Abstract
It is now recognized that a subset of B-cell chronic lymphocytic leukemia (CLL) is familial. The genetic basis of familial CLL is poorly understood, but recently germ line mutations in the Ataxia Telangiectasia (ATM) gene have been proposed to confer susceptibility to CLL. The evidence for this notion is, however, not unequivocal. To examine this proposition further we have screened the ATM gene for mutations in CLLs from 61 individuals in 29 families. Truncating ATM mutations, including a knownATM mutation, were detected in 2 affected individuals, but the mutations did not cosegregate with CLL in the families. In addition, 3 novel ATM missense mutations were detected. Common ATM missense mutations were not overrepresented. The data support previous observations that ATM mutation is associated with B-CLL. However, ATM mutations do not account for familial clustering of the disease.
Introduction
Leukemia affects between 1% and 2% of Western populations.1 B-cell chronic lymphocytic leukemia (CLL) is the most common form of leukemia, accounting for around 30% of cases.2 The incidence rate of CLL increases logarithmically from age 35 with a median age at diagnosis of 65 years.3
Epidemiologic studies and published case reports of families indicate that a subset of the CLL is ascribable to an inherited genetic predisposition. A dominantly acting gene or genes with pleiotropic effects appear to be the most likely genetic model of inheritance since CLL appears to segregate with other lymphoproliferative disorders (LPDs) in many families.4 However, no gene has been shown unequivocally to be causative.
Ataxia Telangiectasia (A-T), is one of a group of recessive syndromes characterized by excessive spontaneous chromosomal breakage associated with an elevated risk of hematologic malignancies.5Specifically, in A-T there is an increased risk of lymphomas and leukemias, including those in the B-cell lineage.6
The Ataxia Telangiectasia (ATM) gene maps to chromosome 11q237 and specifies a 12-kilobase (kb) mRNA encoding an approximately 350-kd protein.8 The 3′ end of the gene has some homology to phosphoinositide 3–kinases and toSaccharomyces cerevisiae telomerase 1 (TEL1) that controls telomere length and maintenance of genome integrity.9 Homozygous germ line mutations in ATMare associated with increased radiosensitivity and genomic instability. The mutation rate is increased, double-strand breaks show reduced rejoining fidelity, there is a high frequency of cytogenetic rearrangements, and homologous recombination is increased and error prone.10-13
A-T cells also show defects in cell-cycle checkpoints. These properties provide the rationale for proposing a model14 in whichATM has a role as a DNA damage-response gene probably responding to a subset of DNA double-strand breaks.
There is evidence that A-T carriers may display an increased cancer risk.15 Some reports have suggested that A-T heterozygotes display an increased chromosomal instability and hypersensitivity to carcinogens, thereby increasing the probability of acquiring oncogenic somatic mutations.16-18 The relationship between cancer risk and A-T heterozygosity has been most extensively studied in relation to breast cancer.19 CLL has been reported in A-T families,15 suggesting that heterozygosity may also confer an increased risk of this disease. In a retrospective study of cancer incidence in 110 A-T families, the risk of hematologic and lymphoid malignancies was increased in blood relatives of A-T patients and CLL accounted for all but one of the leukemias seen in adult blood relatives. However, these observations did not attain statistical significance. It has recently been shown that the ATM gene is mutant in approximately 20% of CLL samples from unrelated patients and that some patients have heterozygous germ line mutations.20-23
The prevalence of B-cell malignancy in A-T coupled with the possible increase in risk of leukemia in relatives of A-T patients has led a number of researchers to question whether germ line ATMmutations are involved in familial cases of CLL. In a small study of 32 CLL patients, Stankovic et al20 reported that 6% of cases harbored germ line ATM mutations.20 Bevan et al24 recently examined the role of ATM in familial CLL through a linkage analysis. While there was no evidence for linkage, the study did not preclude ATM underlying a subset of familial CLL.
ATM represents an attractive candidate CLL predisposition gene but direct evidence for its role is lacking. To investigate further the relationship between ATM and CLL we have screened tumor samples from 61 individuals in 29 CLL families for mutations.
Patients, materials, and methods
Patient selection
Families with 2 or more individuals affected with CLL were ascertained through hematologists in the United Kingdom, Norway, Israel, Italy, Germany, Portugal, and Poland. The diagnosis of CLL in all cases was based on standard hematologic and immunologic criteria. Samples were obtained from family members with informed consent and ethical review board approval from the Royal Marsden Hospital's National Health Service Trust. DNA was salt extracted from peripheral EDTA venous blood samples using a standard sucrose lysis method. No separation of tumor B cells from other cells was undertaken prior to DNA extraction.
Mutation detection
The 63 coding exons and exon 2 of ATM were analyzed by polymerase chain reaction (PCR) amplification of DNA and single-strand conformational polymorphism (SSCP) electrophoresis of32P–deoxycytosine triphosphosphate (dCTP)–labeled products through glycerol polyacrylamide gels as described,25 except that primers were redesigned to ensure that PCR products were approximately 250 base pairs or shorter. Table 1 details the nucleotide sequences of each of the 65 sets of PCR primers. Samples with bandshifts were sequenced with the same PCR primers in forward and reverse directions using the Li-Cor method (MWG, Milton Keynes, United Kingdom; http://www.mwg-biotech.com). Identified nucleotide changes were coded according to the genomic nucleotide sequence at GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) accession no.U82828 for introns and NM 000051 for each exon (renumbered with base 190 corresponding to base 1 in order to provide data in the same form as that used in the Virginia Mason ATM Mutation Database).26
Sequences of primers flanking ATM exons
| ATM exon . | Primer names . | Forward sequence (5′→3′) . | Reverse sequence (5′→3′) . |
|---|---|---|---|
| 3 | AT3F/R | TCC TCT GAG ATA TAT GTT CCC TGA C | GCT CAT TCA CTG ATA GAT GCA AA |
| 4 | AT4F/R | TTC ACA CCT CTT TCT CTC TAT ATA TGC | TCA CAC ATT TCA AGG AAA AAT TG |
| 5 | AT5F/R | TCA ATT TTT CCT TGA AAT GTG TG | TGC AAG GCA TAA TGA TAT ATA GGA A |
| 6 | AT6F/R | GCT CTT TGT GAT GGC ATG AA | AAA AAA AAA AAA AAC TCA CGC G |
| 7 | AT7F/R | AGT TGC CAT TCC AAG TGT CTT | GGT GAA GTT TCA TTT CAT GAG G |
| 8 | AT8IF/R | CCT TTT TCT GTA TGG GAT TAT GGA | CCG TCA GTC TGA GAA CAG CA |
| 8 | AT8IIF/R | CAT GCT GTT ACC AAA GGA TGC | TGA TGG ATC AAT GTT TTA TTT TAA GG |
| 9 | AT9F/R | CCC CCT GTT ATA CCC AGT TG | TGA ATG AAG AAG CAA ATT CAA AA |
| 10 | A10F/R | GGA GCT AGC AGT GTA AAC AGA | AAA TGT GAC ATG ACC TAC TTA CTG |
| 11 | A11F/R | GAT ACG AGA TCG TGC TGT TCC | GGA TTC CAC TGA AAG TTT TCT G |
| 12 | AT12IF/R | TCC TTT TAG TTT GTT AAT GTG ATG GA | CCC TGA ATT ATG GCT CCA AG |
| 12 | AT12IIF/R | TTT GGT GTA TTA CCT TTC GTG GT | TGA AAA TGA TCA GGG ATA TGT GA |
| 13 | AT13F/R | GCT TGC TTT TCA CAA TTG TCC T | AAA CAG CAG CAT GCT AAT GAA C |
| 14 | AT14F/R | TTC TTT ACA TGG CTT TTG GTC T | TAA GAT GCA GCT ACT ACC CAG C |
| 15 | AT15IF/R | GGC AAA GCA TTA GGT ACT TGG | TGG TGG ACA GAG AAG CCA AT |
| 15 | AT15IIF/R | ATT GGC TTC TCT GTC CAC CA | TTC CTG CCC CTA TTT CTC CT |
| 16 | AT16IF/R | CCA GGA TAT GCC ACC TTT AAC | TCC TAC CTT GGC TTT CTG GA |
| 16 | AT16IIF/R | GTC CGG TGT TCA CGT CTT TT | AAG AGA AAG GGT TAA CCT GCA T |
| 17 | AT17F/R | TAA AAA GCA ATA CTA AAC TA | ATG AGT TGT GAC AAT CCC ACT G |
| 18 | AT18F/R | TCT GCC GAG AAT AAT TGT TTT T | TGT TGT GAG ATG CAT CCT TAT T |
| 19 | AT19F/R | TGA CTA CAG CAT GCT CCT GC | CAA TGA GGC CTC TTA TAC TGC C |
| 20 | AT20F/R | TGT GCC CTT CTC TTA GTG TT | TCC AAG AGC TTC TTC ATT TAA C |
| 21 | AT21F/R | AAA TGA TTT GTG GAT AAA CCT GAT T | TGG TCA CGA CGA TAC AAA GAA |
| 22 | AT22F/R | AAT AAC TGA TGT GTT CTG TTA AGC | GCA AAA GAA AAA TAG GAC CA |
| 23 | AT23F/R | TCA GTG AGT TTT CTG AGT GCT TTT | ACT CAT TAA CAA ACA AAG ACT GCT TTA |
| 24 | AT24F/R | TTA ACC ACA GTT CTT TTC CCG TA | CCA TCT GCA GCA TTC CAA ATA |
| 25 | AT25F/R | ATG CTT TGG AAA GTA GGG TTT G | TAT GGG ATA TTC ATA GCA AGC A |
| 26 | AT26F/R | TGG AGT TCA GTT GGG ATT TT | TTC ACA GTG ACC TAA GGA AGC |
| 27 | AT27F/R | TTT TTC TTA ACA CAT TGA CTT TTT GG | CTG GTG AGG GGA CTT GCT AA |
| 28 | AT28IF/R | TTA ATG CTG ATG GTA TTA AAA CAG | CTT TTC CAG TCC TCT TGA ATC |
| 28 | AT28IIF/R | TTC AAG AGG ACT GGA AAA GTC | CAT TCA GGG AAT GAA AAG TAC A |
| 29 | AT29F/R | TGC CTT TTG AGC TGT CTT GA | GAC ATT GAA GGT GTC AAC CAA |
| 30 | AT30F/R | TTT TCA TTT TGG AAG TTC ACT GG | AAA TTT TTC ATT TGT TTA CTT TTC CTC |
| 31 | AT31IF/R | AAG TGT ATT TAT TGT AGC CGA GT | CTC CTC CTA AGC CAC TTT TT |
| 31 | AT31IIF/R | AAA AAG TGG CTT AGG AGG AG | CGG ACA GAG TGA GTC TTT GT |
| 32 | AT32F/R | TTT TCC AGA ACT TAC TGG TTG TTG | CCA TTT TGA AGA TGA GTC AGA AAA |
| 33 | AT33F/R | TGG CTT ACT TTA AAA TTA TTT CTC TCC | TGC TAG AGC ATT ACA GAT TTT TGA A |
| 34 | AT34F/R | AAC CAA TAC GTG TTA AAA GCA AGT T | CAG GTA GAA ATA GCC CAT GTC A |
| 35 | AT35F/R | CAA AAA GTG TTG TCT TCA TGC T | TAT GTG ATC CGC AGT TGA CTG |
| 36 | AT36F/R | TGA TCT CTT ACC TAT GAC TCT ACT GA | TGT GAA GTA TCA TTC TCC ATG A |
| 37 | AT37F/R | TTT GAA ATT TTT TCA GTG GAG G | TTT AAC AGT CAT GAC CCA CAG C |
| 38 | AT38F/R | TAC ATT TTC TAA TCC CTT TCT TTC | TGC AGT ATC ACA GCA CTC TTT |
| 39 | AT39F/R | ACT ATT GGG TGG ATT TGT T | CCA TCT TAA ATC CAT CTT TC |
| 40 | AT40F/R | CTG GGA CTG AGG GGA GAT A | CAT GTT AAA ATT CAG CCG ATA GTT |
| 41 | AT41F/R | CTA CCA TTG TAT TCT ATA TCA ACA TGC | TGC CAA CAT TAA TTT TAA AAA CC |
| 42 | AT42F/R | CAG GAG CTT CCA AAT AGT ATG T | GGC ATC TGT ACA GTG TCT ATA A |
| 43 | AT43F/R | TTG GGA GTT ACA TAT TGG TAA TGA | GCT TTG GGT TTT ACA CAC ACA T |
| 44 | AT44F/R | TTG CAT TTT TCT AAA CAA CGG TA | CCA ACA TAC TGA AAT AAC CTC AGC |
| 45 | AT45F/R | TCT CTG GTT TTC TGT TGA TAT C | CAG TTG TTG TTT AGA ATG AGG A |
| 46 | AT46F/R | AAA TTT TGT CCT TTG GTG AAG C | TTT CAG AAA AGA AGC CAT GAC A |
| 47 | AT47F/R | ATT TCC CTG AAA ACC TCT TCT T | GGT AAC AGA AAA GCT GCA CTT T |
| 48 | AT48F/R | CAT TTC TCT TGC TTA CAT GAA CTC | AGT CAA GTG GTA AGA TGA CAT AGT |
| 49 | AT49F/R | GCT GCT TTC ATT ATT ATT ATT CAT GGT | CAG TAA AAC ACT AAT CCA GCC AAT |
| 50 | AT50F/R | GGG CAG TTG GGT ACA GTC AT | GAT CTT GAT GAA AAG ATG AAG CA |
| 51 | AT51IF/R | AAA TTG GTT GTG TTT TCT TGA | ACT TCC TCT TTG GCT CTT TT |
| 51 | AT51IIF/R | CTC ATT AGC CCG GTT TTC AG | GAC CAA GTC ACT CTT TCT ATG CAA |
| 52 | AT52F/R | TGT TAA AGT TCA TGG CTT TTG TG | TTG GGC TGA GTA ACA CTT GC |
| 53 | AT53F/R | TTG CTT AGA TGT GAG AAT ATT TGA A | TTG TGG TTT GAT TTT CAG GTT T |
| 54 | AT54F/R | TGC AGG CAT ACA CGC TCT AC | TGC ATT TGC TAA GGC CAG TA |
| 54 | AT54IIF/R | GAT CAC CCC CAT CAC ACT TT | CCA GCC TTG AAC CGA TTT TA |
| 55 | AT55F/R | TCT GAG AAG TTT AAA TGT TGG GTA | GAG TAA CAC AGC AAG AAA GTA ACG |
| 56 | AT56F/R | CTT GAC CTT CAA TGC TGT TCC | TGC CAA TAT TTA GCC AAT TTT G |
| 57 | AT57F/R | TTT AAG TGC AAA TAG TGT ATC TGA CC | AAG GGC TAA GCC AGA GAA GG |
| 58 | AT58F/R | GAG TGC CCT TTG CTA TTC TCA | CCA ACC AAA TGG CAT CTT TT |
| 59 | AT59F/R | GAT AGC TGA ATG ATC ATC AAA TG | AGC TGT CAG CTT TAA TAA GCC A |
| 60 | AT60IF/R | CTG TTC ATC TTT ATT GCC CCT A | ACT GCG CGT ATA AGC CAA TC |
| 60 | AT60IIF/R | GAA GTC TTC ATG GAT GTT TGC | ATC CCC CTG CAA CTC AGA AT |
| 61 | AT61F/R | CTC AAC ATG GCC GGT TAT G | CAA ACA ACA TTC CAT GAT GAC C |
| 62 | AT62F/R | AGC TGT CAA ACC TCC TAA CTT CA | CCC AGC CCA TGT AAT TTT GA |
| 63 | AT63F/R | TTG ACA ACA TTG GTG TGT AAC A | GCC ACA TCC CCC TAT GTT AA |
| 64 | AT64F/R | CCA TGT GAC TGG CTT ATT TGT ATG | TGA AAA ACT GAC AAC AGG ACC TT |
| 65 | AT65F/R | TCA CCT CAC TGA AAC CTT TGT G | AAT TTC TAA AGG CTG AAT GAA AGG |
| ATM exon . | Primer names . | Forward sequence (5′→3′) . | Reverse sequence (5′→3′) . |
|---|---|---|---|
| 3 | AT3F/R | TCC TCT GAG ATA TAT GTT CCC TGA C | GCT CAT TCA CTG ATA GAT GCA AA |
| 4 | AT4F/R | TTC ACA CCT CTT TCT CTC TAT ATA TGC | TCA CAC ATT TCA AGG AAA AAT TG |
| 5 | AT5F/R | TCA ATT TTT CCT TGA AAT GTG TG | TGC AAG GCA TAA TGA TAT ATA GGA A |
| 6 | AT6F/R | GCT CTT TGT GAT GGC ATG AA | AAA AAA AAA AAA AAC TCA CGC G |
| 7 | AT7F/R | AGT TGC CAT TCC AAG TGT CTT | GGT GAA GTT TCA TTT CAT GAG G |
| 8 | AT8IF/R | CCT TTT TCT GTA TGG GAT TAT GGA | CCG TCA GTC TGA GAA CAG CA |
| 8 | AT8IIF/R | CAT GCT GTT ACC AAA GGA TGC | TGA TGG ATC AAT GTT TTA TTT TAA GG |
| 9 | AT9F/R | CCC CCT GTT ATA CCC AGT TG | TGA ATG AAG AAG CAA ATT CAA AA |
| 10 | A10F/R | GGA GCT AGC AGT GTA AAC AGA | AAA TGT GAC ATG ACC TAC TTA CTG |
| 11 | A11F/R | GAT ACG AGA TCG TGC TGT TCC | GGA TTC CAC TGA AAG TTT TCT G |
| 12 | AT12IF/R | TCC TTT TAG TTT GTT AAT GTG ATG GA | CCC TGA ATT ATG GCT CCA AG |
| 12 | AT12IIF/R | TTT GGT GTA TTA CCT TTC GTG GT | TGA AAA TGA TCA GGG ATA TGT GA |
| 13 | AT13F/R | GCT TGC TTT TCA CAA TTG TCC T | AAA CAG CAG CAT GCT AAT GAA C |
| 14 | AT14F/R | TTC TTT ACA TGG CTT TTG GTC T | TAA GAT GCA GCT ACT ACC CAG C |
| 15 | AT15IF/R | GGC AAA GCA TTA GGT ACT TGG | TGG TGG ACA GAG AAG CCA AT |
| 15 | AT15IIF/R | ATT GGC TTC TCT GTC CAC CA | TTC CTG CCC CTA TTT CTC CT |
| 16 | AT16IF/R | CCA GGA TAT GCC ACC TTT AAC | TCC TAC CTT GGC TTT CTG GA |
| 16 | AT16IIF/R | GTC CGG TGT TCA CGT CTT TT | AAG AGA AAG GGT TAA CCT GCA T |
| 17 | AT17F/R | TAA AAA GCA ATA CTA AAC TA | ATG AGT TGT GAC AAT CCC ACT G |
| 18 | AT18F/R | TCT GCC GAG AAT AAT TGT TTT T | TGT TGT GAG ATG CAT CCT TAT T |
| 19 | AT19F/R | TGA CTA CAG CAT GCT CCT GC | CAA TGA GGC CTC TTA TAC TGC C |
| 20 | AT20F/R | TGT GCC CTT CTC TTA GTG TT | TCC AAG AGC TTC TTC ATT TAA C |
| 21 | AT21F/R | AAA TGA TTT GTG GAT AAA CCT GAT T | TGG TCA CGA CGA TAC AAA GAA |
| 22 | AT22F/R | AAT AAC TGA TGT GTT CTG TTA AGC | GCA AAA GAA AAA TAG GAC CA |
| 23 | AT23F/R | TCA GTG AGT TTT CTG AGT GCT TTT | ACT CAT TAA CAA ACA AAG ACT GCT TTA |
| 24 | AT24F/R | TTA ACC ACA GTT CTT TTC CCG TA | CCA TCT GCA GCA TTC CAA ATA |
| 25 | AT25F/R | ATG CTT TGG AAA GTA GGG TTT G | TAT GGG ATA TTC ATA GCA AGC A |
| 26 | AT26F/R | TGG AGT TCA GTT GGG ATT TT | TTC ACA GTG ACC TAA GGA AGC |
| 27 | AT27F/R | TTT TTC TTA ACA CAT TGA CTT TTT GG | CTG GTG AGG GGA CTT GCT AA |
| 28 | AT28IF/R | TTA ATG CTG ATG GTA TTA AAA CAG | CTT TTC CAG TCC TCT TGA ATC |
| 28 | AT28IIF/R | TTC AAG AGG ACT GGA AAA GTC | CAT TCA GGG AAT GAA AAG TAC A |
| 29 | AT29F/R | TGC CTT TTG AGC TGT CTT GA | GAC ATT GAA GGT GTC AAC CAA |
| 30 | AT30F/R | TTT TCA TTT TGG AAG TTC ACT GG | AAA TTT TTC ATT TGT TTA CTT TTC CTC |
| 31 | AT31IF/R | AAG TGT ATT TAT TGT AGC CGA GT | CTC CTC CTA AGC CAC TTT TT |
| 31 | AT31IIF/R | AAA AAG TGG CTT AGG AGG AG | CGG ACA GAG TGA GTC TTT GT |
| 32 | AT32F/R | TTT TCC AGA ACT TAC TGG TTG TTG | CCA TTT TGA AGA TGA GTC AGA AAA |
| 33 | AT33F/R | TGG CTT ACT TTA AAA TTA TTT CTC TCC | TGC TAG AGC ATT ACA GAT TTT TGA A |
| 34 | AT34F/R | AAC CAA TAC GTG TTA AAA GCA AGT T | CAG GTA GAA ATA GCC CAT GTC A |
| 35 | AT35F/R | CAA AAA GTG TTG TCT TCA TGC T | TAT GTG ATC CGC AGT TGA CTG |
| 36 | AT36F/R | TGA TCT CTT ACC TAT GAC TCT ACT GA | TGT GAA GTA TCA TTC TCC ATG A |
| 37 | AT37F/R | TTT GAA ATT TTT TCA GTG GAG G | TTT AAC AGT CAT GAC CCA CAG C |
| 38 | AT38F/R | TAC ATT TTC TAA TCC CTT TCT TTC | TGC AGT ATC ACA GCA CTC TTT |
| 39 | AT39F/R | ACT ATT GGG TGG ATT TGT T | CCA TCT TAA ATC CAT CTT TC |
| 40 | AT40F/R | CTG GGA CTG AGG GGA GAT A | CAT GTT AAA ATT CAG CCG ATA GTT |
| 41 | AT41F/R | CTA CCA TTG TAT TCT ATA TCA ACA TGC | TGC CAA CAT TAA TTT TAA AAA CC |
| 42 | AT42F/R | CAG GAG CTT CCA AAT AGT ATG T | GGC ATC TGT ACA GTG TCT ATA A |
| 43 | AT43F/R | TTG GGA GTT ACA TAT TGG TAA TGA | GCT TTG GGT TTT ACA CAC ACA T |
| 44 | AT44F/R | TTG CAT TTT TCT AAA CAA CGG TA | CCA ACA TAC TGA AAT AAC CTC AGC |
| 45 | AT45F/R | TCT CTG GTT TTC TGT TGA TAT C | CAG TTG TTG TTT AGA ATG AGG A |
| 46 | AT46F/R | AAA TTT TGT CCT TTG GTG AAG C | TTT CAG AAA AGA AGC CAT GAC A |
| 47 | AT47F/R | ATT TCC CTG AAA ACC TCT TCT T | GGT AAC AGA AAA GCT GCA CTT T |
| 48 | AT48F/R | CAT TTC TCT TGC TTA CAT GAA CTC | AGT CAA GTG GTA AGA TGA CAT AGT |
| 49 | AT49F/R | GCT GCT TTC ATT ATT ATT ATT CAT GGT | CAG TAA AAC ACT AAT CCA GCC AAT |
| 50 | AT50F/R | GGG CAG TTG GGT ACA GTC AT | GAT CTT GAT GAA AAG ATG AAG CA |
| 51 | AT51IF/R | AAA TTG GTT GTG TTT TCT TGA | ACT TCC TCT TTG GCT CTT TT |
| 51 | AT51IIF/R | CTC ATT AGC CCG GTT TTC AG | GAC CAA GTC ACT CTT TCT ATG CAA |
| 52 | AT52F/R | TGT TAA AGT TCA TGG CTT TTG TG | TTG GGC TGA GTA ACA CTT GC |
| 53 | AT53F/R | TTG CTT AGA TGT GAG AAT ATT TGA A | TTG TGG TTT GAT TTT CAG GTT T |
| 54 | AT54F/R | TGC AGG CAT ACA CGC TCT AC | TGC ATT TGC TAA GGC CAG TA |
| 54 | AT54IIF/R | GAT CAC CCC CAT CAC ACT TT | CCA GCC TTG AAC CGA TTT TA |
| 55 | AT55F/R | TCT GAG AAG TTT AAA TGT TGG GTA | GAG TAA CAC AGC AAG AAA GTA ACG |
| 56 | AT56F/R | CTT GAC CTT CAA TGC TGT TCC | TGC CAA TAT TTA GCC AAT TTT G |
| 57 | AT57F/R | TTT AAG TGC AAA TAG TGT ATC TGA CC | AAG GGC TAA GCC AGA GAA GG |
| 58 | AT58F/R | GAG TGC CCT TTG CTA TTC TCA | CCA ACC AAA TGG CAT CTT TT |
| 59 | AT59F/R | GAT AGC TGA ATG ATC ATC AAA TG | AGC TGT CAG CTT TAA TAA GCC A |
| 60 | AT60IF/R | CTG TTC ATC TTT ATT GCC CCT A | ACT GCG CGT ATA AGC CAA TC |
| 60 | AT60IIF/R | GAA GTC TTC ATG GAT GTT TGC | ATC CCC CTG CAA CTC AGA AT |
| 61 | AT61F/R | CTC AAC ATG GCC GGT TAT G | CAA ACA ACA TTC CAT GAT GAC C |
| 62 | AT62F/R | AGC TGT CAA ACC TCC TAA CTT CA | CCC AGC CCA TGT AAT TTT GA |
| 63 | AT63F/R | TTG ACA ACA TTG GTG TGT AAC A | GCC ACA TCC CCC TAT GTT AA |
| 64 | AT64F/R | CCA TGT GAC TGG CTT ATT TGT ATG | TGA AAA ACT GAC AAC AGG ACC TT |
| 65 | AT65F/R | TCA CCT CAC TGA AAC CTT TGT G | AAT TTC TAA AGG CTG AAT GAA AGG |
Results
Twenty-nine families with 2 or more affected individuals with CLL were studied. Table 2shows the pedigree structure of each of the families and the ages at diagnosis of CLL in family members.
ATM nucleotide changes detected in familial CLL cases
| Family . | CLL case . | Age at diagnosis, y . | ATM nucleotide changes . | |
|---|---|---|---|---|
| Truncating or amino acid substitution . | Other . | |||
| 5 | Father | nk | ne | ne |
| Uncle | nk | ne | ne | |
| Sister | 54 | 5042T>C (Ile1681Thr) | IVS25-13insA†; 4578C>T‡; IVS63+60G/A‡ | |
| Brother | 46 | nd | IVS4-36insTG; IVS25-13insA†; 4578C>T; IVS63+60G/A‡ | |
| Brother | 47 | 5042T>C (Ile1681Thr) | IVS25-13insA†,2-153 | |
| 6 | Mother | 69 | nd | IVS25-13insA†; IVS40+27G/A; IVS63+60G/A‡ |
| Daughter | 42 | 5557G>A (Asp1853Asn)‡ | IVS4-36insTG2-153; IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ | |
| Son | 40 | 5557G>A (Asp1853Asn)‡ | IVS4-36insTG2-153; IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ | |
| 8 | Brother | 70 | 6919C>T (Leu2307Phe) | IVS63+60G/A‡,2-153; IVS25-13insA† |
| Brother | 76 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| Brother | nk | ne | ne | |
| 11 | Sister | 69 | nd | nd |
| Sister | 61 | nd | nd | |
| 13 | Sister | 70 | 2572T>C (Phe858Leu)‡ | IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 58 | nd | 162T>C2-153; IVS25-13insA†; IVS63+60G/A‡ | |
| 15 | Brother | 63 | 8266A>T (Lys2756Xaa)‡ | IVS25-13insA†; 4578C>T; IVS63+60G/A‡ |
| Sister | 61 | nd | IVS25-13insA†,2-153 | |
| 18 | Sister | 74 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 73 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 20 | Brother | 64 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 66 | nd | IVS63+60G/A2-153,‡ | |
| 39 | Sister | 60 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 51 | 4258C>T (Leu1420Phe)‡ | IVS4-36insTG2-153; IVS63+60G/A‡,2-153 | |
| Sister | nk | ne | ne | |
| 40 | Brother | 71 | nd | IVS25-13insA†,2-153 |
| Sister | 48 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 41 | Brother | 35 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 51 | nd | IVS25-13insA†,2-153 | |
| 42 | Uncle | 59 | nd | IVS25-13insA†,2-153 |
| Second cousin | 43 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 43 | Sister | 72 | nd | nd |
| Sister | 67 | 5756delAA; 7271insGT | nd | |
| 45 | Brother | 61 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 52 | nd | IVS25-13insA†,2-153 | |
| 46 | Sister | 61 | nd | IVS62+8A/C |
| Sister | 59 | nd | IVS62+8A/C | |
| 48 | Sister | 45 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 51 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 49 | Brother | 69 | nd | IVS25-13insA†,2-153 |
| Sister | 80 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 60 | Brother | 55 | nd | nd |
| Sister | 49 | nd | nd | |
| 63 | Brother | 57 | 378T>A (Asp126Glu) | IVS25-13insA† |
| Sister | 43 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 64 | Brother | 62 | 5557G>A (Asp1853Asn)‡ | IVS15-48T/C; IVS24-10delT |
| Brother | 64 | 5557G>A (Asp1853Asn)‡,2-153 | IVS16+78G/A | |
| 78 | Uncle | 54 | nd | IVS25-13insA†,2-153 |
| Uncle | 50 | ne | ne | |
| Nephew | 32 | nd | IVS4-36insTG2-153; 4578C>T; IVS63+60G/A‡,2-153 | |
| 85 | Brother | 60 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 49 | 2572T>C (Phe858Leu) | nd | |
| 96 | Brother | 65 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†,2-153 |
| Brother | 61 | nd | IVS4-36insTG2-153; IVS63+60G/A‡,2-153 | |
| 98 | Brother | 67 | 378T>A(Asp126Glu); | IVS16+78G/A; IVS24-10delT; IVS25-13insA† |
| 2572T>C (Phe858Leu) | ||||
| 5557G>A (Asp1853Asn)‡ | ||||
| Brother | 64 | 5557G>A (Asp1853Asn)‡ | IVS4-36insTG; IVS16+78G/A; IVS24-10delT; IVS25-13insA† | |
| 115 | Brother | 53 | 2572T>C (Phe858Leu); | IVS24-10delT |
| 5557G>A (Asp1853Asn)‡ | ||||
| Brother | 51 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS63+60G/A‡,2-153 | |
| 117 | Sister | 65 | nd | nd |
| Sister | 64 | nd | nd | |
| 118 | Father | 79 | nd | nd |
| Daughter | 46 | nd | IVS63+60G/A‡ | |
| 138 | Sister | 64 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†,2-153 |
| Brother | 74 | nd | IVS25-13insA† | |
| 162 | Brother | 54 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 61 | 5557G>A (Asp1853Asn)‡,2-153 | IVS25-13insA†,2-153 | |
| Family . | CLL case . | Age at diagnosis, y . | ATM nucleotide changes . | |
|---|---|---|---|---|
| Truncating or amino acid substitution . | Other . | |||
| 5 | Father | nk | ne | ne |
| Uncle | nk | ne | ne | |
| Sister | 54 | 5042T>C (Ile1681Thr) | IVS25-13insA†; 4578C>T‡; IVS63+60G/A‡ | |
| Brother | 46 | nd | IVS4-36insTG; IVS25-13insA†; 4578C>T; IVS63+60G/A‡ | |
| Brother | 47 | 5042T>C (Ile1681Thr) | IVS25-13insA†,2-153 | |
| 6 | Mother | 69 | nd | IVS25-13insA†; IVS40+27G/A; IVS63+60G/A‡ |
| Daughter | 42 | 5557G>A (Asp1853Asn)‡ | IVS4-36insTG2-153; IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ | |
| Son | 40 | 5557G>A (Asp1853Asn)‡ | IVS4-36insTG2-153; IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ | |
| 8 | Brother | 70 | 6919C>T (Leu2307Phe) | IVS63+60G/A‡,2-153; IVS25-13insA† |
| Brother | 76 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| Brother | nk | ne | ne | |
| 11 | Sister | 69 | nd | nd |
| Sister | 61 | nd | nd | |
| 13 | Sister | 70 | 2572T>C (Phe858Leu)‡ | IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 58 | nd | 162T>C2-153; IVS25-13insA†; IVS63+60G/A‡ | |
| 15 | Brother | 63 | 8266A>T (Lys2756Xaa)‡ | IVS25-13insA†; 4578C>T; IVS63+60G/A‡ |
| Sister | 61 | nd | IVS25-13insA†,2-153 | |
| 18 | Sister | 74 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 73 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 20 | Brother | 64 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 66 | nd | IVS63+60G/A2-153,‡ | |
| 39 | Sister | 60 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 51 | 4258C>T (Leu1420Phe)‡ | IVS4-36insTG2-153; IVS63+60G/A‡,2-153 | |
| Sister | nk | ne | ne | |
| 40 | Brother | 71 | nd | IVS25-13insA†,2-153 |
| Sister | 48 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 41 | Brother | 35 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 51 | nd | IVS25-13insA†,2-153 | |
| 42 | Uncle | 59 | nd | IVS25-13insA†,2-153 |
| Second cousin | 43 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 43 | Sister | 72 | nd | nd |
| Sister | 67 | 5756delAA; 7271insGT | nd | |
| 45 | Brother | 61 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 52 | nd | IVS25-13insA†,2-153 | |
| 46 | Sister | 61 | nd | IVS62+8A/C |
| Sister | 59 | nd | IVS62+8A/C | |
| 48 | Sister | 45 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Sister | 51 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 49 | Brother | 69 | nd | IVS25-13insA†,2-153 |
| Sister | 80 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 60 | Brother | 55 | nd | nd |
| Sister | 49 | nd | nd | |
| 63 | Brother | 57 | 378T>A (Asp126Glu) | IVS25-13insA† |
| Sister | 43 | nd | IVS25-13insA†; IVS63+60G/A‡ | |
| 64 | Brother | 62 | 5557G>A (Asp1853Asn)‡ | IVS15-48T/C; IVS24-10delT |
| Brother | 64 | 5557G>A (Asp1853Asn)‡,2-153 | IVS16+78G/A | |
| 78 | Uncle | 54 | nd | IVS25-13insA†,2-153 |
| Uncle | 50 | ne | ne | |
| Nephew | 32 | nd | IVS4-36insTG2-153; 4578C>T; IVS63+60G/A‡,2-153 | |
| 85 | Brother | 60 | nd | IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 49 | 2572T>C (Phe858Leu) | nd | |
| 96 | Brother | 65 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†,2-153 |
| Brother | 61 | nd | IVS4-36insTG2-153; IVS63+60G/A‡,2-153 | |
| 98 | Brother | 67 | 378T>A(Asp126Glu); | IVS16+78G/A; IVS24-10delT; IVS25-13insA† |
| 2572T>C (Phe858Leu) | ||||
| 5557G>A (Asp1853Asn)‡ | ||||
| Brother | 64 | 5557G>A (Asp1853Asn)‡ | IVS4-36insTG; IVS16+78G/A; IVS24-10delT; IVS25-13insA† | |
| 115 | Brother | 53 | 2572T>C (Phe858Leu); | IVS24-10delT |
| 5557G>A (Asp1853Asn)‡ | ||||
| Brother | 51 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS63+60G/A‡,2-153 | |
| 117 | Sister | 65 | nd | nd |
| Sister | 64 | nd | nd | |
| 118 | Father | 79 | nd | nd |
| Daughter | 46 | nd | IVS63+60G/A‡ | |
| 138 | Sister | 64 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†,2-153 |
| Brother | 74 | nd | IVS25-13insA† | |
| 162 | Brother | 54 | 5557G>A (Asp1853Asn)‡ | IVS24-10delT; IVS25-13insA†; IVS63+60G/A‡ |
| Brother | 61 | 5557G>A (Asp1853Asn)‡,2-153 | IVS25-13insA†,2-153 | |
ne indicates not examined; nd, not detected; and nk, not known.
Nucleotide change reported at Genbank (http://www.ncbi.nlm.nih.gov/).
Nucleotide change reported at Virginia MasonATM mutation database (http://www.vmresearch.org/atm.htm).
Apparent homozygosity.
DNA from blood samples was examined for nucleotide changes in theATM gene by PCR-SSCP. All coding exons were examined using primers in the flanking introns or, where an exon was long, using 2 pairs of primers ensuring the middle 2 primers amplified overlapping segments of the exon.
The ATM nucleotide changes detected in the 61 affected individuals from the 29 CLL families are detailed in Table 2. Allele frequencies estimated for each of the mutations detected in the study are given in Table 3. This table also shows the allele frequencies of each of the mutations reported in published studies together with reported designations of each nucleotide change. In some instances different authors have interpreted the same nucleotide change as a mutation, a polymorphism, or present in a carrier. To be consistent we have classified nucleotide changes as “substitution” or “truncating” (leading to amino acid substitution or to truncation of the ATM polypeptide) or other (outside exons and their splice site consensus sequences or within an exon but not leading to change in amino acid usage).
Allele frequencies of ATM nucleotide changes in this and previous studies and designation of nucleotide changes
| Nucleotide change (amino acid substitution) . | Allele frequencies . | Type of nucleotide change . | Reported designation of nucleotide change . | Reference . | |
|---|---|---|---|---|---|
| This study . | Previous studies3-150 . | ||||
| IVS4-36insTG | 12/122 | — | Intronic | ||
| 162T>C | 2/122 | — | No substitution | ||
| 378T>A (Asp126Glu) | 2/120 | — | Substitution | ||
| IVS15-48T/C | 1/118 | — | Intronic | ||
| IVS16+78G/A | 4/118 | — | Intronic | ||
| 2572T>C (Phe858Leu) | 4/120 | 0.02 (C) | Substitution | Polymorphism carrier mutation | 27-29 |
| IVS24-10delT | 11/122 | 0.18 (delT) | Intronic | Polymorphism | 30 |
| IVS25-13insA | 50/120 | 0.37 (ins A) | Intronic | Polymorphism | 27 |
| 4258C>T (Leu1420Phe) | 1/118 | 0.02 | Substitution | Polymorphism carrier mutation | 28, 29, 31 |
| 4578C>T | 4/120 | — | No substitution | ||
| 5042T>C (Ile1681Thr) | 2/122 | — | Substitution | ||
| 5557G>A (Asp1853Asn) | 15/122 | 0.18(A) | Substitution | Polymorphism risk factor | 27, 32 |
| 5756delAA | 1/122 | NA | Truncating | ||
| IVS40+27G/A | 1/122 | — | Intronic | ||
| 6919C>T (Leu2307Phe) | 1/122 | — | Substitution | ||
| 7271insGT | 1/122 | NA | Truncating | ||
| 8266A>T (Lys2756Xaa) | 1/122 | NA | Truncating | Mutation | 33-35 |
| IVS63+60G/A | 36/122 | 0.37(A) | Intronic | Polymorphism | |
| Nucleotide change (amino acid substitution) . | Allele frequencies . | Type of nucleotide change . | Reported designation of nucleotide change . | Reference . | |
|---|---|---|---|---|---|
| This study . | Previous studies3-150 . | ||||
| IVS4-36insTG | 12/122 | — | Intronic | ||
| 162T>C | 2/122 | — | No substitution | ||
| 378T>A (Asp126Glu) | 2/120 | — | Substitution | ||
| IVS15-48T/C | 1/118 | — | Intronic | ||
| IVS16+78G/A | 4/118 | — | Intronic | ||
| 2572T>C (Phe858Leu) | 4/120 | 0.02 (C) | Substitution | Polymorphism carrier mutation | 27-29 |
| IVS24-10delT | 11/122 | 0.18 (delT) | Intronic | Polymorphism | 30 |
| IVS25-13insA | 50/120 | 0.37 (ins A) | Intronic | Polymorphism | 27 |
| 4258C>T (Leu1420Phe) | 1/118 | 0.02 | Substitution | Polymorphism carrier mutation | 28, 29, 31 |
| 4578C>T | 4/120 | — | No substitution | ||
| 5042T>C (Ile1681Thr) | 2/122 | — | Substitution | ||
| 5557G>A (Asp1853Asn) | 15/122 | 0.18(A) | Substitution | Polymorphism risk factor | 27, 32 |
| 5756delAA | 1/122 | NA | Truncating | ||
| IVS40+27G/A | 1/122 | — | Intronic | ||
| 6919C>T (Leu2307Phe) | 1/122 | — | Substitution | ||
| 7271insGT | 1/122 | NA | Truncating | ||
| 8266A>T (Lys2756Xaa) | 1/122 | NA | Truncating | Mutation | 33-35 |
| IVS63+60G/A | 36/122 | 0.37(A) | Intronic | Polymorphism | |
NA indicates not applicable.
Only provided for ATM polymorphisms/variants defined in the Virginia Mason database (http://www.vmresearch.org/atm.htm).
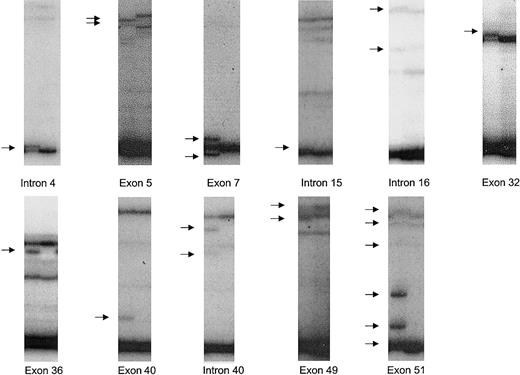
In the samples analyzed, 11 nucleotide changes were identified that have not previously been reported. Altered SSCP patterns corresponding to these nucleotide changes are shown in Figure1. Three of the nucleotide changes lead to amino acid substitutions. Two of the nucleotide changes lead to truncation of ATM due to either an insertion or deletion of 2 base pairs within respective exons of the gene. Of the 7 other nucleotide changes detected, 2 result in no change in amino acid usage and 5 were intronic nucleotide changes residing at least 27 base pairs away from intron-exon boundaries. No nucleotide changes identified created or destroyed consensus splice sites.
SSCP bandshifts corresponding to novel ATM nucleotide changes identified.
Out of 61 samples, 11 novel nucleotide changes were identified. Bandshifts (arrowed) due to these nucleotide changes detected on autoradiograms after SSCP are shown adjacent to normal samples. The nucleotide changes were: IVS4-36insTG (intron 4); 162T>C (exon 5); 378T>A (exon 7); IVS15-48T/C (intron 15); IVS16 + 78G/A (intron 16); 4578C>T (exon 32); 5042T>C (exon 36); 5756delAA (exon 40); IVS40 + 27G/A (intron 40); 6919C>T (exon 49); 7271insGT (exon 51).
SSCP bandshifts corresponding to novel ATM nucleotide changes identified.
Out of 61 samples, 11 novel nucleotide changes were identified. Bandshifts (arrowed) due to these nucleotide changes detected on autoradiograms after SSCP are shown adjacent to normal samples. The nucleotide changes were: IVS4-36insTG (intron 4); 162T>C (exon 5); 378T>A (exon 7); IVS15-48T/C (intron 15); IVS16 + 78G/A (intron 16); 4578C>T (exon 32); 5042T>C (exon 36); 5756delAA (exon 40); IVS40 + 27G/A (intron 40); 6919C>T (exon 49); 7271insGT (exon 51).
Among previously unreported nucleotide changes, intervening sequence (IVS)4-36insTG was the most common in this series, detected in 10% (12/120) of alleles. The abnormal band pattern due to IVS4-36insTG was only detected when autoradiograms were briefly exposed to film (∼15 minutes). The pattern involved a new dark band virtually comigrating with a normal dark band. There were no detectable new bands among the less intense bands. The subtle nature of change in bands associated with IVS4-36insTG may explain why this common polymorphism has not previously been detected. The allelic frequencies of other previously unreported nucleotide changes were less than 4%.
In 61 of the samples analyzed, 9 amino acid substitutions or truncating changes were detected that might affect the function of the ATMprotein. Six of these changes were observed in single cases but for none of these cases was the same change seen in any affected relative. Among these 6 changes, 2 were detected in the same case: 5756delAA and 7271insGT were detected in an affected sibling in family 43. Figure 1 shows the comparative relative intensities of the exon 40 and exon 51 bands corresponding to these mutations. Examination of autoradiograms indicates that 5756delAA constituted around 20% of the sample while 7271insGT constituted about 50% of the sample. Two amino acid substitutions, 378T>A (Asp126Glu) and 5042T>C (Ile1681Thr) were each detected in 2 affected individuals. 378T>A was detected in the index case of family 63, but not in his affected sibling, and in the index case of family 98, but not in his affected sibling. 5042T>C was detected in 2 of the 3 affected siblings in family 5. One mutation, (5557G>A), which leads to an amino acid substitution (Asp1853Asn), was detected in 15 alleles giving a frequency comparable to that previously reported in healthy individuals from the general population.
Discussion
The hypothesis that ATM represents a predisposition locus for CLL is attractive a priori. Strong evidence for such a notion would be provided by cosegregation in families of microsatellite markers and of mutations. However, this depends on ATMmutations conferring a markedly elevated risk. Furthermore, the power of any set of families to demonstrate linkage will be severely reduced if the disease is genetically heterogeneous. If ATMonly confers modest risks, mutations will not generate multiple-case families and linkage will not be detected, especially if phenocopies are common. There should, however, be evidence of overrepresentation of mutations in cases compared to the general population. The mutation analysis reported here on the ATMgene in tumor samples from 61 individuals in 29 CLL families was undertaken to assess further the question of whether ATMrepresents a predisposition locus for CLL.
In our study, 2 of the 61 patients with CLLs in the 29 families had truncating ATM mutations. One patient with CLL had 8266A>T (Lys2756Xaa). This mutation has been reported in A-T families.33-35 The second patient with CLL had 2 truncating mutations, 5766delAA and 7271insGT. It is highly probable that just one of these mutations is somatic. To assess whether this class of ATM mutation confers an increase in risk of CLL per se, the observed number of mutations needs to be compared with that expected by chance. The number of mutations expected is not simply a function of the number of affected individuals analyzed but is also a function of the familial relationship between affected individuals within each family. The expected number of mutations computed on this basis is 0.46. Therefore, observation of 2 mutations equates to a 4.4-fold overrepresentation, albeit, nonsignificant (95% CI, 0.5-15.9). Stankovic et al20 suggested that germ line mutations in ATM confer susceptibility to CLL following the finding that 2 of 32 sporadic CLL cases harbored constitutional mutations. One of the mutations identified was a truncating mutation and the other was a missense mutation (Pro1054Arg, 3161C>G) which the authors designated as pathogenic. The significance of this observation depends critically on the carrier frequency of pathogenic ATMmutations in the general population. Most estimates based on the prevalence of A-T suggest that the population carrier frequency of pathogenic mutations in ATM is around 1%.36Assuming this to be the case, then the observation of 2 mutations in 32 cases is significant (P = .04). Pro1054Arg has however, been designated as a polymorphism by some workers.37 If this is the case, then the observation of 1 pathogenic constitutional mutation in 32 CLL cases does not constitute an overrepresentation (P = .28). However, combining this data with ours, the observation of 3 truncating mutations indicates overrepresentation at the 0.04 level.
It is conceivable that ATM may impact more significantly on the risk of CLL if some nontruncating mutations have pathogenic potential. Gatti et al38 have recently proposed that cancer susceptibility may be associated not with truncating ATMmutations, but rather with missense ATM mutations. In the present study, 6 missense mutations were identified; 3 have been reported previously (Asp1853Asn, Phe858Leu, and Leu1420Phe) and 3 are new (Asp126Glu, Ile1681Thr, and Leu2307Phe). Asp1853Asn (5557G>A) was detected in 13 cases of CLL from 8 of the 29 families studied. Some evidence suggests that 5557A may be associated with a modest increase in cancer risk.39 The evidence from our study does not support such a postulate in the context of CLL. Although in 4 families the CLLs from both affected individuals harbored at least one copy of this allele, in the other 4 families the mutation did not segregate with disease. Furthermore, there was no evidence of overrepresentation of the 5557A allele. Overall the frequency of the 5557A allele was comparable to the population estimate of 18%. 2572T>C (Phe858Leu) was detected in 4 unrelated individuals, failing to segregate with CLL. The frequency of the 2572C allele was also similar to the population estimate of 2% in published reports. The recent proposal40that 2572C only confers an elevated cancer risk in the presence of 5557A is not supported by our study as this combination was not observed to segregate with CLL. Asp126Glu (378T>A) was also not observed to segregate with CLL, being detected in 2 unrelated individuals. While the residue is conserved between human and murine ATM protein, 126Glu is unlikely to confer risk as the amino acid change is conservative. Ile1681Thr (5042T>C) was detected in 2 of the 5 affected individuals in family 5. 5042C is unlikely to confer risk as the amino acid change is conservative and Ile1681 is not conserved between human and murine ATM proteins.
Leu1420Phe (4258C>T) and Leu2307Phe (6919C>T) mutations were detected in single CLLs. Leu1420Phe (4258C>T) has been reported as conferring risk of A-T. Leu2307Phe is a nonconservative change and thus may confer risk of CLL. In the absence of robust functional assays for each domain of the multifunctional ATM protein it is difficult to specify pathogenicity of these variants. However, the absence of cosegregation of amino acid changes in the families we have studied implies that the nonconservative amino acid changes will by themselves confer small genotypic risks. If a sequence variant is detected in just one family member we cannot exclude in the present study the possibility that some mutations are somatic. As a result, the ATMsequence variants we report here may overrepresent the germ line variability of ATM in this collection of CLL samples.
As with most common cancers susceptibility to CLL is likely to exhibit genetic heterogeneity. Furthermore, excluding large multiple-case families segregating CLL in a clear mendelian fashion, nuclear families with small numbers of cases may display a degree of within-family heterogeneity whereby, in addition to phenocopies, affected individuals in the same family may be caused by different susceptibility genes.
We report additional data that provide support for previous observations that ATM mutations are associated with CLL. However, on the basis of the data presented here, ATM is unlikely to represent a major susceptibility gene for familial CLL as the familial relative risk ascribable to mutations is unlikely to be more than approximately 1.1. To determine whether specificATM variants represent predisposition mutations, large cohorts of CLL patients and controls will be required. However, the spectrum and the frequency of ATM variants makes the evaluation of specific nucleotide changes as risk factors for CLL inherently difficult, as has been seen in studies seeking to establishATM as a risk factor for breast cancer.
A complete list of the participating clinicians are included in the at the end of this article. We thank Jenny Burrows, Julie Fuller and Andrea Marossy for their assistance. Finally, we are grateful to the 2 anonymous reviewers for their comments.
We thank the following clinicians and their patients: K. Quabeck, Germany; P. Stark, Israel; E. Gaminara, St Albans, United Kingdom; P. Antunovic, Norway; F. Mauro, Italy; H. Sykes, Kingston Upon Thames, United Kingdom; I. Ribeiro, Portugal; A. Bell, Poole, United Kingdom; M. Auger, Sutton in Ashfield, United Kingdom; S. Rassam, Sidcup, United Kingdom; M. Junior, Portugal; I. Ben-Bassat, Israel; R. M. Stewart, Chesterfield, United Kingdom; J. R. Duncan, Brighton, United Kingdom; L.G. Quaglino, Italy; P.M. Chipping, Stoke on Trent, United Kingdom.
Supported by the Kay Kendall Leukemia Trust and the Leukemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Richard S. Houlston, Section of Cancer Genetics, Institute of Cancer Research, Sutton, Surrey, United Kingdom; e-mail:r.houlston@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal