Rapidly developing technologies such as quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR) and microarrays enable characterization and quantification of nucleic acids in clinical samples. Identification of DNA from pathogens such as bacteria or viruses is already widely used in molecular diagnostics.1 The use of RNA as a diagnostic tool is currently being investigated. Gene expression profiling of tumor samples using microarray technology has become the prototypic application for this new type of molecular diagnostics. Cluster analysis of large-scale microarrays revealed that characterization of only a few genes is sufficient to distinguish different tumors.2 However, levels of gene expression may change between sample acquisition and the beginning of analysis, and these changes are not well understood.
Blood is frequently used for diagnostic sampling and processing. However, differences in blood collection and preparation techniques may cause changes in gene expression levels ex vivo and consequently affect the resulting mRNA expression profiles. For example, several investigators have shown that anticoagulants affect leukocyte viability and cause ex vivo changes in cytokine production.3 4
The extent to which events after blood collection influence gene expression is not known. To our knowledge, changes in gene expression between phlebotomy and the beginning of analysis have not been studied. We evaluated the effect of ex vivo incubation of blood on the gene expression status of human peripheral blood. After informed consent, blood was obtained from healthy donors. Whole blood collected in EDTA (ethylenediaminetetraacetic acid) tubes was stored under sterile conditions at room temperature for different periods of time. Gene expression was monitored by quantitative real-time RT-PCR using TaqMan assays on an ABI PRISM 7700 (Applied Biosystems, Weiterstadt, Germany). Primer and probe sequences are available upon request. mRNA quantity was calculated using the ΔΔCT method (PE Applied Biosystems User Bulletin #2; ABI PRISM 7700 Sequence Detection System, 1997).
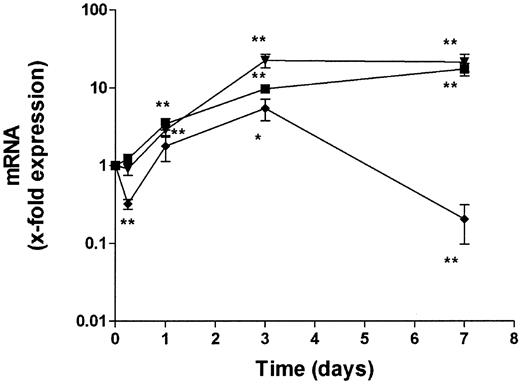
We studied the changes in transcript levels of proinflammatory genes known to be sensitive to extracellular stimuli in whole blood after phlebotomy. Figure 1 shows the changes in levels of IL-1β, IL-6, and TNFα transcripts up to 7 days. Levels of IL-6 and TNFα transcripts increased over the entire observation period, reaching maximum levels of about 20-fold higher than levels measured immediately after blood collection. IL-1β transcripts showed a different pattern. After 6 hours a decrease in expression was observed, followed by an increase after 1 and 3 days. Seven days after phlebotomy, IL-1β mRNA levels again decreased to below the original level. Since changes in mRNA levels after blood collection affect accurate measurement of gene expression, we tested 2 methods to stabilize RNA after phlebotomy. First we tried addition of a mixture of acidic phenol and guanidine isothiocyanate to blood immediately after collection. Then we used the PAXgene blood RNA tube (PreAnalytix, Hombrechtikon, Switzerland), a blood collection tube containing an additive that stabilizes cellular RNA. RNA was prepared at different time points from these stabilized samples and compared with whole blood samples collected in conventional EDTA tubes. Both stabilization methods preserved the RNA of all 3 cytokines for up to 7 days. No significant changes in mRNA levels compared to levels at phlebotomy were observed.
Induction of proinflammatory cytokines in whole blood.
Fresh blood samples were incubated at ambient temperature for up to 7 days. Levels of TNFα (▾), IL-1β (♦), and IL-6 (▪) mRNA were determined using real-time RT-PCR. Data are expressed as arbitrary units normalized to β-actin to correct for RNA quantity and integrity. RNA levels at the time of phlebotomy were used as a reference and set to 1. Data points are mean ± SE (mean of 10 donors). *P < .05, **P < .01 compared with mRNA levels at phlebotomy (day 0).
Induction of proinflammatory cytokines in whole blood.
Fresh blood samples were incubated at ambient temperature for up to 7 days. Levels of TNFα (▾), IL-1β (♦), and IL-6 (▪) mRNA were determined using real-time RT-PCR. Data are expressed as arbitrary units normalized to β-actin to correct for RNA quantity and integrity. RNA levels at the time of phlebotomy were used as a reference and set to 1. Data points are mean ± SE (mean of 10 donors). *P < .05, **P < .01 compared with mRNA levels at phlebotomy (day 0).
These results highlight the need for standardization and stabilization of patient samples after phlebotomy. In blood, cells are exposed to a variety of extracellular agents, which may influence their activation status. The change of environment after phlebotomy and the exposure to other surfaces during and after blood collection is likely to affect gene expression. Both methods tested for stabilization of RNA worked equally well in preserving the transcript levels of the genes analyzed. However, immediate addition of a toxic solution to blood is impracticable in the clinic. The use of a collection device containing stabilizing reagents, which preserves gene expression profiles at the time of phlebotomy, is preferable.
Studies using gene expression profiling of clinical samples must be aware of these events and implement routines to standardize and stabilize clinical samples before RNA isolation and analysis. Neglecting nucleic acid stabilization may lead to artifactual results in gene expression profiling.
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) (grant no. 312242).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal