Triton CF-54, and protease inhibitor cocktail. After 4 hours at 37°C the reaction mixtures were diluted with water to a final volume of 800 μL and extracted with 800 μL chloroform-methanol (2:1). The aqueous phase was dried under vacuum, dissolved in water, and the radiolabeled glycopeptide products were separated from GDP-[3H]Fuc using Sep-Pak Vac C18 (1 mL) cartridges (Waters Corporation). The bound glycopeptide samples were eluted with methanol, and the radioactivities of the eluted samples were measured.
Analysis of α1-3 fucosyltransferase reaction products
Radiolabeled glycopeptide products were filtered on a Spin-X membrane (Corning Costar) and subjected to reversed phase C18 high-performance liquid chromatography (HPLC; Beckman System Gold). The following solvent system was used at 1 mL/min flow rate: 1-10 minutes, isocratic 20% aqueous acetonitrile containing 0.1% trifluoroacetic acid (TFA); 10-70 minutes, linear gradient of 20% to 45% aqueous acetonitrile containing 0.1% TFA. The radioactivity of collected fractions (1 minute) was measured. As a positive control, [3H]fucosylated glycopeptide samples were synthesized by incubating GP-4 or GP-5 (6 μM) as acceptors and GDP-[3H]Fuc (1100 cpm/pmol) as donor with 2 mU α1-3-FTVI (Calbiochem) for 15 hours. The reaction products were analyzed by HPLC.
Cell rolling under flow
P-selectin, E-selectin, or control CD45 IgM chimeras were captured on goat anti–human IgM antibodies immobilized in a parallel-plate flow chamber. Selectin site densities were measured by binding of 125I-labeled anti– P-selectin mAb RB40.34 or anti–E-selectin mAb 10E9.6.26 Untreated or FTVI-treated WEHI-3 cells or murine leukocytes (106/mL in HBSS/HSA) were perfused over the chimeras at the indicated wall shear stresses. The accumulated number of rolling cells was measured after 4 minutes of perfusion with a videomicroscopy system coupled to an image analysis system.27 For each experiment, adherent cells in 10 to 12 × 20 fields were counted. In some experiments, cells were pretreated with pronase or perfused in buffer containing 10 mM EDTA (ethylenediaminetetraacetic acid).
Reverse transcriptase–polymerase chain reaction
Total RNA was isolated from murine lung or spleen or from WEHI-3 or HL-60 cells using Trizol (GibcoBRL). Aliquots (5-μg) of RNA were treated with or without reverse transcriptase (RT) for first-strand cDNA synthesis in a total volume of 20 μL using the SuperScript Preamplification System (GibcoBRL). Aliquots (2-μL) from this reaction were subjected to polymerase chain reaction (PCR) for 30 cycles with 2.5 units of Taq polymerase, primers for murine FTVII or FTIV, and nucleotides in a final volume of 50 μL. PCR primers derived from cDNA sequences for murine FTVII and FTIV were as follows: FTVII: sense 5′-GTG GTC TTC CAC CAC CGT GAG-3′, antisense 5′-AGC AGC AGG AGT TCA AGC CTG-3′; FTIV: sense 5′-GTC CGT TAC TAC CAC CAG CTG-3′; antisense 5′-TCG CTG GAA CCA GTC TGC CAA-3′.28PCR products were resolved in 2% agarose and stained with ethidium bromide.
Results
Differential protease sensitivity of ligands for P- or E-selectin on murine and human leukocytes
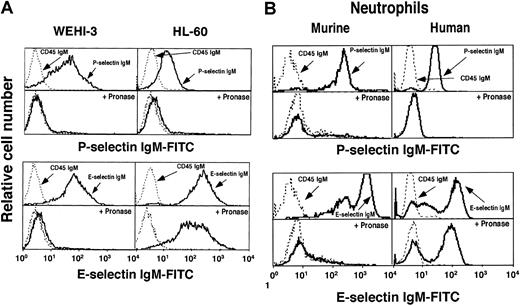
We used flow cytometry to compare binding of murine P- and E-selectin IgM chimeras to murine monocytic WEHI-3 cells, human promyelocytic HL-60 cells, and murine and human neutrophils (Figure1). Both selectins bound to each cell type. A CD45 IgM chimera, used as a negative control, did not bind to either cell. As expected for selectin-dependent interactions, chelation of Ca++ ions with EDTA or treatment of cells with sialidase eliminated binding of both selectins (data not shown). P-selectin bound preferentially to the N terminus of PSGL-1 on both cells, because a mAb to the N-terminal region of murine PSGL-1 (4RA10) or human PSGL-1 (PL1) blocked binding to the respective cells. The anti–PSGL-1 mAbs did not inhibit binding of E-selectin (data not shown). Pretreatment of WEHI-3 and HL-60 cells with pronase prevented binding of P-selectin, confirming previous studies that PSGL-1 is sensitive to proteases.20,29,30 Pronase did not affect binding of E-selectin to HL-60 cells or human neutrophils, consistent with earlier studies.31 In marked contrast, pronase eliminated binding sites for E-selectin on WEHI-3 cells and murine neutrophils (Figure 1). Pronase also markedly reduced rolling adhesion of WEHI-3 cells, but not HL-60 cells, on E-selectin in flow (data not shown). These data demonstrate that most or all of the ligands for E-selectin on WEHI-3 cells and murine neutrophils are on glycoproteins. In contrast, the resistance of E-selectin ligands on HL-60 cells and human neutrophils to pronase, a mixture of many proteases, suggests that many are on glycolipids or on unusually protease-resistant glycoprotein(s).
P-selectin and E-selectin bind to murine WEHI-3 cells, human HL-60 cells, and murine and human neutrophils.
Cell lines (A) or neutrophils (B) were incubated with saturating concentrations of murine P-selectin or E-selectin IgM chimera or control CD45 IgM chimera. Bound chimera was detected by incubation with FITC-conjugated goat anti–human IgM, followed by flow cytometry. Some cells were pretreated with pronase. The data are representative of at least 10 experiments.
P-selectin and E-selectin bind to murine WEHI-3 cells, human HL-60 cells, and murine and human neutrophils.
Cell lines (A) or neutrophils (B) were incubated with saturating concentrations of murine P-selectin or E-selectin IgM chimera or control CD45 IgM chimera. Bound chimera was detected by incubation with FITC-conjugated goat anti–human IgM, followed by flow cytometry. Some cells were pretreated with pronase. The data are representative of at least 10 experiments.
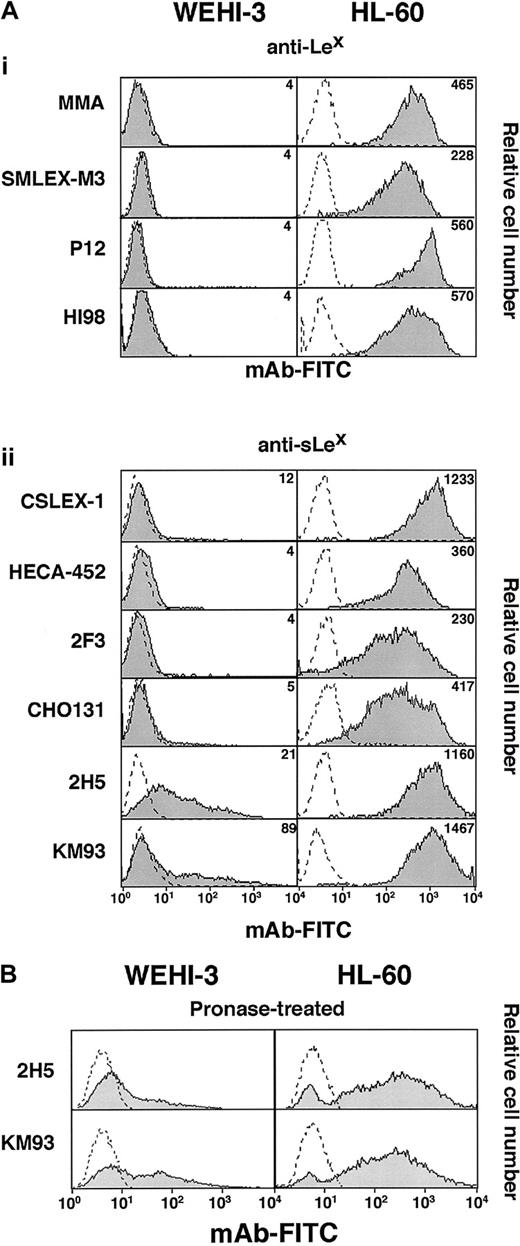
Discordance between expression of selectin ligands and epitopes for anti-sLex or anti-Lex mAbs on WEHI-3 cells
We used flow cytometry to compare binding of a panel of mAbs to sLex or related glycans on WEHI-3 and HL-60 cells. Six mAbs to sLex and 4 mAbs to Lex bound to many sites on HL-60 cells (Figure 2A). In contrast, the anti-sLex mAbs CSLEX-1, HECA-452, CHO131, and 2F3 did not bind to WEHI-3 cells; the apparent low-level binding observed in Figure 2A was not reproducible and was not eliminated by sialidase treatment of the cells, suggesting that it was not specific. The anti-sLex mAbs 2H5 and KM93 bound reproducibly to WEHI-3 cells but only at low levels. Sialidase treatment eliminated binding of 2H5 and KM93 to both WEHI-3 and HL-60 cells, confirming the sialic acid–dependent nature of the binding (data not shown). The anti-Lex mAbs MMA, SMLEX-M3, P12, and HI98 did not bind to WEHI-3 cells (Figure 2A). The anti-Lex mAbs M-G1120 and V1MC6 also failed to bind to WEHI-3 cells, although they bound to many sites on HL-60 cells (data not shown). Finally, mAbs to sLea and Lea, the isomers of sLexand Lex, and a mAb to an internally fucosylated variant of sLex (VIM-2) did not bind to WEHI-3 cells (data not shown).
mAbs to sLex or Lex bind differently to WEHI-3 and HL-60 cells.
(A) Cells were incubated with saturating concentrations of mAbs to Lex or sLex or with a nonbinding control IgM mAb. Bound antibody was detected with FITC-conjugated goat F(ab)′2 fragments to murine IgM. (B) Cells pretreated with pronase were analyzed for binding of mAbs 2H5 and KM93. The data are representative of at least 15 experiments.
mAbs to sLex or Lex bind differently to WEHI-3 and HL-60 cells.
(A) Cells were incubated with saturating concentrations of mAbs to Lex or sLex or with a nonbinding control IgM mAb. Bound antibody was detected with FITC-conjugated goat F(ab)′2 fragments to murine IgM. (B) Cells pretreated with pronase were analyzed for binding of mAbs 2H5 and KM93. The data are representative of at least 15 experiments.
Previously shown to bind to murine or rat leukocytes or leukocyte cell lines were 2H5 and KM93, which also were thought to identify the sLex-related ligands for P- and E-selectin on these cells.32 33 However, treatment of WEHI-3 cells with the broad range of proteases in pronase did not detectably diminish the epitopes for these mAbs (Figure 2B), whereas pronase treatment eliminated binding sites for P- and E-selectin (Figure 1A). Furthermore, 2H5 and KM93, like other mAbs to sLex or Lex, did not detectably bind to glycoproteins in Western blots of WEHI-3 cell lysates, although they bound to numerous glycoproteins in HL-60 cell lysates (Figure3). Finally, 2H5 and KM93 did not inhibit binding of E-selectin to WEHI-3 cells (data not shown). Thus, the epitopes on WEHI-3 cells defined by 2H5 and KM93 are primarily on pronase-resistant glycoconjugates, which are likely to be glycolipids, and these epitopes do not represent the major ligands for P- and E-selectin. These results demonstrate that expression of selectin ligands on WEHI-3 cells does not correlate with expression of epitopes for mAbs to sLex-related glycans.
mAbs to sLex or Lex bind differently to immunoblots of proteins from WEHI-3 and HL-60 cells.
Cell extracts were resolved by SDS-PAGE, transferred to membranes, and probed with the indicated mAb. Bound mAb was identified by peroxidase-conjugated goat anti–murine IgM coupled to a chemiluminescence detection procedure. The data are representative of at least 6 experiments. DF indicates dye front.
mAbs to sLex or Lex bind differently to immunoblots of proteins from WEHI-3 and HL-60 cells.
Cell extracts were resolved by SDS-PAGE, transferred to membranes, and probed with the indicated mAb. Bound mAb was identified by peroxidase-conjugated goat anti–murine IgM coupled to a chemiluminescence detection procedure. The data are representative of at least 6 experiments. DF indicates dye front.
Modification of sialic acids, sulfation, or α1-3-galactosylation does not mask epitopes for mAbs to Lex or sLex on WEHI-3 cells
Modifications of sialic acid can mask epitopes for some mAbs to sLex.34 Sulfation of the GlcNAc residue in the sLex structure also prevents binding of some mAbs to sLex.35 Conceivably, these or related modifications might also mask epitopes for mAbs to Lex. We treated WEHI-3 and HL-60 cells with sialidase to remove sialic acids from cell-surface glycoconjugates. We cultured other cells in chlorate, an inhibitor of sulfation, for 3 days to allow turnover of previously sulfated molecules. Chlorate is a selective inhibitor of ATP sulfurylase (ATP sulfate adenylyltransferase), which is required for formation of phosphoadenosine phosphosulfate (PAPS), the donor for sulfation reactions. Control and treated cells were then analyzed by flow cytometry for binding of CSLEX-1, a mAb to sLex or MMA, a mAb to Lex (Figure4A). Consistent with previous studies, sialidase treatment of HL-60 cells markedly reduced binding of the anti-sLex mAb, whereas it increased binding of the anti-Lex mAb. However, neither mAb bound to sialidase-treated WEHI-3 cells. Chlorate treatment of WEHI-3 cells did not enhance the binding sites for either mAb, although chlorate treatment of HL-60 cells slightly increased binding of the anti-sLex and anti-Lex mAbs (Figure 4A). Sialidase digestion or chlorate treatment of WEHI-3 cells also failed to unmask binding sites for other mAbs to Lex or sLex (data not shown). Sialidase digestion of WEHI-3 cells was effective, because it prevented binding of P- and E-selectin IgM chimeras to the cells, as it did to HL-60 cells (Figure 4B). Chlorate treatment of WEHI-3 cells also was effective, because it prevented binding to P-selectin, but not E-selectin, consistent with the requirement that PSGL-1 be tyrosine sulfated to bind to P-selectin.36-38 These data demonstrate that neither sialic acid modifications nor sulfation on WEHI-3 cells prevent binding of mAbs to sLex or Lex.
Sialidase, chlorate, or α-galactosidase treatment of WEHI-3 cells does not expose epitopes for mAbs to Lex or sLex.
Cells were untreated (control) or pretreated with sialidase, chlorate, both sialidase and chlorate, or α-galactosidase. (A) The cells were incubated with a nonbinding control IgM or with anti-sLexmAb CSLEX-1 or anti-Lex mAb MMA. Bound mAb was detected with FITC-conjugated goat F(ab)′2 fragments to murine IgM. (B) The cells were incubated with murine P- or E-selectin IgM chimera, followed by FITC-conjugated goat anti–human IgM. (C) The cells were incubated with FITC-conjugated GSI-B4 lectin, which recognizes α1-3–linked galactose residues, or with a nonbinding control IgM or anti-Lex mAb MMA. The data are representative of at least 4 experiments.
Sialidase, chlorate, or α-galactosidase treatment of WEHI-3 cells does not expose epitopes for mAbs to Lex or sLex.
Cells were untreated (control) or pretreated with sialidase, chlorate, both sialidase and chlorate, or α-galactosidase. (A) The cells were incubated with a nonbinding control IgM or with anti-sLexmAb CSLEX-1 or anti-Lex mAb MMA. Bound mAb was detected with FITC-conjugated goat F(ab)′2 fragments to murine IgM. (B) The cells were incubated with murine P- or E-selectin IgM chimera, followed by FITC-conjugated goat anti–human IgM. (C) The cells were incubated with FITC-conjugated GSI-B4 lectin, which recognizes α1-3–linked galactose residues, or with a nonbinding control IgM or anti-Lex mAb MMA. The data are representative of at least 4 experiments.
On murine leukocytes, α1-3–linked galactose may substitute for α2-3–linked sialic acid as a capping group on lactosamine to produce the structure Galα1-3Galβ1-4GlcNAcβ1-R. α1-3-fucosylation of the GlcNAc on this structure might yield a form of Lex that is not recognized by mAbs.39 We used an α-galactosidase to remove α1-3–linked galactose, which was confirmed by markedly reduced binding of GSI-B4, a lectin from Griffonia simplicifolia that recognizes this modification.40This treatment, however, did not unmask binding sites for a mAb to Lex (Figure 4C). Thus, alternative capping with galactose rather than sialic acid does not explain the lack of binding of mAbs to Lex to WEHI-3 cells.
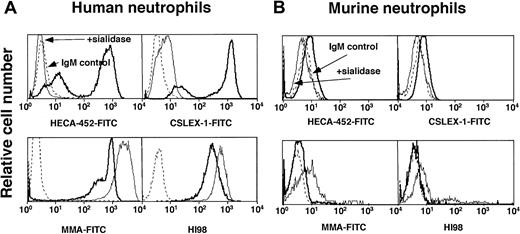
Murine neutrophils express low but detectable levels of epitopes for mAbs to sLex and Lex
To determine whether the data obtained for WEHI-3 and HL-60 cells reflected that for primary myeloid cells, we compared binding of mAbs to sLex or Lex on murine and human neutrophils (Figure 5). Anti-sLex mAbs HECA-452 and CSLEX-1 and anti-Lex mAbs MMA and HI98 bound at high levels to human neutrophils (Figure 5A). Binding was specific, because sialidase treatment of human neutrophils markedly reduced binding of the anti-sLex mAbs and increased binding of the anti-Lex mAbs. The mAbs also bound detectably to murine neutrophils, although at very low levels (Figure 5B). This binding also was specific, as sialidase treatment eliminated binding of the anti-sLex mAbs and slightly increased binding of the anti-Lex mAbs. Thus, unlike WEHI-3 cells, murine neutrophils express detectable epitopes for Lex-related epitopes, although the levels are much lower than those on human neutrophils.
mAbs to sLex and Lex bind at very low levels to murine neutrophils.
Human or murine neutrophils were untreated or pretreated with sialidase. The cells were incubated with a nonbinding control IgM or with anti-sLex mAb HECA-452 or CSLEX-1 or with anti-Lex mAb MMA or HI98. Bound mAb was detected with FITC-conjugated goat F(ab)′2 fragments to murine IgM. The data are representative of at least 3 experiments.
mAbs to sLex and Lex bind at very low levels to murine neutrophils.
Human or murine neutrophils were untreated or pretreated with sialidase. The cells were incubated with a nonbinding control IgM or with anti-sLex mAb HECA-452 or CSLEX-1 or with anti-Lex mAb MMA or HI98. Bound mAb was detected with FITC-conjugated goat F(ab)′2 fragments to murine IgM. The data are representative of at least 3 experiments.
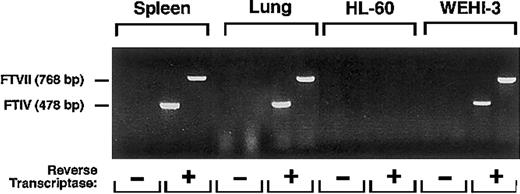
WEHI-3 cells and neutrophils express very low levels of α1-3-fucosyltransferase activity
The low-to-undetectable binding of mAbs to sLex and Lex to murine neutrophils and WEHI-3 cells could be explained if the cells expressed extremely low levels of α1-3-fucosylated glycans. As assessed by RT-PCR, WEHI-3 cells, like cells in murine spleen and lung, expressed mRNA transcripts for FTVII and FTIV, the 2 α1-3-fucosyltransferases known to be expressed in leukocytes41 42 (Figure 6). RT-PCR did not amplify transcripts for murine FTVII and FTIV in human HL-60 cells, demonstrating that the primers were specific for the murine mRNA sequences.
RT-PCR reveals mRNA for FTVII and FTIV in WEHI-3 cells.
Total RNA from murine spleen and lung, used as positive controls, and human HL-60 cells, used as a negative control, and from WEHI-3 cells were incubated in the presence or absence of reverse transcriptase. The samples were then amplified by PCR using primers for murine FTVII or FTIV. The reaction products were resolved on an agarose gel and stained with ethidium bromide. The data are representative of 5 experiments.
RT-PCR reveals mRNA for FTVII and FTIV in WEHI-3 cells.
Total RNA from murine spleen and lung, used as positive controls, and human HL-60 cells, used as a negative control, and from WEHI-3 cells were incubated in the presence or absence of reverse transcriptase. The samples were then amplified by PCR using primers for murine FTVII or FTIV. The reaction products were resolved on an agarose gel and stained with ethidium bromide. The data are representative of 5 experiments.
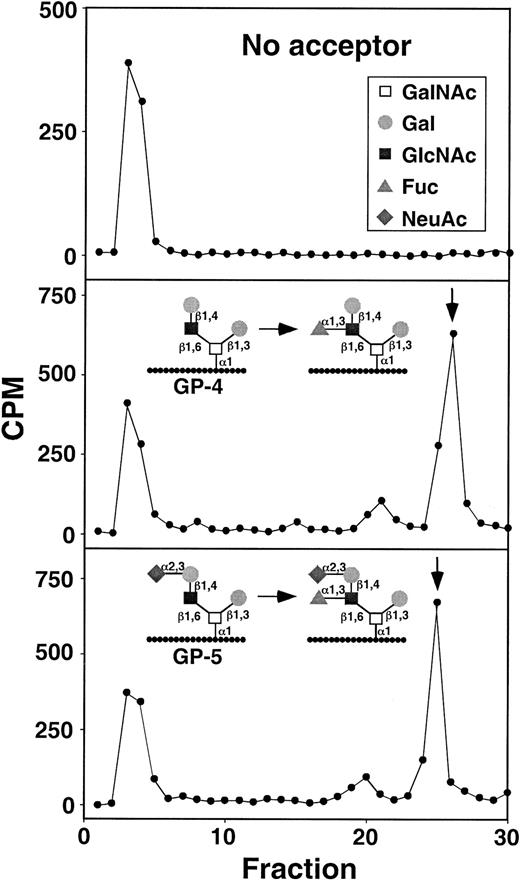
To determine whether WEHI-3 cells and murine leukocytes expressed functional FTVII and FTIV, we measured their enzymatic activities in cell lysates and compared them with the activities in lysates of HL-60 cells and human leukocytes. FTIV preferentially fucosylates nonsialylated acceptors, whereas FTVII preferentially fucosylates sialylated acceptors.43 44 We used sensitive glycopeptide acceptors bearing a core-2 O-glycan on a short peptide derived from the human PSGL-1 sequence to monitor activity of each enzyme. GP-4 capped with lactosamine was used to measure FTIV activity and ability to synthesize the Lex antigen, whereas GP-5 capped with sialyllactosamine was used to measure FTVII activity and ability to synthesize the sLex antigen. Cell lysates were incubated with GDP-[3H]Fuc with no acceptor or with GP-4 or GP-5. Free GDP-[3H]Fuc was separated from3H-labeled glycopeptide products on a C18 reversed phase cartridge. WEHI-3 cells expressed detectable but very small amounts of activity for FTIV and FTVII, which were 20- to 25-fold lower than the activities measured in human HL-60 cells (Table1). Product analysis by HPLC confirmed that the activities in WEHI-3 cells, although very low, specifically generated the Lex and sLex structures from GP-4 and GP-5, respectively (Figure 7). Similar low α1-3-fucosyltransferase activities were observed in murine leukocytes or purified murine neutrophils and, surprisingly, in human leukocytes or purified human neutrophils (Table 1). These data indicate that the α1-3-fucosyltransferase activities measured in disrupted cells correlate poorly with the levels of Lex or sLex epitopes on cell surfaces.
α1-3-fucosyltransferase activities in human and murine leukocytes
| Cells . | α1-3-fucosyltransferase activity (pmol/h/mg) . | |
|---|---|---|
| GP-4 . | GP-5 . | |
| Murine leukocytes | 4.9 ± 0.5 | 8.6 ± 1.7 |
| Murine neutrophils | 6.4 ± 1.9 | 13.7 ± 1.7 |
| WEHI-3 cells | 6.0 ± 0.5 | 5.6 ± 0.6 |
| Human leukocytes | 7.3 ± 0.2 | 12.9 ± 5.1 |
| Human neutrophils | 6.7 ± 1.0 | 27.9 ± 5.6 |
| HL-60 cells | 125.0 ± 3.0 | 254.0 ± 20.8 |
| Cells . | α1-3-fucosyltransferase activity (pmol/h/mg) . | |
|---|---|---|
| GP-4 . | GP-5 . | |
| Murine leukocytes | 4.9 ± 0.5 | 8.6 ± 1.7 |
| Murine neutrophils | 6.4 ± 1.9 | 13.7 ± 1.7 |
| WEHI-3 cells | 6.0 ± 0.5 | 5.6 ± 0.6 |
| Human leukocytes | 7.3 ± 0.2 | 12.9 ± 5.1 |
| Human neutrophils | 6.7 ± 1.0 | 27.9 ± 5.6 |
| HL-60 cells | 125.0 ± 3.0 | 254.0 ± 20.8 |
Enzyme activities were measured using the nonsialylated glycopeptide GP-4 or the sialylated glycopeptide GP-5 as acceptor. Results are expressed as the mean ± range of 2 determinations for HL-60 cells and the mean ± SD of 3 determinations for all other cells.
HPLC characterization of α1-3-fucosyltransferase reaction products formed by WEHI-3 cell lysates.
The arrows indicate the retention times of fucosylated control reaction products formed with FTVI using either GP-4 or GP-5 as acceptor. The radioactivity eluting at fractions 3-4 contains an unidentified nonpeptide contaminant.
HPLC characterization of α1-3-fucosyltransferase reaction products formed by WEHI-3 cell lysates.
The arrows indicate the retention times of fucosylated control reaction products formed with FTVI using either GP-4 or GP-5 as acceptor. The radioactivity eluting at fractions 3-4 contains an unidentified nonpeptide contaminant.
Forced fucosylation of the surfaces of WEHI-3 cells or murine neutrophils by an exogenous α1-3-fucosyltransferase creates epitopes for mAbs to sLex and Lex but does not increase selectin ligands
Low levels of FTIV and FTVII in WEHI-3 cells or murine neutrophils might be sufficient to preferentially fucosylate selectin ligands but not to create many epitopes for mAbs to sLex or Lex. Because of the low activities of the α1-3-fucosyltransferases in murine cells, the mature glycan structures might terminate in simple lactosamine and sialyllactosamine sequences. Such structures could be potential acceptors for α1-3-fucosyltransferase action. To determine whether an exogenous α1-3-fucosyltransferase could create epitopes for mAbs to Lex-related glycans, we treated intact WEHI-3 cells or murine neutrophils with GDP-fucose and α1-3-fucosyltransferase VI. FTVI was used because, unlike FTVII or FTIV, it adds fucose to both sialylated and nonsialylated acceptors and is not known to demonstrate specificity for particular glycoproteins or glycolipids. Treatment with FTVI created many epitopes for the anti-sLex mAb HECA-452 (Figure 8) and for other mAbs to sLex and Lex (data not shown). Sialidase treatment eliminated the binding sites for HECA-452 and increased the binding sites for anti-Lex mAb MMA, consistent with the specificities of these mAbs (Figure 8 and data not shown). Western blot analysis revealed that FTVI treatment created epitopes for HECA-452, CSLEX-1, and MMA on many glycoproteins. Pronase treatment of FTVI-treated cells did not remove all the epitopes, suggesting that some might be on glycolipids or protease-resistant glycoproteins (data not shown).
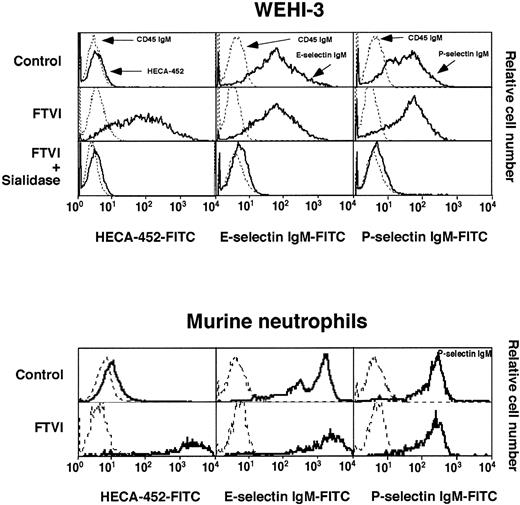
Forced α1-3-fucosylation of WEHI-3 cells or murine neutrophils with FTVI and GDP-fucose creates many sLex and Lex epitopes but does not significantly increase selectin ligands.
WEHI-3 cells or murine neutrophils were incubated with or without FTVI and GDP-fucose. Some of the FTVI-treated WEHI-3 cells were subsequently treated with sialidase. The cells were analyzed for binding of control IgM or the anti-sLex mAb HECA-452 or for binding of control CD45 IgM chimera or P- or E-selectin IgM chimera. The data are representative of at least 8 experiments.
Forced α1-3-fucosylation of WEHI-3 cells or murine neutrophils with FTVI and GDP-fucose creates many sLex and Lex epitopes but does not significantly increase selectin ligands.
WEHI-3 cells or murine neutrophils were incubated with or without FTVI and GDP-fucose. Some of the FTVI-treated WEHI-3 cells were subsequently treated with sialidase. The cells were analyzed for binding of control IgM or the anti-sLex mAb HECA-452 or for binding of control CD45 IgM chimera or P- or E-selectin IgM chimera. The data are representative of at least 8 experiments.
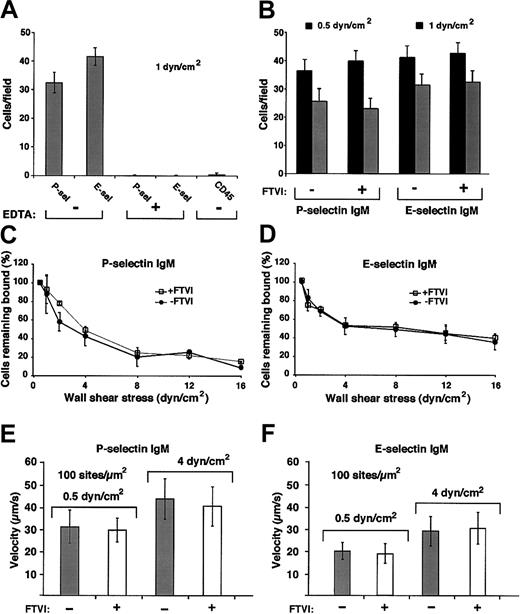
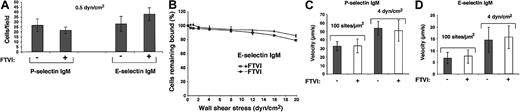
Despite the large increase in sLex and Lexepitopes, forced fucosylation with FTVI either failed to increase or only modestly increased binding of fluid-phase P-selectin or E-selectin to WEHI-3 cells or murine neutrophils (Figure 8). We also compared rolling of control and FTVI-treated cells on immobilized P- or E-selectin in shear flow. WEHI-3 cells rolled in a specific Ca++-dependent manner on murine P- or E-selectin (Figure9A), and similar numbers of control and FTVI-treated cells rolled at all wall shear stresses examined (Figure9B and data not shown). Control and FTVI-treated cells, after accumulation on P- or E-selectin at low shear stress, similarly resisted detachment as wall shear stress was increased (Figure 9C-D). Furthermore, the mean rolling velocities were equivalent for control and FTVI-treated cells (Figure 9E-F). Wild-type and FTVI-treated WEHI-3 cells rolled similarly even at the lowest selectin densities that supported rolling, which should increase the probability of observing differences in rolling behavior (data not shown). Control and FTVI-treated murine neutrophils also rolled similarly on P- and E-selectin (Figure 10A), resisted detachment from E-selectin equivalently as wall shear stress was increased (Figure 10B), and rolled with similar velocities on P- and E-selectin (Figure 10C-D). Thus, the FTVI-mediated addition of epitopes for sLex and Lex to the surfaces of WEHI-3 cells or murine neutrophils did not significantly augment interactions with P- or E-selectin.
Forced α1-3-fucosylation of WEHI-3 cells with FTVI and GDP-fucose does not augment cell rolling on P-selectin or E-selectin in shear flow.
(A) WEHI-3 cells were perfused over immobilized murine P-selectin IgM chimera (90 sites/μm2) or E-selectin IgM chimera (140 sites/μm2) or control CD45 IgM chimera in the presence or absence of EDTA. After 4 minutes, the accumulated number of rolling cells was quantified by counting all cells in each of 4 randomly selected × 40 fields. The data represent the means ± SDs of 4 experiments. (B) Control or FTVI-treated WEHI-3 cells were perfused over P-selectin IgM (90 sites/μm2) or E-selectin IgM (140 sites/μm2) at 0.5 or 1 dyn/cm2. After 4 minutes, the accumulated number of rolling cells was quantified. The data represent the means ± SDs of 4 experiments. (C-D) Control or FTVI-treated cells were allowed to accumulate at 0.5 dyn/cm2, and cell-free buffer was then introduced. Wall shear stress was increased every 30 seconds, and the percentage of remaining adherent cells was determined. The data represent the means ± SDs from 3 experiments. (E-F) Control or FTVI-treated cells were perfused over P- or E-selectin IgM at 0.5 or 4 dyn/cm2. The data represent the means ± SDs for rolling velocities measured for 50 cells from 3 experiments.
Forced α1-3-fucosylation of WEHI-3 cells with FTVI and GDP-fucose does not augment cell rolling on P-selectin or E-selectin in shear flow.
(A) WEHI-3 cells were perfused over immobilized murine P-selectin IgM chimera (90 sites/μm2) or E-selectin IgM chimera (140 sites/μm2) or control CD45 IgM chimera in the presence or absence of EDTA. After 4 minutes, the accumulated number of rolling cells was quantified by counting all cells in each of 4 randomly selected × 40 fields. The data represent the means ± SDs of 4 experiments. (B) Control or FTVI-treated WEHI-3 cells were perfused over P-selectin IgM (90 sites/μm2) or E-selectin IgM (140 sites/μm2) at 0.5 or 1 dyn/cm2. After 4 minutes, the accumulated number of rolling cells was quantified. The data represent the means ± SDs of 4 experiments. (C-D) Control or FTVI-treated cells were allowed to accumulate at 0.5 dyn/cm2, and cell-free buffer was then introduced. Wall shear stress was increased every 30 seconds, and the percentage of remaining adherent cells was determined. The data represent the means ± SDs from 3 experiments. (E-F) Control or FTVI-treated cells were perfused over P- or E-selectin IgM at 0.5 or 4 dyn/cm2. The data represent the means ± SDs for rolling velocities measured for 50 cells from 3 experiments.
Forced α1-3-fucosylation of murine neutrophils with FTVI and GDP-fucose does not augment cell rolling on P-selectin or E-selectin in shear flow.
(A) Control or FTVI-treated neutrophils were perfused over P-selectin IgM (90 sites/μm2) or E-selectin IgM (140 sites/μm2) at 0.5 or 1 dyn/cm2. After 4 minutes, the accumulated number of rolling cells was quantified. The data represent the mean ± SD of 3 experiments. (B) Control or FTVI-treated cells were allowed to accumulate on E-selectin at 0.5 dyn/cm2, and cell-free buffer was then introduced. Wall shear stress was increased every 30 seconds, and the percentage of remaining adherent cells was determined. The data represent the means ± SDs from 3 experiments. (C-D) Control or FTVI-treated cells were perfused over P- or E-selectin IgM at 0.5 or 4 dyn/cm2. The data represent the means ± SDs for rolling velocities measured for 50 cells from 3 experiments.
Forced α1-3-fucosylation of murine neutrophils with FTVI and GDP-fucose does not augment cell rolling on P-selectin or E-selectin in shear flow.
(A) Control or FTVI-treated neutrophils were perfused over P-selectin IgM (90 sites/μm2) or E-selectin IgM (140 sites/μm2) at 0.5 or 1 dyn/cm2. After 4 minutes, the accumulated number of rolling cells was quantified. The data represent the mean ± SD of 3 experiments. (B) Control or FTVI-treated cells were allowed to accumulate on E-selectin at 0.5 dyn/cm2, and cell-free buffer was then introduced. Wall shear stress was increased every 30 seconds, and the percentage of remaining adherent cells was determined. The data represent the means ± SDs from 3 experiments. (C-D) Control or FTVI-treated cells were perfused over P- or E-selectin IgM at 0.5 or 4 dyn/cm2. The data represent the means ± SDs for rolling velocities measured for 50 cells from 3 experiments.
Discussion
A large body of evidence suggests that sLex or related glycans are essential components of selectin ligands on human leukocytes. However, the contribution of sLex to selectin ligands on murine leukocytes must still be inferred from indirect evidence, of which the most important is the absence of selectin ligands on leukocytes from mice that are genetically deficient in FTVII and/or FTIV.12-14 Yet mAbs to sLex or Lex, which bind to human leukocytes or leukocyte cell lines, reportedly do not bind to murine leukocytes or leukocyte cell lines. Here we show that murine monocytic WEHI-3 cells and murine neutrophils have very low α1-3-fucosyltransferase activities, measured with acceptors used by FTVII or FTIV. These activities are apparently sufficient for WEHI-3 cells and murine neutrophils to create selectin ligands but few or no detectable epitopes for mAbs to sLex or Lex. Forced fucosylation with an exogenous α1-3-fucosyltransferase creates many sLexepitopes but does not substantially increase selectin ligands. These data suggest that WEHI-3 cells and murine neutrophils selectively fucosylate a subset of glycoproteins that function as selectin ligands. The discordant expression of selectin ligands and sLexepitopes emphasizes that these epitopes may not correlate with or identify these ligands.
P-selectin bound to PSGL-1, a protease-sensitive mucin, on murine and human leukocytes. The major binding sites for E-selectin on HL-60 cells and other human leukocytes are resistant to digestion with chymotrypsin or trypsin.31,45 Here we show that these ligands are resistant even to pronase, a mixture of many proteases fromStreptomyces griseus. Pronase is commonly used for structural characterization of oligosaccharides because it digests the polypeptide backbone of most glycoproteins at virtually every peptide bond.46-49 The pronase resistance of E-selectin ligands on HL-60 cells and human neutrophils suggests that they are primarily on glycolipids or on glycoproteins with features that resist digestion, for example, unusual mucins with many closely spaced O-glycans. In contrast, pronase cleaved all of the E-selectin ligands on WEHI-3 cells and murine neutrophils, indicating that the ligands on these cells are glycoproteins. The physiological significance of this differential protease sensitivity is not known.
WEHI-3 cells did not interact with a large panel of mAbs to sLex, Lex, or related glycans. The only exceptions were the anti-sLex mAbs 2H5 and KM93, which bound at relatively low levels to WEHI-3 cells, as previously shown for other murine leukocytes.32,33 However, the epitopes identified by these mAbs did not identify ligands for P- or E-selectin because the epitopes, unlike the ligands, were resistant to pronase digestion. Furthermore, recent data indicate that these mAbs do not specifically recognize α1-3-fucose-dependent epitopes. 2H5 binds to some human and murine cell lines that lack detectable expression of α1-3-fucosyltransferase mRNA or enzymatic activity and that do not bind to E-selectin.50 2H5 and KM93 bind equivalently to granulocytes from wild-type mice and from mice deficient in FTIV and FTVII, and KM93 binds to Chinese hamster ovary cells that lack the ability to synthesize GDP-fucose and fucosylated glycans (John B. Lowe, written communication, June 2001). Given the lack of specificity of 2H5 and KM93, there is no definitive immunological evidence that WEHI-3 cells express α1-3-fucosylated glycans, even though they clearly express sialidase-sensitive selectin ligands. Murine neutrophils did express detectable epitopes recognized by mAbs to Lex or sLex, but even these epitopes were far less abundant than those on human leukocytes.
WEHI-3 cells expressed mRNA for FTVII and FTIV, but activities for these enzymes were much lower in WEHI-3 cells and murine neutrophils than in HL-60 cells. Thus, WEHI-3 cells and murine neutrophils may display few or no detectable sLex epitopes because they express very few fucosylated glycans. In some cells, the level of α1-3-fucosyltransferase activity correlates with the expression of selectin ligands and epitopes for anti-sLexmAbs.50,51 Higher expression of FTVII has been suggested to be particularly important to create sLex epitopes for mAbs such as HECA-452,9,15 and these epitopes have been further interpreted to represent E-selectin ligands.9However, we observed that human neutrophils had very low α1-3-fucosyltransferase activities, even though these cells express abundant sLex epitopes and selectin ligands. Furthermore, the uncoupling of expression of selectin ligands and sLexepitopes in WEHI-3 cells and murine neutrophils suggests that modest levels of enzymatic activity are sufficient to fucosylate ligands for both P- and E-selectin. Thus, FTVII and FTIV may preferentially modify specific glycoproteins that function as selectin ligands. Consistent with this notion, structural analysis indicates that HL-60 cells fucosylate a portion of the O-glycans on PSGL-1 but fucosylate none of the O-glycans on CD43, a mucin of similar size.52Immunological data suggest that FTVII preferentially fucosylates PSGL-1 and FTIV preferentially fucosylates ESL-1 in murine leukocytes and in some transfected Chinese hamster ovary cells.53
We found no evidence that sialylation, sulfation, or α1-3-galactosylation masked epitopes for mAbs to sLex or Lex on WEHI-3 cells. Undersialylation can prevent significant expression of sLex epitopes but still allow synthesis of selectin ligands.11 Such cells, however, express significant levels of Lex epitopes, whereas murine leukocytes expressed low-to-undetectable Lex epitopes even after treatment with sialidase. Lack of core-2 β1-6-N-acetylglucosaminyltransferase activity might prevent synthesis of core-2 O-glycans that could be subsequently fucosylated.54 However, preliminary analysis indicates that the O-glycans isolated from PSGL-1 on WEHI-3 cells are predominantly disialylated core-2 structures with little or no detectable fucose (M. Kudo and R.D.C., unpublished data, July 2000). It is possible that other undefined modifications of fucosylated glycans prevent mAb binding without affecting selectin binding. Yet it is striking that addition of FTVI and GDP-fucose created many sLex epitopes but did not significantly increase selectin ligands. Thus, low levels of endogenous FTVII and FTIV may have already fucosylated most of the relevant selectin ligands. Forced fucosylation with FTVI and GDP-fucose created sLex epitopes on additional glycans that did not function well as selectin ligands. Elucidation of the glycan structures on WEHI-3 cells or primary murine leukocytes will provide definitive information as to why these cells express putative fucosylated selectin ligands that do not react with mAbs to sLex or Lex. Here we show that murine leukocytes employ low levels of FTVII and FTIV to create selectin ligands that appear to be displayed on a small subset of protease-sensitive glycoconjugates. It is noteworthy that only a small minority of O-glycans on PSGL-1 from HL-60 cells are fucosylated.52 Both human and murine leukocytes may employ limited but specific α1-3-fucosylation to synthesize glycoconjugates that bind to selectins.
We thank Cindy Carter, Kelsey Kennedy, Todd Walker, and Nici Barnard for technical assistance; Tadayuki Yago for assistance with site density measurements; Janos Kappelmayer for assistance with flow cytometry; John Lowe, Dietmar Vestweber, Bruce Walcheck, and Kwame Nyame for providing reagents; and Jari Helin for MALDI-TOF analysis of glycopeptide samples.
Prepublished online as Blood First Edition Paper, August 29, 2002; DOI 10.1182/blood-2002-06-1799.
Supported by National Institutes of Health grants HL 54304, AI 44902, and AI 48075. V.R. was the recipient of a postdoctoral fellowship from the Heartland Affiliate of the American Heart Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Rodger P. McEver, Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, 825 NE 13th Street, Oklahoma City, OK 73104; e-mail:rodger-mcever@omrf.ouhsc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal