The ATM serine-threonine kinase plays a central role in the cellular response to DNA damage. Germ-line mutations in theATM gene cause ataxia-telangiectasia (A-T), a multisystem disorder associated with predisposition to lymphoma and acute leukemia. Moreover, somatic ATM mutations have been identified in T-cell prolymphocytic leukemia, mantle cell lymphoma, and B-cell chronic lymphocytic leukemia. In this study, the entire ATMcoding sequence was examined in genomic DNA from 120 lymphoid neoplasms. Novel mutations and mutations implicated in cancer and/or A-T were found in 9 of 45 diffuse large B-cell lymphomas (DLBCLs), 2 of 24 follicular lymphomas, and 1 of 27 adult acute lymphoblastic leukemias, whereas no such mutations were detected among 24 peripheral T-cell lymphomas. The mutational spectrum consisted of 2 nonsense mutations, 1 mutation affecting RNA splicing, and 10 missense variants. Most of these mutations were associated with loss or mutation of the paired ATM allele, consistent with biallelic inactivation of ATM. Of the 9 DLBCLs with ATM mutations, 7 also carried TP53 mutations and/or deletions of theINK4a/ARF locus (P = .003). TheATM 735C>T substitution previously considered a rare normal variant was found to be 5.6 times more frequent in individuals with DLBCL than in random individuals (P = .026), suggesting that it may predispose to B-cell lymphoma. Our data suggest that ATM mutations contribute to the development of DLBCL, and that ATM and the ARF-p53 tumor suppressor pathway may cooperate in the pathogenesis of this malignancy.
Introduction
Germ-line mutations in the ATM gene are the cause of ataxia-telangiectasia (A-T; MIM 208900), an autosomal recessive disorder characterized by progressive cerebellar ataxia, oculocutaneous telangiectasias, hypersensitivity to ionizing radiation, immunodeficiency, chromosomal instability, and cancer susceptibility.1 Malignancies occur in childhood or early adulthood in 30% to 40% of all A-T patients, and approximately 85% of the tumors are of lymphoid origin.2 Although it is still a matter of controversy, A-T heterozygotes have been suggested to have a reduced life expectancy owing to a higher frequency of ischemic heart disease3 and an increased risk for cancer, particularly carcinoma of the breast.4,5 Several studies have indicated that ATM may also be involved in the development of some subtypes of sporadic lymphoma and leukemia: missense and loss-of-function mutations in the ATM gene have been demonstrated in T-cell prolymphocytic leukemia (T-PLL),6-9 mantle cell lymphoma (MCL),10,11and B-cell chronic lymphocytic leukemia (B-CLL).12-15Taken together, these epidemiologic and molecular data strongly implicate ATM as a tumor suppressor involved in the control of lymphoproliferation.
The ATM protein is a pleiotropic molecule whose activities are induced by chromosomal double-strand breaks that arise endogenously or after exposure to DNA-damaging agents, including ionizing radiation and radiomimetic drugs.16,17 ATM protects the integrity of the genome at different levels: (1) it mediates arrest of the cell cycle at G1/S, S, and G2/M to prevent the processing of damaged DNA; (2) it activates DNA-repair pathways; and (3) it induces apoptosis if the DNA damage is so detrimental that normal cell function can no longer be rescued.17 Many of these effects are mediated via a phosphatidylinositol-3 kinase (PI-3K) domain in the C-terminus of the ATM protein (residues 2656-3056). After radiation-induced DNA damage, the kinase activity of ATM increases severalfold, leading to phosphorylation of downstream effectors involved in the regulation of cell-cycle checkpoints, DNA repair, and apoptosis. Recent work has identified p53, nibrin, CHK2, MDM2, c-Abl, replication protein A, BRCA1, E2F1, and BLM as direct targets of ATM activity.16-19 In addition to the PI-3K domain, ATM comprises a number of specific amino-acid motifs, including a p53-binding region (residues 1-246), a β-adaptin-binding region (residues 812-948), a leucine zipper (residues 1217-1238), a c-Abl-binding region (residues 1373-1382), and a Rad3-homology region (residues 1443-2428).16 17
Several lines of evidence suggest that alterations of ATM in lymphoid neoplasias are not confined to MCL, T-PLL, and CLL but may be involved in a broader range of histologic subtypes. First, several cases of centroblastic B-cell lymphomas have been observed in A-T patients.2 Second, hemizygous deletions of theATM locus at 11q23 were identified in 13% to 24% of sporadic diffuse large B-cell lymphomas (DLBCLs),20,2112% of follicular center cell lymphomas (FLs),20 and 28% of adult acute lymphoblastic leukemias (ALLs).22 Third, Vorechovsky et al6 demonstrated missense variants in theATM gene in 2 of 32 non-Hodgkin lymphomas. Here, we have studied in detail the spectrum of ATM mutations in 120 lymphoid neoplasms of 4 histological subtypes. Because mutation ofTP53 is well established as a pathogenic factor in non-Hodgkin lymphoma,23 and because ATM acts upstream of p53 in the cellular response to ionizing radiation,24,25we specifically focused on the relationship between ATM andTP53 mutations. We also examined the status of the gene encoding ARF, a tumor suppressor that binds to and inactivates MDM2, thereby stabilizing p53.26 ARF is activated by a variety of stimuli, including DNA damage,27 and loss of ARF may in some aspects be functionally equivalent to loss of p53.
Materials and methods
Tissue samples
A total of 120 Danish cases of lymphoid malignancy were included in this study. The samples were classified according to the World Health Organization classification28 and included 45 DLBCLs, 24 FLs, 24 peripheral T-cell lymphomas of the NOS/unspecified category (PTLs), and 27 adult ALLs of the precursor B-cell type. All lymphoma samples were obtained fresh and immediately frozen in either liquid N2 or a mixture of 2-methyl butane and dry ice. Samples were stored at −80°C until use. Mononuclear cells from ALL patients were cryopreserved in 10% fetal calf serum and 10% dimethyl sulfoxide (DMSO). In all cases, diagnosis was based on both histology and phenotypic criteria, using immunohistochemistry or flow cytometry. In 5 of the ATM-mutant cases, samples of uninvolved normal tissue were available as paraffin-embedded sections. To determine the frequency of ATM variants in the general population of Denmark, a series of 100 genomic DNA samples were collected from random, anonymous blood donors. Approval of this study was obtained from the local ethical committee.
DNA isolation, mutation analysis, and direct sequencing
Genomic DNA was isolated after proteinase K digestion with the Purescript DNA Isolation Kit (Gentra Systems, Minneapolis, MN). Paraffin-embedded tissue was treated with xylene prior to DNA extraction. Mutations in the TP53 gene (exons 2-11) and deletions of the INK4a/ARF locus (exons 1β and 2) had been previously determined in most of the DLBCL and FL cases.29The remaining cases were analyzed as described.29,30 The entire coding sequence and all splice sites of the ATM gene (exons 4-65)31 were scanned for mutations using a combination of polymerase chain reaction (PCR) and denaturing gradient gel electrophoresis (DGGE). The melting characteristics of each exonic region were calculated by means of the MELT87 computer algorithm,32 and appropriate GC- and AT-clamps were included in the PCR primers to modulate the melting properties into a 2-domain profile.33 The 75 sets of primers for mutation analysis of ATM are listed in Table1. PCR was performed in 15-μL reactions containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.5% DMSO, 0.2 mM cresol red, 12% sucrose, 10 pmol of each primer, 100 μM each dNTP, 20 ng of genomic DNA, and 0.8 units of AmpliTaq polymerase (Perkin-Elmer Cetus, Emeryville, CA). PCR conditions were 38 cycles at 94°C for 20 seconds, 50°C to 55°C for 20 seconds, and 72°C for 30 seconds. For amplicons 8, 10, 13II, 26, 28I, 31II, and 35, Taq polymerase was added to the reaction in the first cycle at an 80°C step between the denaturation and annealing steps (“hot start”). PCR products were analyzed in a 0% denaturant/6% polyacrylamide–70% denaturant/12% polyacrylamide double-gradient gel.34 The gels were run at 80 V for 16 hours in 1 × TAE buffer kept at a constant temperature of 54°C, then stained with ethidium bromide and photographed under ultraviolet (UV) transillumination.
Primers for DGGE analysis of the ATM gene
| . | Forward primer sequence (5′ → 3′) . | Reverse primer sequence (5′ → 3′) . | Product length, bp . |
|---|---|---|---|
| Primers for mutation analysis (exons) | |||
| 4 | TGATGTGTGTTCTGAAATTGT | [40GC]-CAAGGAAAAATTGAATTTAAGTA | 170 |
| 5 | [8GC]-CCCATTATTATTTCCTTTTTA | [40GC]-ACAGTAAAATAAATTTGAATAGAAT | 213 |
| 6 | [40GC]-AAGTATTCAACGAGTTTCTGA | [11GC]-TTTATTTATAATTTATAACATCACTT | 269 |
| 7 | [40GC]-TTTTCTTTATTTGTTTATTTTGA | AAACAACAACCTTCAAAACA | 256 |
| 8 | [40GC]-TGACTAATAATTTTgTTTTTTT | ACAAAAGAAAAAGAGATTAGATT | 259 |
| 9I | AGTTGAGCTTGTTTGTTTCTT | [40GC]-TGAAATAATTCAATAATGACTTCT | 255 |
| 9II | [40GC]-AACATAGGCTTAATGATTCTTTA | CTATGTTTGAATGAAGAAGCA | 195 |
| 10 | [40GC]-ATTACATTTTAATTTTTTGGAT | TGTGACATGACCTACTTACTGT | 255 |
| 11 | [40GC]-AAAAAAAGAAAAAAGTGGATT | GAATGAGAAAATGGTAACAC | 271 |
| 12I | [40GC]-TCAAATTATCCTTTTTTgTTTTTT | ACACATATGGTGTACGTTCCC | 200 |
| 12II | GATGATACTATCTCAGCTTCTACCC | [40GC]-CCAAATTTTATTCCAGAGTTTT | 200 |
| 12III | [40GC]-CCTAGAAAGCTCACAAAAGTC | ACATAATGCATGCTGAACTT | 257 |
| 13I | CAATTGTCCTTTGTTTTGT | [40GC]-TAGAATAAGAGCCATTTCATT | 205 |
| 13II | [13GC]-CTTTTCTTTAAAGGAATCAAT | [40GC]-aaaaATAAAACAAACAAAAAAAACA | 198 |
| 14 | [5GC]-GTGAAGCTTTTTGTTTTTCTT | [40GC]-ATAGCTTAAAATGATAAAGAGCA | 206 |
| 15I | [40GC]-TTTATATATTAAAGATCTTACTTTCT | GGACAGAGAAGCCAATAC | 220 |
| 15II | ATAGAAAAGCACCAGTCCA | [40GC]-TAAGCTTAGTTCTTTTTTTAAAA | 185 |
| 16I | [40GC]-AATATATATATTTTTATTTGTGGTT | ACCAAAAGACGTGAACAC | 118 |
| 16II | CAAATTCAGAAACTCTTGTCC | [40GC]-CGAAACTTTATTAGTATAAATTCTC | 190 |
| 17 | [40GC]-ATTTTAACTGGAATTTGCATT | [5GC]-ACAAAACAAGAAGAAGAAAATCT | 238 |
| 18 | TTGGTTCTTTGTTTGTCTTAA | [40GC]-AATTATGCCTATTAGAATCAAAA | 209 |
| 19 | AACATCTTTGTTTCTCTTCCT | [40GC]-AAGAAGTAAAAGAAATCCCAA | 278 |
| 20 | [40GC]-TTAGTGTTAATGAGTGCTTTTT | GCTTCTTCATTTAACGTAACT | 294 |
| 21 | [40GC]-ATTTTTTTCCCTCCTACCA | [10GC]-ACATAACATAAGGTTTTAATTTCTT | 185 |
| 22 | [40GC]-ttttttttAGTTGAACTTTTTTTgTTTTTTTAC | GACCACAAAATAGACTGTACC | 263 |
| 23 | [40GC]-ATCAGAATGATTATTTAACTTTG | ACTAAAAAAATGCAAAAACTC | 182 |
| 24 | [40GC]-TTTTTCTCTATTTCATATTTAACC | [7GC]-TCATGAATATTTGACCCATT | 242 |
| 25 | [5GC]-TGTTTGTTTGCTTGCTTGT | [40GC]-CAGCAAATACATGTTACTTAAAAT | 215 |
| 26 | [40GC]-CATTTTgTTTTTAATTTCTTT | [8GC]-TCTAATAAAATACTCATCCATAT | 275 |
| 27 | CACATTGACTTTTTGGTTC | [40GC]-TCACATATGTCATGTATAAACTT | 259 |
| 28I | [40GC]-ACCTGTATTTTAAATTccgcTTTCTATT | [6GC]-GGCAAAATAAGGAAGAATAT | 230 |
| 28II | CTTTCCAAAGATTCTTGTAA | [40GC]-TACATAAAAATTGAAGCCAT | 198 |
| 29 | [40GC]-TATATTTTAATTTTGTGCCCT | [10GC]-TAGTTCTCTAAGTTTAAAATGTACA | 218 |
| 30 | CTGTATTTTTTCCCTTAACTC | [40GC]-ATTATTAAACTCATCATGTATACTT | 221 |
| 31I | [9GC]-AAGTTTTTACTAAATCTGTTTATT | [40GC]-GCCACTTTTTATATCTTTCA | 211 |
| 31II | [11GC]-TCACCTGTTTGTTAGTTTATT | [40GC]-ATATATTGGTCTAcccAATATGTTATTT | 195 |
| 32 | [13GC]-TTGTTTTTTTTTCTCCCTAT | [40GC]-TTTTAAATACCATTTTGAAGAT | 280 |
| 33 | [40GC]-CCTTATAATTTTTTCTTTTTAAA | AATAGTAGATGATGAAATTTTTAT | 275 |
| 34 | [40GC]-AAAGCAAGTTACATTTTCTC | CCATGTCATTTAAAATTAGC | 221 |
| 35 | [40GC]-AAACTAATTTTTAAAAAATTATT | AGCGCATGACTGTAGTTT | 200 |
| 36 | [7GC]-ACCTATGACTCTACTGAAATAGA | [40GC]-TACATATAAGGTATCAATAAATACT | 284 |
| 37 | ATAGGCATTTGAATTGTTTTT | [40GC]-CTGGGTATTCAATAAACATTTAT | 243 |
| 38 | [8GC]-ACATTTTCTAATCCCTTTCT | [40GC]-TTATATCAGACTAATTAATCTTCTT | 278 |
| 39I | [40GC]-TGGGTGGATTTGTTTGTATAT | TCGTTTGCGAGAAGTGTC | 211 |
| 39II | [10GC]-TCTACACATGTTCAGGGAT | [40GC]-TAATAGAATACCTGAATCCAA | 149 |
| 39III | GATCCACAACCCCTGCAA | [40GC]-CATCTTTCTCTAGAACTGAGTTTACA | 125 |
| 40 | [5GC]-TGAGGGGAGATATTTTTGTT | [40GC]-AAGAAGCAACACTCATTACATT | 183 |
| 41 | [40GC]-AAAGTGAATGACATTATATCTCA | [10GC]-CCAAAAATTCTAAATTCCAT | 263 |
| 42 | CCAAATAGTATGTTCTCATTAAA | [40GC]-TGTCATAAATAATAGAGCCTCTA | 204 |
| 43 | [40GC]-AATTTTTGTTGTTTCCATGTT | [7GC]-GTTGTTTAGAAAAATGCAATTT | 186 |
| 44 | [40GC]-AGCTATTTTCACAATCTTTTC | [7GC]-CTTTACCAAATCTGGGAAA | 204 |
| 45 | [40GC]-aaaaaaTTAAAAACAAAATAACTCCTG | AAAAATCAAGTCAAATTTCTT | 244 |
| 46 | GTATATTTTTTTCTTTGACTTATCT | [40GC]-CAGTAACTTTGTCTTTTCATAAT | 200 |
| 47 | [40GC]-GAGTGTCTTTTCTTTTTTGCT | [10GC]-GGATAACAAAGTCATACGAATTAC | 224 |
| 48 | [4GC]-CATGAACTCTATGTCGTGGC | [40GC]-CAAGAGGTAAGATGACATAGTTTTA | 340 |
| 49I | [10GC]-TTATTCCCATATGTCATTTTC | [40GC]-AGACTCCACAGCTAACTGAA | 146 |
| 49II | [40GC]-AATATTTCAAATTAAACAGTACAA | CAGCCAATAAAAAAAACAA | 215 |
| 50 | [40GC]-TATTTTAAGATTTTGCCTTTC | [7GC]-GGTTGCTCCAAAAATCTT | 210 |
| 51 | [40GC]-GTAAATATAATTTAAATTGGTTGT | [5GC]-TTGTAGAAACCTAGTT | 315 |
| 52I | [40GC]-TTGTGTTTTACCTTAATTATTCTAT | [11GC]-TATAATTTTCAACTGCTTTACA | 185 |
| 52II | [40GC]-AGAGGATCGTAAACGCTTCTT | AGAATATTGGGCTGAGTAACACT | 214 |
| 53 | [40GC]-CTTAATTTTGTGTCTTTTTTTTAAT | [5GC]-GGTTTGATTTTCAGGTTTACT | 213 |
| 54 | [40GC]-TGCATAAATCTAATAGTTCTTTTCT | [4GC]-GGTACGTATGTTTAATCCAAA | 257 |
| 55 | [40GC]-TTGGGTAGTTCCTTATGTAA | [8GC]-TTTCTTCTTTAAAAAAAACAT | 253 |
| 56 | [10GC]-TTTGTCACTAAAATCTCTTCAT | [40GC]-AATCAAGAGTTAATTGCAAATT | 191 |
| 57 | [40GC]-TATTATCAATCATGTTTATACTTTT | CAGAGAAGGGAAGGCTC | 230 |
| 58 | CTTGTTTATTCATGCTTAATTATT | [40GC]-GTGAACTAACAGAAGTACAAATAGT | 223 |
| 59 | TAATGGCCTTTTAAAATTAAAA | [40GC]-CTTTAATTTTGGGTGTCACTC | 262 |
| 60I | [40GC]-GAAAAATAATTATATATATTCTCTATT | GATCCAAGAATTTTTCCA | 178 |
| 60II | [5GC]-GTTTTCCGTTACTTCTGC | [40GC]-CAATCTACTATATGTACAAGAAGAT | 170 |
| 61 | [3GC]-CAGATTGTTTGTTTCTTTTTT | [40GC]-AAAGAATAGATACATAGATTTTATTA | 186 |
| 62 | [40GC]-CTCCTAACTTCACTGTATTCTTT | [5GC]-CCCTCCTTTACTTCATATCACT | 215 |
| 63 | AAATCCGTATTTATAATGTGTTTGA | [40GC]-CCTGGTAAGAAAAAAAATAAAGTCT | 139 |
| 64 | [40GC]-TCTTTTAATACATATGTTCTCTCTG | AGGACCTTCTTAAAATACTGC | 232 |
| 65 | [40GC]-CACCTCACTGAAACCTTTGTGT | AAGGCTGAATGAAAGGGTAATT | 293 |
| Primers for deletion analysis (introns) | |||
| 4 | [40GC]-TGATAGAGCTACAGAACGAAA | TTTCAGGATCTCGAATCAG | 184 |
| 7 | [40GC]-AGTCATGAATAATATATCAGGTGC | GAACTCTGGTTTACTTGGGTT | 157 |
| 17 | [40GC]-GTTATTATAACTCTAAGAAAAGATGTG | TAACAATCGCAGGAAAAAG | 250 |
| 22 | TTTGGCAAGGTGAGTATGTTG | [40GC]-GCAATTTACTAGGGCCATTCT | 250 |
| 48 | TTGGAATTAGATCTGACTTTT | [40GC]-ATGTATGTGTATCTGTGTGTACAT | 200 |
| 62 | [40GC]-ATGCTATTTTGAGATACAGATATGT | AGAGTTTCTCATCACTTCCATGGTTT | 212 |
| 63 | [40GC]-AGACTTTATTTTTTTTCTTACCAG | TAAGTGCACAATCTCCTCCT | 169 |
| . | Forward primer sequence (5′ → 3′) . | Reverse primer sequence (5′ → 3′) . | Product length, bp . |
|---|---|---|---|
| Primers for mutation analysis (exons) | |||
| 4 | TGATGTGTGTTCTGAAATTGT | [40GC]-CAAGGAAAAATTGAATTTAAGTA | 170 |
| 5 | [8GC]-CCCATTATTATTTCCTTTTTA | [40GC]-ACAGTAAAATAAATTTGAATAGAAT | 213 |
| 6 | [40GC]-AAGTATTCAACGAGTTTCTGA | [11GC]-TTTATTTATAATTTATAACATCACTT | 269 |
| 7 | [40GC]-TTTTCTTTATTTGTTTATTTTGA | AAACAACAACCTTCAAAACA | 256 |
| 8 | [40GC]-TGACTAATAATTTTgTTTTTTT | ACAAAAGAAAAAGAGATTAGATT | 259 |
| 9I | AGTTGAGCTTGTTTGTTTCTT | [40GC]-TGAAATAATTCAATAATGACTTCT | 255 |
| 9II | [40GC]-AACATAGGCTTAATGATTCTTTA | CTATGTTTGAATGAAGAAGCA | 195 |
| 10 | [40GC]-ATTACATTTTAATTTTTTGGAT | TGTGACATGACCTACTTACTGT | 255 |
| 11 | [40GC]-AAAAAAAGAAAAAAGTGGATT | GAATGAGAAAATGGTAACAC | 271 |
| 12I | [40GC]-TCAAATTATCCTTTTTTgTTTTTT | ACACATATGGTGTACGTTCCC | 200 |
| 12II | GATGATACTATCTCAGCTTCTACCC | [40GC]-CCAAATTTTATTCCAGAGTTTT | 200 |
| 12III | [40GC]-CCTAGAAAGCTCACAAAAGTC | ACATAATGCATGCTGAACTT | 257 |
| 13I | CAATTGTCCTTTGTTTTGT | [40GC]-TAGAATAAGAGCCATTTCATT | 205 |
| 13II | [13GC]-CTTTTCTTTAAAGGAATCAAT | [40GC]-aaaaATAAAACAAACAAAAAAAACA | 198 |
| 14 | [5GC]-GTGAAGCTTTTTGTTTTTCTT | [40GC]-ATAGCTTAAAATGATAAAGAGCA | 206 |
| 15I | [40GC]-TTTATATATTAAAGATCTTACTTTCT | GGACAGAGAAGCCAATAC | 220 |
| 15II | ATAGAAAAGCACCAGTCCA | [40GC]-TAAGCTTAGTTCTTTTTTTAAAA | 185 |
| 16I | [40GC]-AATATATATATTTTTATTTGTGGTT | ACCAAAAGACGTGAACAC | 118 |
| 16II | CAAATTCAGAAACTCTTGTCC | [40GC]-CGAAACTTTATTAGTATAAATTCTC | 190 |
| 17 | [40GC]-ATTTTAACTGGAATTTGCATT | [5GC]-ACAAAACAAGAAGAAGAAAATCT | 238 |
| 18 | TTGGTTCTTTGTTTGTCTTAA | [40GC]-AATTATGCCTATTAGAATCAAAA | 209 |
| 19 | AACATCTTTGTTTCTCTTCCT | [40GC]-AAGAAGTAAAAGAAATCCCAA | 278 |
| 20 | [40GC]-TTAGTGTTAATGAGTGCTTTTT | GCTTCTTCATTTAACGTAACT | 294 |
| 21 | [40GC]-ATTTTTTTCCCTCCTACCA | [10GC]-ACATAACATAAGGTTTTAATTTCTT | 185 |
| 22 | [40GC]-ttttttttAGTTGAACTTTTTTTgTTTTTTTAC | GACCACAAAATAGACTGTACC | 263 |
| 23 | [40GC]-ATCAGAATGATTATTTAACTTTG | ACTAAAAAAATGCAAAAACTC | 182 |
| 24 | [40GC]-TTTTTCTCTATTTCATATTTAACC | [7GC]-TCATGAATATTTGACCCATT | 242 |
| 25 | [5GC]-TGTTTGTTTGCTTGCTTGT | [40GC]-CAGCAAATACATGTTACTTAAAAT | 215 |
| 26 | [40GC]-CATTTTgTTTTTAATTTCTTT | [8GC]-TCTAATAAAATACTCATCCATAT | 275 |
| 27 | CACATTGACTTTTTGGTTC | [40GC]-TCACATATGTCATGTATAAACTT | 259 |
| 28I | [40GC]-ACCTGTATTTTAAATTccgcTTTCTATT | [6GC]-GGCAAAATAAGGAAGAATAT | 230 |
| 28II | CTTTCCAAAGATTCTTGTAA | [40GC]-TACATAAAAATTGAAGCCAT | 198 |
| 29 | [40GC]-TATATTTTAATTTTGTGCCCT | [10GC]-TAGTTCTCTAAGTTTAAAATGTACA | 218 |
| 30 | CTGTATTTTTTCCCTTAACTC | [40GC]-ATTATTAAACTCATCATGTATACTT | 221 |
| 31I | [9GC]-AAGTTTTTACTAAATCTGTTTATT | [40GC]-GCCACTTTTTATATCTTTCA | 211 |
| 31II | [11GC]-TCACCTGTTTGTTAGTTTATT | [40GC]-ATATATTGGTCTAcccAATATGTTATTT | 195 |
| 32 | [13GC]-TTGTTTTTTTTTCTCCCTAT | [40GC]-TTTTAAATACCATTTTGAAGAT | 280 |
| 33 | [40GC]-CCTTATAATTTTTTCTTTTTAAA | AATAGTAGATGATGAAATTTTTAT | 275 |
| 34 | [40GC]-AAAGCAAGTTACATTTTCTC | CCATGTCATTTAAAATTAGC | 221 |
| 35 | [40GC]-AAACTAATTTTTAAAAAATTATT | AGCGCATGACTGTAGTTT | 200 |
| 36 | [7GC]-ACCTATGACTCTACTGAAATAGA | [40GC]-TACATATAAGGTATCAATAAATACT | 284 |
| 37 | ATAGGCATTTGAATTGTTTTT | [40GC]-CTGGGTATTCAATAAACATTTAT | 243 |
| 38 | [8GC]-ACATTTTCTAATCCCTTTCT | [40GC]-TTATATCAGACTAATTAATCTTCTT | 278 |
| 39I | [40GC]-TGGGTGGATTTGTTTGTATAT | TCGTTTGCGAGAAGTGTC | 211 |
| 39II | [10GC]-TCTACACATGTTCAGGGAT | [40GC]-TAATAGAATACCTGAATCCAA | 149 |
| 39III | GATCCACAACCCCTGCAA | [40GC]-CATCTTTCTCTAGAACTGAGTTTACA | 125 |
| 40 | [5GC]-TGAGGGGAGATATTTTTGTT | [40GC]-AAGAAGCAACACTCATTACATT | 183 |
| 41 | [40GC]-AAAGTGAATGACATTATATCTCA | [10GC]-CCAAAAATTCTAAATTCCAT | 263 |
| 42 | CCAAATAGTATGTTCTCATTAAA | [40GC]-TGTCATAAATAATAGAGCCTCTA | 204 |
| 43 | [40GC]-AATTTTTGTTGTTTCCATGTT | [7GC]-GTTGTTTAGAAAAATGCAATTT | 186 |
| 44 | [40GC]-AGCTATTTTCACAATCTTTTC | [7GC]-CTTTACCAAATCTGGGAAA | 204 |
| 45 | [40GC]-aaaaaaTTAAAAACAAAATAACTCCTG | AAAAATCAAGTCAAATTTCTT | 244 |
| 46 | GTATATTTTTTTCTTTGACTTATCT | [40GC]-CAGTAACTTTGTCTTTTCATAAT | 200 |
| 47 | [40GC]-GAGTGTCTTTTCTTTTTTGCT | [10GC]-GGATAACAAAGTCATACGAATTAC | 224 |
| 48 | [4GC]-CATGAACTCTATGTCGTGGC | [40GC]-CAAGAGGTAAGATGACATAGTTTTA | 340 |
| 49I | [10GC]-TTATTCCCATATGTCATTTTC | [40GC]-AGACTCCACAGCTAACTGAA | 146 |
| 49II | [40GC]-AATATTTCAAATTAAACAGTACAA | CAGCCAATAAAAAAAACAA | 215 |
| 50 | [40GC]-TATTTTAAGATTTTGCCTTTC | [7GC]-GGTTGCTCCAAAAATCTT | 210 |
| 51 | [40GC]-GTAAATATAATTTAAATTGGTTGT | [5GC]-TTGTAGAAACCTAGTT | 315 |
| 52I | [40GC]-TTGTGTTTTACCTTAATTATTCTAT | [11GC]-TATAATTTTCAACTGCTTTACA | 185 |
| 52II | [40GC]-AGAGGATCGTAAACGCTTCTT | AGAATATTGGGCTGAGTAACACT | 214 |
| 53 | [40GC]-CTTAATTTTGTGTCTTTTTTTTAAT | [5GC]-GGTTTGATTTTCAGGTTTACT | 213 |
| 54 | [40GC]-TGCATAAATCTAATAGTTCTTTTCT | [4GC]-GGTACGTATGTTTAATCCAAA | 257 |
| 55 | [40GC]-TTGGGTAGTTCCTTATGTAA | [8GC]-TTTCTTCTTTAAAAAAAACAT | 253 |
| 56 | [10GC]-TTTGTCACTAAAATCTCTTCAT | [40GC]-AATCAAGAGTTAATTGCAAATT | 191 |
| 57 | [40GC]-TATTATCAATCATGTTTATACTTTT | CAGAGAAGGGAAGGCTC | 230 |
| 58 | CTTGTTTATTCATGCTTAATTATT | [40GC]-GTGAACTAACAGAAGTACAAATAGT | 223 |
| 59 | TAATGGCCTTTTAAAATTAAAA | [40GC]-CTTTAATTTTGGGTGTCACTC | 262 |
| 60I | [40GC]-GAAAAATAATTATATATATTCTCTATT | GATCCAAGAATTTTTCCA | 178 |
| 60II | [5GC]-GTTTTCCGTTACTTCTGC | [40GC]-CAATCTACTATATGTACAAGAAGAT | 170 |
| 61 | [3GC]-CAGATTGTTTGTTTCTTTTTT | [40GC]-AAAGAATAGATACATAGATTTTATTA | 186 |
| 62 | [40GC]-CTCCTAACTTCACTGTATTCTTT | [5GC]-CCCTCCTTTACTTCATATCACT | 215 |
| 63 | AAATCCGTATTTATAATGTGTTTGA | [40GC]-CCTGGTAAGAAAAAAAATAAAGTCT | 139 |
| 64 | [40GC]-TCTTTTAATACATATGTTCTCTCTG | AGGACCTTCTTAAAATACTGC | 232 |
| 65 | [40GC]-CACCTCACTGAAACCTTTGTGT | AAGGCTGAATGAAAGGGTAATT | 293 |
| Primers for deletion analysis (introns) | |||
| 4 | [40GC]-TGATAGAGCTACAGAACGAAA | TTTCAGGATCTCGAATCAG | 184 |
| 7 | [40GC]-AGTCATGAATAATATATCAGGTGC | GAACTCTGGTTTACTTGGGTT | 157 |
| 17 | [40GC]-GTTATTATAACTCTAAGAAAAGATGTG | TAACAATCGCAGGAAAAAG | 250 |
| 22 | TTTGGCAAGGTGAGTATGTTG | [40GC]-GCAATTTACTAGGGCCATTCT | 250 |
| 48 | TTGGAATTAGATCTGACTTTT | [40GC]-ATGTATGTGTATCTGTGTGTACAT | 200 |
| 62 | [40GC]-ATGCTATTTTGAGATACAGATATGT | AGAGTTTCTCATCACTTCCATGGTTT | 212 |
| 63 | [40GC]-AGACTTTATTTTTTTTCTTACCAG | TAAGTGCACAATCTCCTCCT | 169 |
[40GC] indicates CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG. Lowercase characters represent nucleotides incorporated into the primer to modulate the melting characteristics of the amplified segment.
Direct sequence analysis of aberrant DGGE bands was performed with a nonclamped, 32P–end-labeled primer, using the ThermoPrime Cycle Sequencing Kit (Amersham Life Science, Cleveland, OH) according to the manufacturer's instructions.
Detection of allelic loss of ATM
To detect allelic loss of ATM, we designed primers to amplify regions encompassing the biallelic polymorphisms in introns 4 (72+36insAA),35 7 (496+221T/C),35 17 (2377−56G/A),35 22 (3078−77T/C),35 48 (6807+239 C/G),36 62 (8787−56T/C),35 and 63 (8850+60G/A)35 (Table 1) . Primers for detection of biallelic polymorphisms in exons 32 (4578C/T)37 and 39 (5557G/A)35 were the same as used for mutation analysis. The individual alleles of the 9 polymorphisms were resolved by DGGE as described above, except that analysis of 6807 + 239C/G was performed in a 15% denaturant/6% polyacrylamide–55% denaturant/9% polyacrylamide gel at 52°C. A tagged image file format (TIFF) image of the gel was generated after ethidium bromide staining and UV transillumination, and the intensities of the 2 homoduplex bands in informative cases were determined with 1D Gel Analysis Phoretix Software (Phoretix, Newcastle, United Kingdom). A case was also considered to harbor an ATM deletion if the aberrant band at the mutational site was more intense than the wild-type band.
Detection of ATM promoter hypermethylation
The methylation status of the ATM promoter CpG island was examined by methylation-specific PCR.38 This method is based on initial treatment of genomic DNA with sodium bisulfite to convert unmethylated cytosine to uracil. The bisulfite reaction was performed essentially as described.39 In summary, approximately 1 μg of genomic DNA was denatured in 0.3 M NaOH, followed by the addition of sodium bisulfite (Sigma Chemical, St Louis, MO) to a final concentration of 3.1 M and hydroquinone (Sigma Chemical) to a final concentration of 2.5 mM. After incubation at 55°C for 16 hours, the DNA was recovered with the GeneClean II Kit (Bio 101, Vista, CA), desulfonated in 0.3 M NaOH, and ethanol-precipitated. DNA treated in vitro with Sss I methyltransferase (New England Biolabs, Beverly, MA) was used as a positive control for methylatedATM alleles. Primers were 5′-GGTATGTTTATGCGTATTTAGTATTACGC-3′ (sense) and 5′-AACGCTAAATCGCTAACCATTAATAA-3′ (antisense) for methylated ATMalleles (GenBank accession no. D83244; positions 607-728), and 5′-TGGTATGTTTATGTGTATTTAGTATTATGT-3′ (sense) and 5′-AAACACTAAATCACTAACCATTAATAAA-3′ (antisense) for unmethylatedATM alleles (GenBank accession no. D83244, positions 606-729). PCR was performed in 25-μL volumes containing 1 × buffer and 1 unit of Taq polymerase (HotStarTaq Kit; Qiagen, Hilden, Germany), 0.2 mM cresol red, 12% sucrose, 10 pmol of each primer, 100 μM each dNTP, and 1 μL of bisulfite-treated DNA. Reactions were started by initial denaturation at 95°C for 15 minutes, followed by 40 cycles at 95°C for 30 seconds, 56°C (for unmethylated ATM) or 60°C (for methylated ATM) for 30 seconds, and 72°C for 30 seconds. The amplification products were resolved in a 2% agarose gel.
Reverse transcriptase polymerase chain reaction (RT-PCR) analysis
RNA was extracted using the Purescript Isolation Kit (Gentra Systems, Minneapolis, MN). cDNA synthesis was carried out using M-MLV SuperScript II reverse transcriptase (Gibco-BRL, Life Technologies, Gaithersburg, MD) in a total volume of 20 μL of 1 × buffer (Gibco-BRL, Life Technologies) containing 10 mM dithiothreitol (DTT). Incubations were performed at 42°C for 50 minutes, followed by 72°C for 5 minutes. Skipping of exon 9 of the ATM gene was detected using primers 5′-GAATAATTCATGCTGTTACCA-3′ (exon 8) and 5′-GAATACTTTCCTCTACTTCCTATAT-3′ (exon 10), and skipping of exon 46 was detected using primers 5′-GAATACTTTCCTCTACTTCCTATAT-3′ (exon 45) and 5′-GATAGAGCGAATACACAGACTC-3′ (exon 47). Amplifications were carried out in a total volume of 15 μL containing 1 × buffer and 1 unit of Taq polymerase (HotStarTaq Kit; Qiagen), 0.2 mM cresol red, 12% sucrose, 10 pmol of each primer, 100 μM each dNTP, and 0.5 μL of cDNA. Reactions were started by initial denaturation at 95°C for 15 minutes, followed by 35 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 40 seconds. Products were analyzed in a 2% agarose gel.
Statistical analysis
The χ2 test was used to compare the total frequencies of ATM class II mutations in DLBCL, FL, and random individuals. All other comparisons were made with the 2-tailed Fisher exact test. The level of significance was set at .05.
Results
Mutation analysis of the ATM gene in lymphoid neoplasms
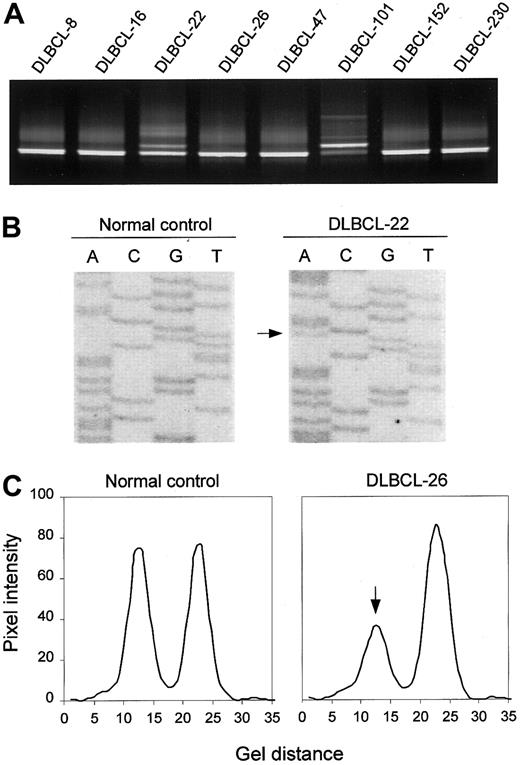
Detection of ATM mutations is complicated by the large size of the gene (> 150 kilobases (kb), 62 coding exons), and by the fact that disease-related mutations are spread throughout the coding sequence. To enable the study of a large series of lymphoid neoplasms from which only crude DNA was available, we adopted a mutation-scanning strategy based on PCR in combination with DGGE. This method provides an effective means of detecting low-abundant mutant alleles in samples where mutant DNA from tumor cells is contaminated with wild-type DNA from noncancerous cells, and of enriching mutant DNA for sequence analysis.33 DGGE analysis of 75 PCR-amplified segments covering all coding exons (4-65) and corresponding splice sites of the ATM gene revealed sequence variants in 18 (40%) of 45 DLBCLs, 9 (38%) of 24 FLs, 4 (15%) of 27 ALLs, and 4 (17%) of 24 PTLs (Figure 1A shows examples). Sequence analysis of DNA recovered from aberrant heteroduplex or homoduplex bands led to the identification of 20 different ATM sequence variants, including 16 missense variants, 2 mutations affecting normal splicing, and 2 mutations introducing premature termination signals (Figure 1B; Table2). The N1853D and P1526P variants, both of which have been established as genuine polymorphisms, were found in all 4 tumor types at frequencies not different from those found in samples from random individuals (Table 2). In addition, 3 DLBCLs each carried one of 3 novel variants, S1987S, R2461R, and V3005V. Examination of mRNA by RT-PCR analysis revealed no aberrant RNA splicing associated with any of these variants (data not shown), suggesting that they represent rare silent variants.
DGGE-based detection of ATM variants and allelic loss.
(A) DGGE analysis of a region spanning ATM exon 65, showing aberrant bands in samples DLBCL-22 and DLBCL-101. In the latter case, the mutant band is more intense than the wild-type band, suggesting loss of the wild-type allele. (B) Direct sequence analysis of exon 65, showing the R3008H (9023G>A) mutation in DLBCL-22. (C) Detection of anATM deletion in DLBCL-26 as manifested by loss of one of the alleles of the 8850 + 60G/A marker by DGGE analysis. Pixel intensity and gel distance are in arbitrary units.
DGGE-based detection of ATM variants and allelic loss.
(A) DGGE analysis of a region spanning ATM exon 65, showing aberrant bands in samples DLBCL-22 and DLBCL-101. In the latter case, the mutant band is more intense than the wild-type band, suggesting loss of the wild-type allele. (B) Direct sequence analysis of exon 65, showing the R3008H (9023G>A) mutation in DLBCL-22. (C) Detection of anATM deletion in DLBCL-26 as manifested by loss of one of the alleles of the 8850 + 60G/A marker by DGGE analysis. Pixel intensity and gel distance are in arbitrary units.
ATM sequence variants in 120 lymphoid neoplasms and samples from 100 random individuals
| Variant . | Nucleotide change . | Localization . | No. of tumors . | No. of random individuals . |
|---|---|---|---|---|
| S49C | C>G at 146 | Exon 5 | 2 | 2 |
| V245V* | C>T at 735 | Exon 9 | 10 | 2 |
| Q466X | C>T at 1396 | Exon 12 | 1 | 0 |
| F570S | T>C at 1709 | Exon 13 | 1 | 0 |
| P604S | C>T at 1810 | Exon 14 | 1 | 0 |
| S707P | T>C at 2119 | Exon 15 | 4 | 3 |
| F858L | T>C at 2572 | Exon 19 | 4 | 3 |
| H996Q | T>G at 2988 | Exon 22 | 1 | 0 |
| P1054R | C>G at 3161 | Exon 24 | 6 | 4 |
| H1380Y | C>T at 4138 | Exon 30 | 1 | 0 |
| I1407S | T>G at 4220 | Exon 30 | 1 | 0 |
| L1420F | C>T at 4258 | Exon 31 | 2 | 1 |
| N1853V | A>T at 5558 | Exon 39 | 2 | 0 |
| G2023R | G>A at 6067 | Exon 43 | 1 | 0 |
| G2063E | G>A at 6188 | Exon 44 | 1 | 0 |
| R2151G† | A>G at 6450 | Exon 46 | 1 | 0 |
| Q2593X | C>T at 7776 | Exon 54 | 1 | 0 |
| M2616I | G>T at 7847 | Exon 55 | 1 | 0 |
| R3008H | G>A at 9023 | Exon 65 | 1 | 0 |
| G3051V | G>T at 9252 | Exon 65 | 1 | 0 |
| N1853D | G>A at 5557 | Exon 39 | 21 | 20 |
| P1526P‡ | C/T at 4578 | Exon 32 | 10 | 7 |
| S1987S‡ | T/C at 5961 | Exon 42 | 1 | 0 |
| R2461R‡ | C/T at 7383 | Exon 52 | 1 | 0 |
| V3005V‡ | A/T at 9015 | Exon 65 | 1 | 0 |
| Variant . | Nucleotide change . | Localization . | No. of tumors . | No. of random individuals . |
|---|---|---|---|---|
| S49C | C>G at 146 | Exon 5 | 2 | 2 |
| V245V* | C>T at 735 | Exon 9 | 10 | 2 |
| Q466X | C>T at 1396 | Exon 12 | 1 | 0 |
| F570S | T>C at 1709 | Exon 13 | 1 | 0 |
| P604S | C>T at 1810 | Exon 14 | 1 | 0 |
| S707P | T>C at 2119 | Exon 15 | 4 | 3 |
| F858L | T>C at 2572 | Exon 19 | 4 | 3 |
| H996Q | T>G at 2988 | Exon 22 | 1 | 0 |
| P1054R | C>G at 3161 | Exon 24 | 6 | 4 |
| H1380Y | C>T at 4138 | Exon 30 | 1 | 0 |
| I1407S | T>G at 4220 | Exon 30 | 1 | 0 |
| L1420F | C>T at 4258 | Exon 31 | 2 | 1 |
| N1853V | A>T at 5558 | Exon 39 | 2 | 0 |
| G2023R | G>A at 6067 | Exon 43 | 1 | 0 |
| G2063E | G>A at 6188 | Exon 44 | 1 | 0 |
| R2151G† | A>G at 6450 | Exon 46 | 1 | 0 |
| Q2593X | C>T at 7776 | Exon 54 | 1 | 0 |
| M2616I | G>T at 7847 | Exon 55 | 1 | 0 |
| R3008H | G>A at 9023 | Exon 65 | 1 | 0 |
| G3051V | G>T at 9252 | Exon 65 | 1 | 0 |
| N1853D | G>A at 5557 | Exon 39 | 21 | 20 |
| P1526P‡ | C/T at 4578 | Exon 32 | 10 | 7 |
| S1987S‡ | T/C at 5961 | Exon 42 | 1 | 0 |
| R2461R‡ | C/T at 7383 | Exon 52 | 1 | 0 |
| V3005V‡ | A/T at 9015 | Exon 65 | 1 | 0 |
Associated with skipping of exon 9.
Associated with skipping of exon 46.
Synonymous substitutions.
Considering that at least some missense variants in the ATMgene have been suggested to represent rare normal variants with no phenotypic implications, we divided the lymphoma-related ATMsequence variants into 2 classes. Sequence variants were assigned to class I if they were first identified in this study or had previously been demonstrated only in cancer and/or A-T, and if they were not identified among 100 random individuals from the same population (Table2). Sequence variants were assigned to class II if they have been identified in tissue from apparently healthy individuals and thus may represent normal variants.
ATM class I variants
A total of 13 ATM class I variants were identified in 12 tumors, including 9 DLBCLs (20%), 2 FLs (8%), and 1 ALL (4%) (Table 3). No class I variants were found in the series of PTLs. The spectrum of class I variants consisted of 10 missense variants, 2 nonsense mutations, and 1 mutation affecting RNA splicing. The 6450A>G variant is predicted to affect a residue within the Rad3-homology region (R2151G); however, this transition alters the penultimate nucleotide of exon 46, suggesting that it may also be involved in aberrant RNA splicing. Indeed, RT-PCR analysis of a region encompassing exons 45 through 47 revealed a shorter band from this tumor (data not shown). Direct sequence analysis of this band showed a sequence lacking the 105 bases corresponding to exon 46, resulting in an in-frame deletion of 35 amino acids (codons 2117-2151).
ATM allelic status, TP53 mutations, and INK4a/ARF deletions in lymphoid neoplasms withATM class I and class II variants
| Tumor . | ATMvariant . | ATM allelic status . | TP53mutation3-150 . | INK4a/ARF deletion3-151 . |
|---|---|---|---|---|
| Class I variants | ||||
| DLBCL-22 | R3008H | NI | — | — |
| DLBCL-26 | R2151G3-152 | LOH | — | — |
| DLBCL-47 | H1380Y | LOH | R158C | — |
| DLBCL-70 | P604S | NI | IVS4nt-1G>A | — |
| DLBCL-101 | G3051V | LOH | — | Exons 1β and 2 |
| DLBCL-141 | H996Q | NI | R273H | — |
| 735C>T3-153,3-155 | ||||
| DLBCL-144 | I1407S | LOH | Y220C | Exons 1β and 2 |
| DLBCL-230 | Q2593X | LOH | P151S | — |
| DLBCL-274 | E466X | — | — | Exons 1β and 2 |
| G2063E | ||||
| FL-18 | M2616I | — | I254N | — |
| P1054R3-155 | ||||
| FL-134 | G2023R | — | — | — |
| F858L3-155 | ||||
| ALL-A22 | F570S | — | — | ND |
| Class II variants | ||||
| DLBCL-2 | 735C>T3-153 | NI | — | — |
| L1420F | ||||
| DLBCL-43 | N1853V | — | — | — |
| DLBCL-133 | 735C>T3-153 | NI | — | Exons 1β and 2 |
| DLBCL-139 | S49C | NI | — | — |
| DLBCL-152 | 735C>T3-153 | LOH | — | — |
| DLBCL-157 | P1054R | — | — | — |
| F858L | ||||
| DLBCL-159 | N1853V | — | R248Q | — |
| DLBCL-234 | S49C | NI | R248W | ND |
| 735C>T3-153 | ||||
| DLBCL-270 | L1420F | — | — | — |
| FL-4 | S707P | — | — | — |
| FL-45 | 735C>T3-153 | NI | — | — |
| FL-49 | S707P | — | — | — |
| FL-123 | P1054R | — | — | — |
| FL-142 | 735C>T3-153 | — | — | — |
| FL-218 | S707P | — | — | ND |
| FL-220 | P1054R | — | — | — |
| PTL-12 | 735C>T3-153 | — | — | — |
| PTL-160 | S707P | — | — | — |
| PTL-244 | 735C>T3-153 | — | — | — |
| PTL-256 | F858L | — | ND | — |
| P1054R | ||||
| ALL-A3 | F858L | NI | — | ND |
| ALL-A27 | P1054R | — | — | ND |
| ALL-A28 | 735C>T3-153 | — | R175H | ND |
| Tumor . | ATMvariant . | ATM allelic status . | TP53mutation3-150 . | INK4a/ARF deletion3-151 . |
|---|---|---|---|---|
| Class I variants | ||||
| DLBCL-22 | R3008H | NI | — | — |
| DLBCL-26 | R2151G3-152 | LOH | — | — |
| DLBCL-47 | H1380Y | LOH | R158C | — |
| DLBCL-70 | P604S | NI | IVS4nt-1G>A | — |
| DLBCL-101 | G3051V | LOH | — | Exons 1β and 2 |
| DLBCL-141 | H996Q | NI | R273H | — |
| 735C>T3-153,3-155 | ||||
| DLBCL-144 | I1407S | LOH | Y220C | Exons 1β and 2 |
| DLBCL-230 | Q2593X | LOH | P151S | — |
| DLBCL-274 | E466X | — | — | Exons 1β and 2 |
| G2063E | ||||
| FL-18 | M2616I | — | I254N | — |
| P1054R3-155 | ||||
| FL-134 | G2023R | — | — | — |
| F858L3-155 | ||||
| ALL-A22 | F570S | — | — | ND |
| Class II variants | ||||
| DLBCL-2 | 735C>T3-153 | NI | — | — |
| L1420F | ||||
| DLBCL-43 | N1853V | — | — | — |
| DLBCL-133 | 735C>T3-153 | NI | — | Exons 1β and 2 |
| DLBCL-139 | S49C | NI | — | — |
| DLBCL-152 | 735C>T3-153 | LOH | — | — |
| DLBCL-157 | P1054R | — | — | — |
| F858L | ||||
| DLBCL-159 | N1853V | — | R248Q | — |
| DLBCL-234 | S49C | NI | R248W | ND |
| 735C>T3-153 | ||||
| DLBCL-270 | L1420F | — | — | — |
| FL-4 | S707P | — | — | — |
| FL-45 | 735C>T3-153 | NI | — | — |
| FL-49 | S707P | — | — | — |
| FL-123 | P1054R | — | — | — |
| FL-142 | 735C>T3-153 | — | — | — |
| FL-218 | S707P | — | — | ND |
| FL-220 | P1054R | — | — | — |
| PTL-12 | 735C>T3-153 | — | — | — |
| PTL-160 | S707P | — | — | — |
| PTL-244 | 735C>T3-153 | — | — | — |
| PTL-256 | F858L | — | ND | — |
| P1054R | ||||
| ALL-A3 | F858L | NI | — | ND |
| ALL-A27 | P1054R | — | — | ND |
| ALL-A28 | 735C>T3-153 | — | R175H | ND |
NI indicates not informative; ND, not done; LOH, loss of heterozygosity.
Three DLBCLs and 1 FL carried TP53 mutations without detectable ATM sequence variants.
Three DLBCLs and 1 FL carried INK4a/ARFdeletions without detectable ATM sequence variants.
Associated with skipping of exon 46.
Associated with skipping of exon 9.
Class II variant.
Among the 9 DLBCLs with class I variants, one carried an additional class I variant and one carried a class II variant, but the allelic distribution of these sequence alterations could not be established. Five of the remaining DLBCLs were informative at the mutational site or at one or more polymorphic sites, and all of these showed evidence of loss of the paired ATM allele (Figure 1C; Table 3). Both FLs carrying class I variants also carried class II variants (Table 3). No evidence of additional mutations or allelic loss was found in the single ALL case carrying a class I variant.
Normal tissue was available from 5 cases with class I variants (DLBCL-22, -26, -70, and -274, and FL-18; Table 3). The 6450A>G splicing variant found in DLBCL-26 was also found in nontumorous tissue from this patient. Considering that the paired ATM allele was found to be deleted in the tumor, it is likely that 6450A>G in the germ-line of this patient was a predisposing factor for DLBCL development. Sequence analysis of the remaining 4 samples showed no evidence of mutations, suggesting that the mutations detected in the lymphoma specimens had arisen somatically.
ATM class II variants
A total of 7 ATM sequence variants identified among the 120 lymphoid neoplasms have also been found among random individuals and, accordingly, were assigned to class II. These variations included 6 missense variants and the 735C>T variant. The latter sequence change does not alter the coding sequence of ATM (V245V) but has been associated with aberrant splicing in A-T families.40In agreement with this observation, RT-PCR analysis of RNA from 6 tumors without 735C>T showed only a single band corresponding to the wild-type sequence, whereas 6 735C>T-positive tumors all showed an additional shorter band. Sequence analysis of this band confirmed skipping of exon 9 (data not shown). One or 2 class II variants were identified in 10 DLBCLs (22%), 9 FLs (38%), 3 ALLs (11%), and 4 PTLs (17%) (Table 3).
Analysis of samples from 100 random individuals from the same population revealed class II variants in 13 cases (13%). The higher frequency of class II variants in FL (P < .001) and a trend toward a higher frequency in DLBCLs (P = .065) compared with samples from random individuals suggest that mutations from this group may also contribute to B-cell lymphomagenesis. For most of the class II variants, the frequencies were too low for statistical analysis. One exception was the 735C>T variant, which was found to be 5.6 times more frequent in DLBCLs than in samples from random individuals (5 of 45 vs 2 of 100; P = .026). The S707P variant was found to be 4.2 times more frequent in FLs than in samples from random individuals, but this association did not reach statistical significance (3 of 24 vs 3 of 100; P = .073).
Of the 23 tumors carrying ATM class II variants, 17 were informative at one or more polymorphic markers, but only one (DLBCL-152) showed evidence of allelic loss.
Analysis of hypermethylation of the ATMpromoter
The ATM promoter contains an approximately 800–base pair (bp) region that has a GC content of 62% and a CpG:GpC ratio of 90% and therefore meets the criteria for a CpG island.41To examine the methylation status of this CpG island, SssI-methylated DNA and DNA from 45 DLBCLs and normal peripheral blood lymphocytes were treated with sodium bisulfite and amplified with primer pairs specific for methylated and unmethylated ATMalleles. None of the tumors contained methylated ATMalleles (data not shown). These observations, together with previous studies in T-PLL,42 suggest that inactivation of theATM gene by de novo methylation is a rare event in lymphoid neoplasia.
ATM mutation in relation to theARF-TP53 pathway
Among the B-cell lymphomas included in this study,TP53 mutations were identified in 10 (22%) of the 45 DLBCLs and in 2 (8%) of the 24 FLs. Surprisingly, more than half (5 of 9) of the DLBCLs with ATM class I variants harbored concomitant mutation of TP53 (Table 3), suggesting a nonrandom coexistence of these genetic alterations (P = .015). Despite the low frequencies of ATM and TP53mutations in FLs, we also identified 1 case with both alterations (Table 3). Among the 9 DLBCLs with ATM class II variants, 2 harbored TP53 mutations (P = .34), suggesting a random distribution. None of the 7 FLs with class II variants showed concomitant mutation of TP53 (Table 3). Three DLBCLs and 1 FL carried TP53 mutations in the absence of detectable ATM sequence variants.
To further study the possible association between ATMmutation and inactivation of the ARF-TP53pathway, exons 1β and 2 of the INK4a/ARF locus were examined for deletions in the DLBCL and FL series. Both exons were deleted in 6 DLCBLs (13%) and one of the FLs (4%). Three of the DLBCLs with INK4a/ARF deletions had ATM class I variants (P = .074), and one of these also had aTP53 mutation (Table 3). Overall, TP53 mutations and/or deletions of the INK4a/ARF locus were found in 7 (78%) of 9 DLBCLs with ATM class I variants and in 8 (22%) of 36 DLBCLs with no detectable ATM class I variants (P = .003).
Clinical features
The 9 DLBCL patients with ATM class I variants in their tumors were 5 men and 4 women, aged 39 to 76 years (median 62 years) at diagnosis. The patient who carried the 6450A>G splice mutation in the germ-line was aged 42 years at diagnosis. Eight of theATM-mutant biopsies were sampled at diagnosis. Seven of these patients presented with advanced disease (clinical stage IV) at diagnosis, and the median survival time was 10 months (0-77 months). The 3 patients with the shortest survival times also harbored deletions of the INK4a/ARF locus, which is a strong negative prognostic factor in DLBCL.29 One sample taken at relapse showed concomitant mutation of ATM, mutation ofTP53, and deletion of ARF, and this patient survived only 1 month after biopsy. Although the size of the sample is small, these data suggest that the low median survival time in patients with ATM-mutated DLBCLs is at least in part due to the relatively high frequency of concurrent ARF deletions in this group, and that ATM mutation in itself may not be associated with a particularly poor outcome.
Discussion
We examined the entire coding sequence and all consensus splice sequences of the ATM gene for mutations in 120 lymphoid neoplasms representing 4 histological subtypes (DLBCL, FL, ALL, and PTL). A total of 20 sequence variants were identified. For simplicity, we assigned these variants to 2 classes to distinguish between mutations with a putative pathogenic effect (class I) and sequence variants that have been identified in the germ-line of random individuals and thus may represent silent variants (class II). Class I variants included tumor-specific mutations first identified in this study, mutations previously implicated in sporadic cancers, typical loss-of-function mutations, and mutations found in the germ-line of A-T patients. The highest frequency of class I variants was found in DLBCLs (20%), with markedly lower frequencies in FLs (8%) and ALLs (4%). In DLBCLs, the majority of these mutations were associated with a second mutation or deletion of the normal allele, consistent with biallelic inactivation of ATM. Novel missense mutations outside the PI-3K domain, but affecting evolutionary conserved residues within or close to putative functional domains of ATM, were found in both DLBCLs and FLs. Thus, the overall spectrum of ATM mutations in these tumors resembles the spectra observed in T-PLLs6-9and B-CLLs,12-15 whereas the ATM variants reported in MCLs are mainly truncating mutations and missense mutations affecting the PI-3K domain.10,11 Together with previous demonstrations of common loss of heterozygosity at 11q23 in MCLs,10 DLBCLs,20,21 and FLs,20and a high frequency of ATM mutations in MCLs,10,11 our data suggest that ATM may be involved in a considerable proportion of B-cell lymphomas. On the other hand, the absence of ATM mutations in the current PTL series suggests that the involvement of ATM in T-cell neoplasia may be confined to T-PLL, a malignancy similar to the T-cell leukemias observed in A-T patients. Considering that the estimated carrier frequency for A-T in the Danish population is 1 in 183,5the presence of an A-T-associated variant (S570P) in one case of ALL may be considered coincidental.
Accumulating evidence suggests that the spectrum of ATMmutations in some sporadic cancers may be composed predominantly of missense mutations and thus differs from that observed in A-T, where more than 80% of the mutations are of the truncating type. This has led Meyn43 and Gatti et al44 to propose the existence of 2 distinct populations of ATM heterozygotes: carriers of truncating ATM mutations associated with A-T and carriers of missense ATM mutations, who may be more prone to carcinogenesis. Of the 20 sequence variants identified in B-cell tumors in the present study, 7 were also found as germ-line variants among 100 random individuals from the same population (class II variants). Interestingly, however, the frequency of these variants as a group was severalfold higher in DLBCLs and FLs than in samples from random individuals, suggesting that one or more of these variants may contribute to the etiology and/or progression of B-cell lymphoma. Because of the relatively small sample sizes and the low frequencies of individual ATM sequence variants, the possible clinical implications of class II variants remain to be determined. Nevertheless, one compelling observation was a 5.6-fold excess of one of these variants, 735C>T, in DLBCLs compared with samples from random individuals. Because this nucleotide substitution does not alter the coding sequence of ATM (V245V), it has generally been regarded as a neutral variant. However, Laake et al40showed that this alteration coincides with skipping of exon 9, an observation confirmed in the present study. Further support for a pathogenic effect of 735C>T was provided by the demonstration of this alteration as a possible disease-causing allele in 2 Finnish A-T families.40 Another variant, S707P, was found to be 4 times more frequent in FL cases than in the control population. This variant was recently found to be 5 times more frequent in breast cancer patients with bilateral disease than in random individuals.45 Large-scale studies are needed to establish whether ATM class II variants alone or together with as yet unknown factors may confer an increased risk of developing B-cell lymphoma.
A remarkable finding in the present study was the occurrence ofTP53 mutations and/or deletions of the INK4a/ARFlocus in 7 of 9 DLBCLs with ATM class I variants. This nonrandom coexistence of ATM mutations and genetic events in the ARF-TP53 pathway may seem paradoxical in view of the fact that p53 is a downstream effector of ATM, and that loss of ATM function compromises activation of p53 in response to ionizing radiation. Indeed, the idea that loss of ATM and loss of p53 may in some aspects be functionally equivalent in human cancer cells was supported by recent studies by Pettitt et al46 and Stankovic et al,15 who showed that a defective p53 signal transduction pathway in B-CLL cases with wild-type TP53 may be attributable to mutation of the ATM gene. However, while CLL cells with mutant p53 were profoundly radioresistant, CLL cells with ATM mutations were only partially resistant to irradiation-induced apoptosis.46 This is consistent with observations that p53 is still induced, albeit with delayed kinetics, in γ-irradiated A-T cells and mouse Atm-null thymocytes,47 probably owing to the activity of ATM-related proteins such as ATR (ATM- and Rad3-related).48 Also considering that loss ofATM compromises a wide range of cellular functions that are less likely to depend on p53 activity,48 it is conceivable that the ATM and p53 effector pathways are overlapping, but not congruent, in suppressing tumorigenesis, and that the combined status of these pathways may be an important determinant of the malignant phenotype.
The consequences of loss of p53 or ARF in cells lacking ATM have been studied extensively in cell culture and animal models. Like fibroblasts from A-T patients, embryonic fibroblasts from Atm-null mice proliferate slowly in culture and soon reach a growth-arrested state resembling senescence,49-52 probably because of their increased genomic instability. These cells express elevated levels of several proteins capable of inhibiting cell-cycle progression, including p53, Arf, p21Cip1, and p16INK4a. By contrast, fibroblasts fromAtm/Trp5351 orAtm/Arf52 double-mutant mice proliferate rapidly in culture and do not arrest prematurely. Thus, loss of p53 or Arf may effectively reverse the growth arrest associated with the Atm-null state.
Although the association between ATM mutations and inactivation of the ARF-TP53 pathway in DLBCL was found to be statistically highly significant, most of theATM missense variants in these tumors resided outside the PI-3K domain, and their functional significance remains uncertain. It is tempting to speculate that these mutations may leave the kinase activity of ATM unaffected and select for functions other than p53 inactivation. However, previous studies of missense mutations outside the PI-3K domain of ATM showed depletion of functional ATM protein,53 and functional studies in B-CLLs with missense mutations in the N-terminal part of ATM showed defective induction of downstream components in the p53 pathway following ionizing radiation.15,46 Furthermore, a TP53mutation was also identified in a DLBCL specimen carrying a truncatingATM mutation, suggesting that inactivation of the p53 checkpoint may provide an additional or even necessary tumorigenic effect in some ATM-null cells. Thus, it is possible that the effects of ATM mutations and inactivation of theARF-TP53 pathway may act as synergistic layers in DLBCL. This situation is different from B-CLL, where ATM andTP53 mutations occur in a mutually exclusive pattern15,46; T-PLL, where TP53 mutations have not been demonstrated6; and MCL, where ATMmutations seem to occur independently of TP53 mutations and inactivation of ARF.11 The biologic basis of these diverse patterns of ATM andTP53/ARF inactivation in hematologic malignancies is currently unknown and is a topic for future studies.
We thank Anni Aggerholm, Dorrit Lüzthøft, and Tina Seremet for help with preparation of samples.
Prepublished online as Blood First Edition Paper, April 30, 2002; DOI 10.1182/blood-2002-02-0382.
Supported by grants from the Danish Cancer Society, the Danish Cancer Research Foundation, Gerda and Aage Haensch's Foundation, the Novo Nordisk Foundation, the Danish Medical Association Research Fund, and Desireé and Niels Yde's Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Per Guldberg, Institute of Cancer Biology, Danish Cancer Society, Strandboulevarden 49, DK-2100 Copenhagen, Denmark; e-mail: perg@cancer.dk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal